Structural Homology Fails to Predict Secretion Efficiency in Pichia pastoris: Divergent Responses of Architecturally Similar scFvs to Multi-Parametric Genetic Engineering
Abstract
1. Introduction
2. Results
2.1. Comparative Analysis of PR961 and PR953 scFv Variants
2.2. The Limited Effect of Gene Dosage Optimization on Enhancement of PR961 and PR953 Secretion
2.3. VH-Linker-VL Construction Enhanced scFv Secretion Compared to VL-Linker-VH
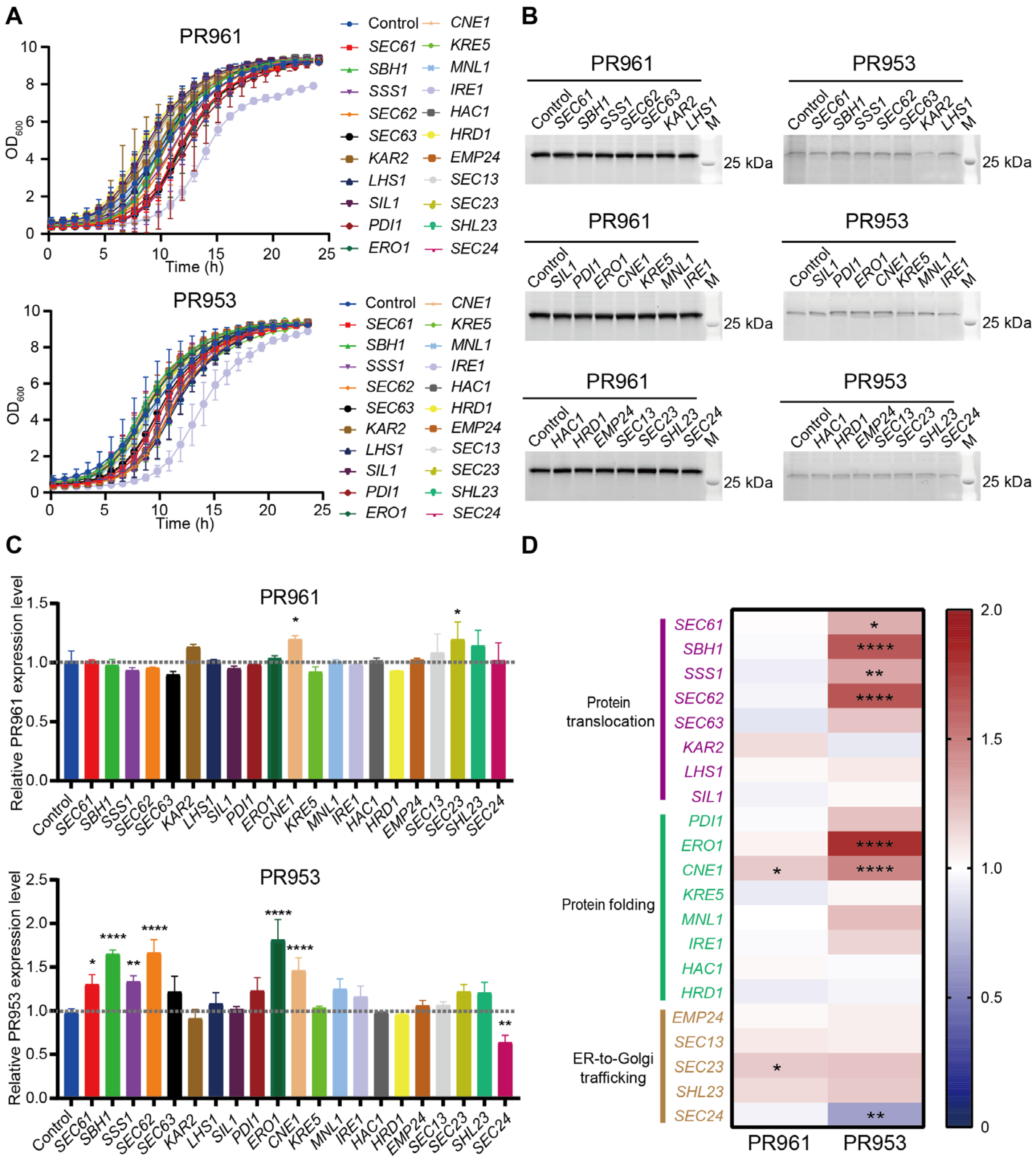
2.4. Disrupting Key ER Components Downregulated scFv Secretion
2.5. Overexpression of ER Components Enhanced PR953 Secretion More Compared to PR961
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, Reagents, and Media
4.2. Construction and Screening of Recombinant P. pastoris Strains
4.2.1. Construction and Screening of Strains with Optimized Gene Dosage
4.2.2. Construction and Screening of Strains with Different VH-VL Orientations
4.2.3. Construction and Screening of Gene Knockout Strains
4.2.4. Construction and Screening of Gene Over-Expression Strains
4.3. Evaluation of PR961 and PR953 Gene Copy Numbers by Digital PCR
4.4. Inducible Expression and Growth Curves of Recombinant P. pastoris Strains
4.5. Quantify of Secreted Proteins with SYPRO Ruby Staining
4.6. Sequence and Structure Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ER | Endoplasmic reticulum |
| scFv | Single-chain variable fragment |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| Pdi | Protein disulfide isomerase |
| UPR | Unfolded protein reaction |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| VL | Variable light chain |
| VH | Variable heavy chain |
| Cas9 | CRISPR-associated protein 9 |
| ERAD | Endoplasmic reticulum-associated degradation |
| ddPCR | Droplet-digital PCR |
References
- Radziwon, K.; Weeks, A.M. Protein engineering for selective proteomics. Curr. Opin. Chem. Biol. 2021, 60, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Lakhawat, S.S.; Chandel, S.; Jaswal, S.K.; Sharma, P.K. Protein Engineering, a Robust Tool to Engineer Novel Functions in Protein. Protein Pept. Lett. 2023, 30, 541–551. [Google Scholar] [CrossRef]
