Identification of Cancer Associated Fibroblasts Related Genes Signature to Facilitate Improved Prediction of Prognosis and Responses to Therapy in Patients with Pancreatic Cancer
Abstract
1. Introduction
2. Results