- Meinen, B.A.; Bahl, C.D. Breakthroughs in computational design methods open up new frontiers for de novo protein engineering. Protein Eng. Des. Sel. 2021, 34, gzab007. [Google Scholar] [CrossRef]
- Linsky, T.W.; Noble, K.; Tobin, A.R.; Crow, R.; Carter, L.; Urbauer, J.L.; Baker, D.; Strauch, E.-M. Sampling of structure and sequence space of small protein folds. Nat. Commun. 2022, 13, 7151. [Google Scholar] [CrossRef] [PubMed]
- Orsi, E.; Schada von Borzyskowski, L.; Noack, S.; Nikel, P.I.; Lindner, S.N. Automated in vivo enzyme engineering accelerates biocatalyst optimization. Nat. Commun. 2024, 15, 3447. [Google Scholar] [CrossRef]
- Nammi, B.; Jayasinghe-Arachchige, V.M.; Madugula, S.S.; Artiles, M.; Radler, C.N.; Pham, T.; Liu, J.; Wang, S. CasGen: A Regularized Generative Model for CRISPR Cas Protein Design with Classification and Margin-Based Optimization. bioRxiv 2025, in press. [Google Scholar] [CrossRef]
- Moraes, L.M.P.d.; Marques, H.F.; Reis, V.C.B.; Coelho, C.M.; Leitão, M.d.C.; Galdino, A.S.; Porto de Souza, T.P.; Piva, L.C.; Perez, A.L.A.; Trichez, D. Applications of the Methylotrophic Yeast Komagataella phaffii in the Context of Modern Biotechnology. J. Fungi 2024, 10, 411. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Barone, G.D.; Emmerstorfer-Augustin, A.; Biundo, A.; Pisano, I.; Coccetti, P.; Mapelli, V.; Camattari, A. Industrial Production of Proteins with Pichia pastoris—Komagataella phaffii. Biomolecules 2023, 13, 441. [Google Scholar] [CrossRef]
- Wang, P.; Huang, L.; Jiang, H.; Tian, J.; Chu, X.; Wu, N. Improving the secretion of a methyl parathion hydrolase in Pichia pastoris by modifying its N-terminal sequence. PLoS ONE 2014, 9, e96974. [Google Scholar] [CrossRef]
- Zhao, F.; Yu, C.-h.; Liu, Y. Codon usage regulates protein structure and function by affecting translation elongation speed in Drosophila cells. Nucleic Acids Res. 2017, 45, 8484–8492. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Guerriero, C.J.; Brodsky, J.L. Substrate ubiquitination retains misfolded membrane proteins in the endoplasmic reticulum for degradation. Cell Rep. 2021, 36, 109717. [Google Scholar] [CrossRef]
- Delic, M.; Valli, M.; Graf, A.B.; Pfeffer, M.; Mattanovich, D.; Gasser, B. The secretory pathway: Exploring yeast diversity. FEMS Microbiol. Rev. 2013, 37, 872–914. [Google Scholar] [CrossRef] [PubMed]
- Werten, M.W.; Eggink, G.; Stuart, M.A.C.; de Wolf, F.A. Production of protein-based polymers in Pichia pastoris. Biotechnol. Adv. 2019, 37, 642–666. [Google Scholar] [CrossRef] [PubMed]
- Suarez, S.; Martiny, A.; Asempa, T. Gene amplification uncovers large previously unrecognized cryptic antibiotic resistance potential in E. coli, Microbiol. Spectrum 2021, 9, e00289-21. [Google Scholar] [CrossRef]
- Zheng, X.; Zhang, Y.; Liu, X.; Li, C.; Lin, Y.; Liang, S. High-level expression and biochemical properties of a thermo-alkaline pectate lyase from Bacillus sp. RN1 in Pichia pastoris with potential in ramie degumming. Front. Bioeng. Biotechnol. 2020, 8, 850. [Google Scholar] [CrossRef]
- Wang, X.; Goldstein, D.B. Enhancer domains predict gene pathogenicity and inform gene discovery in complex disease. Am. J. Hum. Genet. 2020, 106, 215–233. [Google Scholar] [CrossRef]
- Bakoulis, S.; Krautz, R.; Alcaraz, N.; Salvatore, M.; Andersson, R. Endogenous retroviruses co-opted as divergently transcribed regulatory elements shape the regulatory landscape of embryonic stem cells. Nucleic Acids Res. 2022, 50, 2111–2127. [Google Scholar] [CrossRef]
- Ben Azoun, S.; Belhaj, A.E.; Göngrich, R.; Gasser, B.; Kallel, H. Molecular optimization of rabies virus glycoprotein expression in Pichia pastoris. Microb. Biotechnol. 2016, 9, 355–368. [Google Scholar] [CrossRef]
- Datta, S. Optimizing Granulocyte Colony-Stimulating Factor Transcript for Enhanced Expression in Escherichia coli. Front. Bioeng. Biotechnol. 2021, 9, 630367. [Google Scholar] [CrossRef]
- Gidalevitz, T.; Stevens, F.; Argon, Y. Orchestration of secretory protein folding by ER chaperones. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2410–2424. [Google Scholar] [CrossRef]
- Du, H.; Zheng, C.; Aslam, M.; Xie, X.; Wang, W.; Yang, Y.; Liu, X. Endoplasmic reticulum-mediated protein quality control and endoplasmic reticulum-associated degradation pathway explain the reduction of N-glycoprotein level under the lead stress. Front. Plant Sci. 2021, 11, 598552. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Tyo, K.E.J.; Liu, Z.; Petranovic, D.; Nielsen, J. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 491–510. [Google Scholar] [CrossRef]
- Raschmanová, H.; Weninger, A.; Knejzlík, Z.; Melzoch, K.; Kovar, K. Engineering of the unfolded protein response pathway in Pichia pastoris: Enhancing production of secreted recombinant proteins. Appl. Microbiol. Biotechnol. 2021, 105, 4397–4414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-S.; Gong, J.-S.; Jiang, J.-Y.; Xu, Z.-H.; Shi, J.-S. Engineering protein translocation and unfolded protein response enhanced human PH-20 secretion in Pichia pastoris. Appl. Microbiol. Biotechnol. 2024, 108, 54. [Google Scholar] [CrossRef]
- Zahrl, R.J.; Prielhofer, R.; Ata, Ö.; Baumann, K.; Mattanovich, D.; Gasser, B. Pushing and pulling proteins into the yeast secretory pathway enhances recombinant protein secretion. Metab. Eng. 2022, 74, 36–48. [Google Scholar] [CrossRef]
- Fu, D.; Zhang, G.; Wang, Y.; Zhang, Z.; Hu, H.; Shen, S.; Wu, J.; Li, B.; Li, X.; Fang, Y. Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes. PLoS Biol. 2021, 19, e3001209. [Google Scholar] [CrossRef]
- Karlsson, J.; Larsson, E. FocalScan: Scanning for altered genes in cancer based on coordinated DNA and RNA change. Nucleic Acids Res. 2016, 44, e150. [Google Scholar] [CrossRef]
- Tang, X.; Kadara, H.; Behrens, C.; Liu, D.D.; Xiao, Y.; Rice, D.; Gazdar, A.F.; Fujimoto, J.; Moran, C.; Varella-Garcia, M. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: Implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 2011, 17, 2434–2443. [Google Scholar] [CrossRef]
- Bezrookove, V.; De Semir, D.; Nosrati, M.; Tong, S.; Wu, C.; Thummala, S.; Dar, A.A.; Leong, S.P.; Cleaver, J.E.; Sagebiel, R.W. Prognostic impact of PHIP copy number in melanoma: Linkage to ulceration. J. Investig. Dermatol. 2014, 134, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Satheeshkumar, P.K. Expression of single chain variable fragment (scFv) molecules in plants: A comprehensive update. Mol. Biotechnol. 2020, 62, 151–167. [Google Scholar] [CrossRef]
- Liu, C.; Gong, J.-S.; Su, C.; Li, H.; Li, H.; Rao, Z.-M.; Xu, Z.-H.; Shi, J.-S. Pathway engineering facilitates efficient protein expression in Pichia pastoris. Appl. Microbiol. Biotechnol. 2022, 106, 5893–5912. [Google Scholar] [CrossRef]
- Santiago-Tirado, F.H.; Hurtaux, T.; Geddes-McAlister, J.; Nguyen, D.; Helms, V.; Doering, T.L.; Römisch, K. The ER protein translocation channel subunit Sbh1 controls virulence of Cryptococcus neoformans. mBio 2023, 14, e03384. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.S.; Linxweiler, J.; Radosa, J.C.; Linxweiler, M.; Zimmermann, R. The endoplasmic reticulum membrane protein Sec62 as potential therapeutic target in SEC62 overexpressing tumors. Front. Physiol. 2022, 13, 1014271. [Google Scholar] [CrossRef]
- Frand, A.R.; Kaiser, C.A. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1998, 1, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Esaki, M.; Liu, Y.; Glick, B.S. The budding yeast Pichia pastoris has a novel Sec23p homolog. FEBS Lett. 2006, 580, 5215–5221. [Google Scholar] [CrossRef]
- Parlati, F.; Dominguez, M.; Bergeron, J.J.M.; Thomas, D.Y. Saccharomyces cerevisiae CNE1 Encodes an Endoplasmic Reticulum (ER) Membrane Protein with Sequence Similarity to Calnexin and Calreticulin and Functions as a Constituent of the ER Quality Control Apparatus. J. Biol. Chem. 1995, 270, 244–253. [Google Scholar] [CrossRef]
- Kozlov, G.; Gehring, K. Calnexin cycle–structural features of the ER chaperone system. FEBS J. 2020, 287, 4322–4340. [Google Scholar] [CrossRef]
- Jing, J.; Wang, B.; Liu, P. The functional role of SEC23 in vesicle transportation, autophagy and cancer. Int. J. Biol. Sci. 2019, 15, 2419. [Google Scholar] [CrossRef]
- Bujotzek, A.; Dunbar, J.; Lipsmeier, F.; Schäfer, W.; Antes, I.; Deane, C.M.; Georges, G. Prediction of VH–VL domain orientation for antibody variable domain modeling. Proteins Struct. Funct. Bioinform. 2015, 83, 681–695. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Ye, Y.; Chen, W.; Wang, X.; Chen, F. Expression of VH-linker-VL orientation-dependent single-chain Fv antibody fragment derived from hybridoma 2E6 against aflatoxin B1 in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2015, 42, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Influence of mRNA secondary structure on binding and migration of 40S ribosomal subunits. Cell 1980, 19, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Lee, G.; Jeon, E.; Sohn, E.j.; Lee, Y.; Kang, H.; Lee, D.w.; Kim, D.H.; Hwang, I. The immediate upstream region of the 5′-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana. Nucleic Acids Res. 2013, 42, 485–498. [Google Scholar] [CrossRef]
- Simpson, E.R.; Herold, E.M.; Buchner, J. The folding pathway of the antibody VL domain. J. Mol. Biol. 2009, 392, 1326–1338. [Google Scholar] [CrossRef]
- Besada-Lombana, P.B.; Da Silva, N.A. Engineering the early secretory pathway for increased protein secretion in Saccharomyces cerevisiae. Metab. Eng. 2019, 55, 142–151. [Google Scholar] [CrossRef]
- Ellgaard, L.; McCaul, N.; Chatsisvili, A.; Braakman, I. Co-and post-translational protein folding in the ER. Traffic 2016, 17, 615–638. [Google Scholar] [CrossRef]
- Sevier, C.S.; Kaiser, C.A. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2008, 1783, 549–556. [Google Scholar] [CrossRef]
- Chen, X.; Deng, J.; Zheng, X.; Zhang, J.; Zhou, Z.; Wei, H.; Zhan, C.-G.; Zheng, F. Development of a long-acting Fc-fused cocaine hydrolase with improved yield of protein expression. Chem. Biol. Interact. 2019, 306, 89–95. [Google Scholar] [CrossRef]
- Song, X.; Shao, C.; Guo, Y.; Wang, Y.; Cai, J. Improved the expression level of active transglutaminase by directional increasing copy of mtg gene in Pichia pastoris. BMC Biotechnol. 2019, 19, 54. [Google Scholar] [CrossRef]
- Xie, S.; Sun, S.; Lin, F.; Li, M.; Pu, Y.; Cheng, Y.; Xu, B.; Liu, Z.; da Costa Sousa, L.; Dale, B.E.; et al. Mechanism-Guided Design of Highly Efficient Protein Secretion and Lipid Conversion for Biomanufacturing and Biorefining. Adv. Sci. 2019, 6, 1801980. [Google Scholar] [CrossRef] [PubMed]
- Proshkin, S.; Rahmouni, A.R.; Mironov, A.; Nudler, E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 2010, 328, 504–508. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.A.; Akkermans, S.; Van Impe, J.F. Effects of temperature and pH on recombinant Thaumatin II production by Pichia pastoris. Foods 2022, 11, 1438. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiong, F.; Lin, Q.; d’Anjou, M.; Daugulis, A.J.; Yang, D.S.; Hew, C.L. Low-temperature increases the yield of biologically active herring antifreeze protein in Pichia pastoris. Protein Expr. Purif. 2001, 21, 438–445. [Google Scholar] [CrossRef]
- Faber, K.N.; Haima, P.; Harder, W.; Veenhuis, M.; AB, G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 1994, 25, 305–310. [Google Scholar] [CrossRef]
- Besleaga, M.; Vignolle, G.A.; Kopp, J.; Spadiut, O.; Mach, R.L.; Mach-Aigner, A.R.; Zimmermann, C. Evaluation of reference genes for transcript analyses in Komagataella phaffii (Pichia pastoris). Fungal Biol. Biotechnol. 2023, 10, 7. [Google Scholar] [CrossRef]
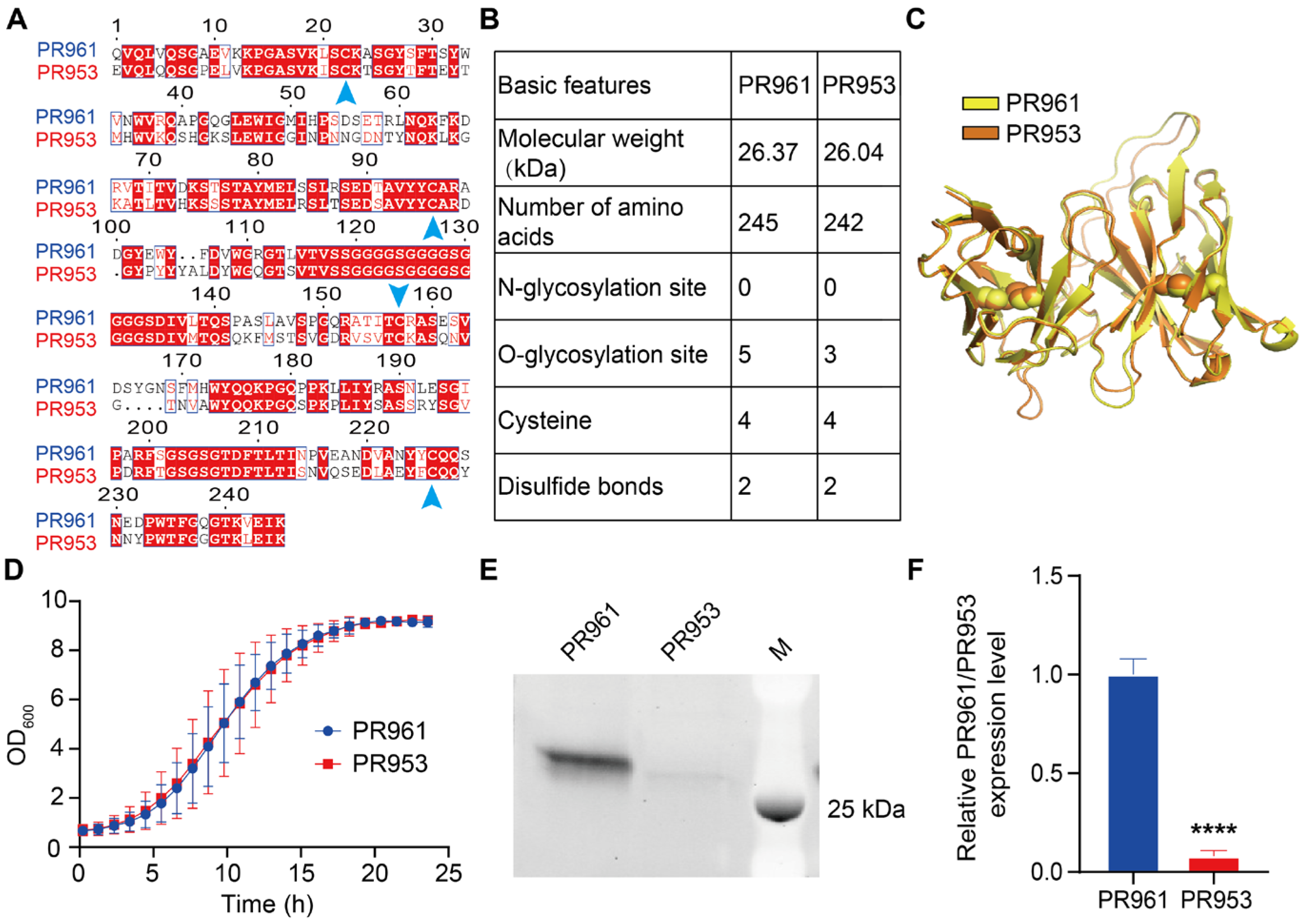
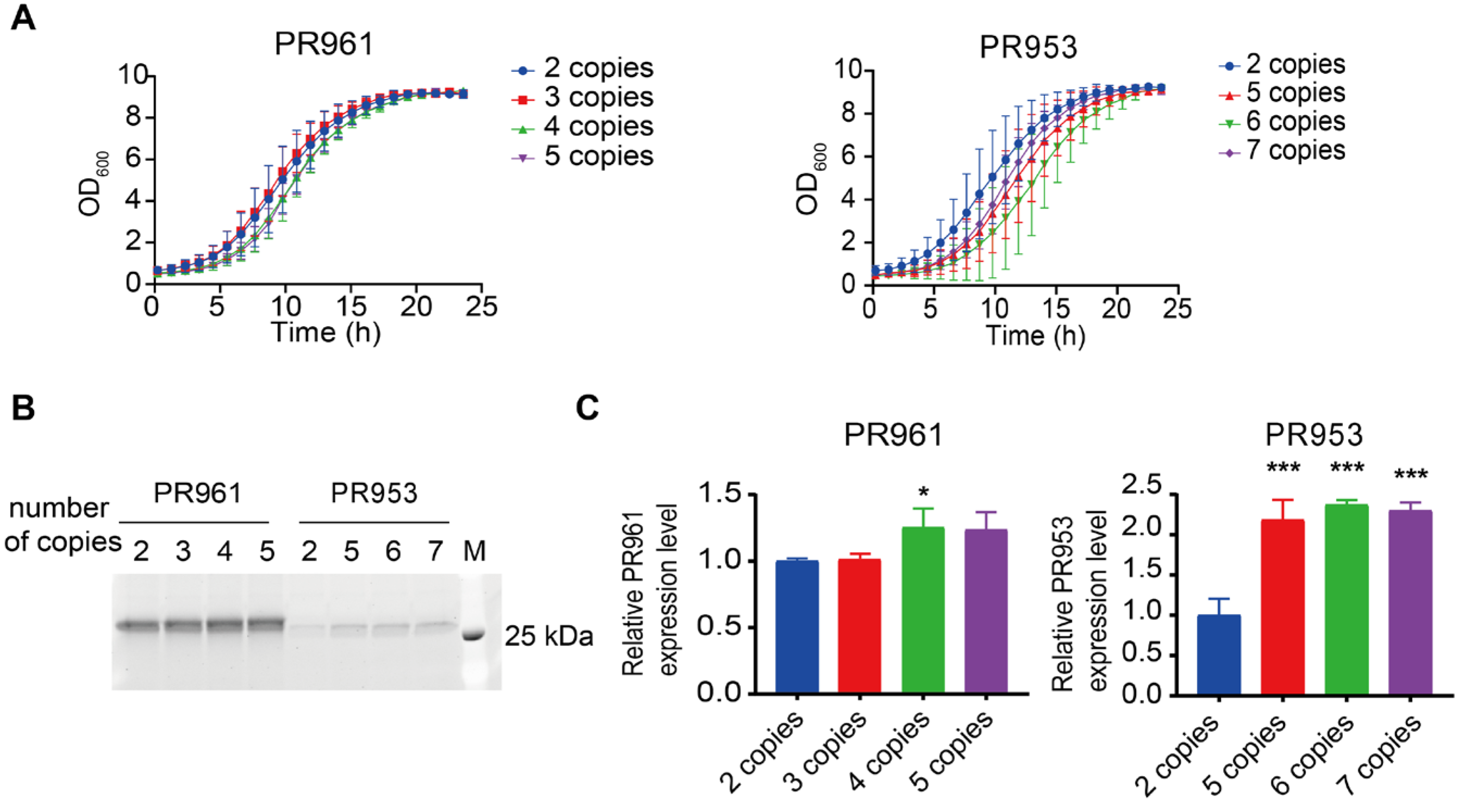
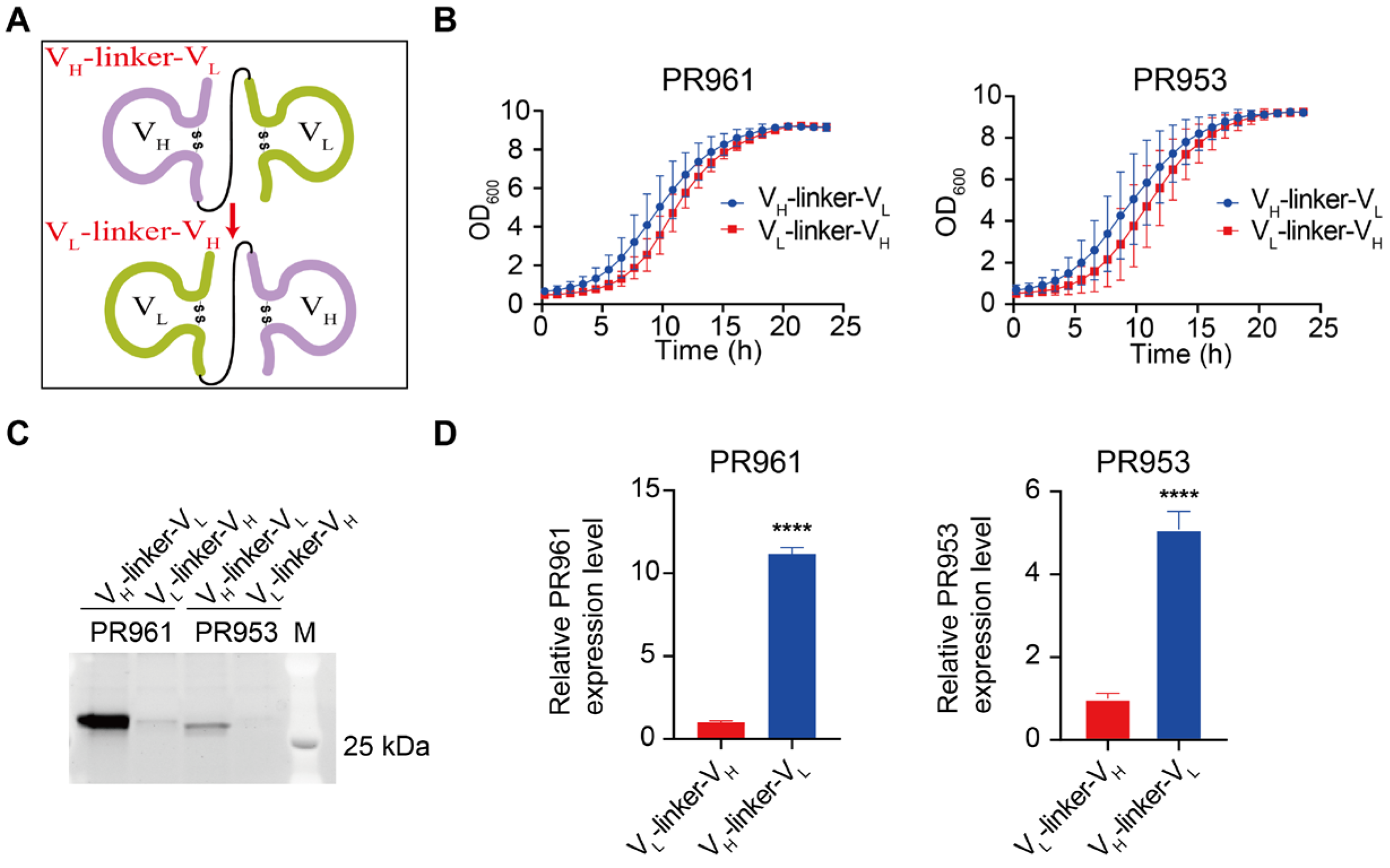
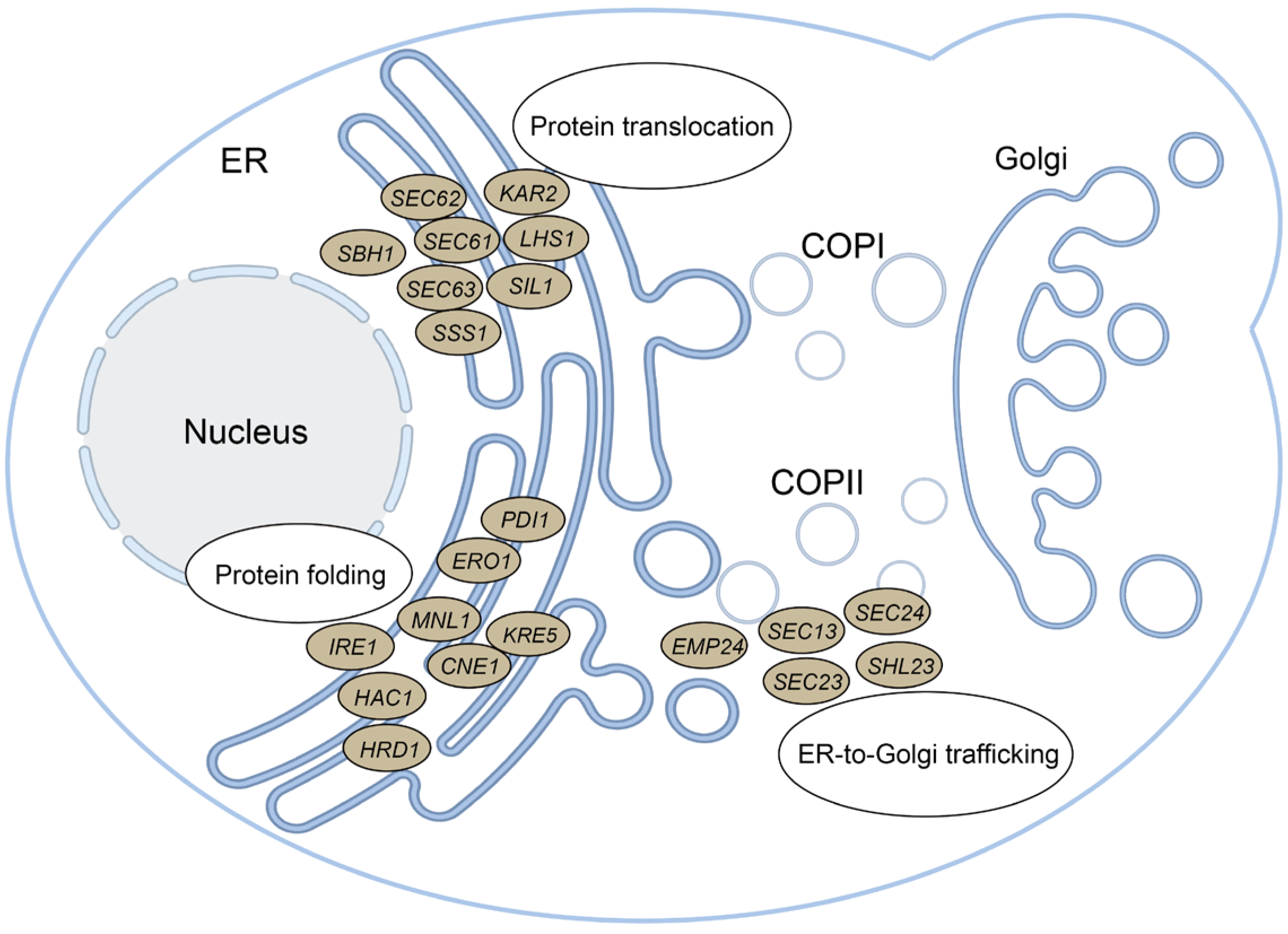

| Engineering Strategy | Fold Change Engineered vs. Parental | ||||
|---|---|---|---|---|---|
| PR961 | PR953 | ||||
| gene dosage optimization | 2 | - | - | ||
| 4 | 1.25 * ± 0.14 | - | |||
| 5 | ns | 2.18 * ± 0.25 | |||
| 6 | - | 2.37 * ± 0.06 | |||
| 7 | - | 2.30 * ± 0.10 | |||
| cassette architecture engineering | VL-linker-VH | - | - | ||
| VH-linker-VL | 11.18 * ± 0.37 | 5.09 * ± 0.43 | |||
| ER secretory pathway reprogramming | Protein translocation | SEC61 | + | ns | 1.30 * ± 0.11 |
| SBH1 | + | ns | 1.65 * ± 0.05 | ||
| SSS1 | + | ns | 1.33 * ± 0.07 | ||
| SEC62 | + | ns | 1.66 * ± 0.15 | ||
| SIL1 | - | 0.83 * ± 0.03 | 0.70 * ± 0.05 | ||
| Protein folding | ERO1 | + | ns | 1.81 * ± 0.23 | |
| CNE1 | + | 1.20 * ± 0.03 | 1.46 * ± 0.15 | ||
| - | 0.71 * ± 0.01 | 0.48 * ± 0.03 | |||
| KRE5 | - | ns | 0.60 * ± 0.04 | ||
| MNL1 | - | 0.72 * ± 0.01 | ns | ||
| IRE1 | - | 0.76 * ± 0.02 | ns | ||
| HAC1 | - | 0.70 * ± 0.03 | 0.75 * ± 0.05 | ||
| HRD1 | - | 0.91 * ± 0.01 | 0.52 * ± 0.01 | ||
| ER-to-Golgi trafficking | SEC23 | + | 1.20 * ± 0.15 | ns | |
| SEC24 | + | ns | 0.64 * ± 0.08 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Xiao, Y.; Liu, X.; Li, Y.; Yu, D.; Guo, J.; Lu, P.; Zhang, X. Structural Homology Fails to Predict Secretion Efficiency in Pichia pastoris: Divergent Responses of Architecturally Similar scFvs to Multi-Parametric Genetic Engineering. Int. J. Mol. Sci. 2025, 26, 4922. https://doi.org/10.3390/ijms26104922
Wang N, Xiao Y, Liu X, Li Y, Yu D, Guo J, Lu P, Zhang X. Structural Homology Fails to Predict Secretion Efficiency in Pichia pastoris: Divergent Responses of Architecturally Similar scFvs to Multi-Parametric Genetic Engineering. International Journal of Molecular Sciences. 2025; 26(10):4922. https://doi.org/10.3390/ijms26104922
Chicago/Turabian StyleWang, Ningning, Yang Xiao, Xiyu Liu, Yuanqing Li, Dehua Yu, Jia Guo, Ping Lu, and Xiaopeng Zhang. 2025. "Structural Homology Fails to Predict Secretion Efficiency in Pichia pastoris: Divergent Responses of Architecturally Similar scFvs to Multi-Parametric Genetic Engineering" International Journal of Molecular Sciences 26, no. 10: 4922. https://doi.org/10.3390/ijms26104922
APA StyleWang, N., Xiao, Y., Liu, X., Li, Y., Yu, D., Guo, J., Lu, P., & Zhang, X. (2025). Structural Homology Fails to Predict Secretion Efficiency in Pichia pastoris: Divergent Responses of Architecturally Similar scFvs to Multi-Parametric Genetic Engineering. International Journal of Molecular Sciences, 26(10), 4922. https://doi.org/10.3390/ijms26104922






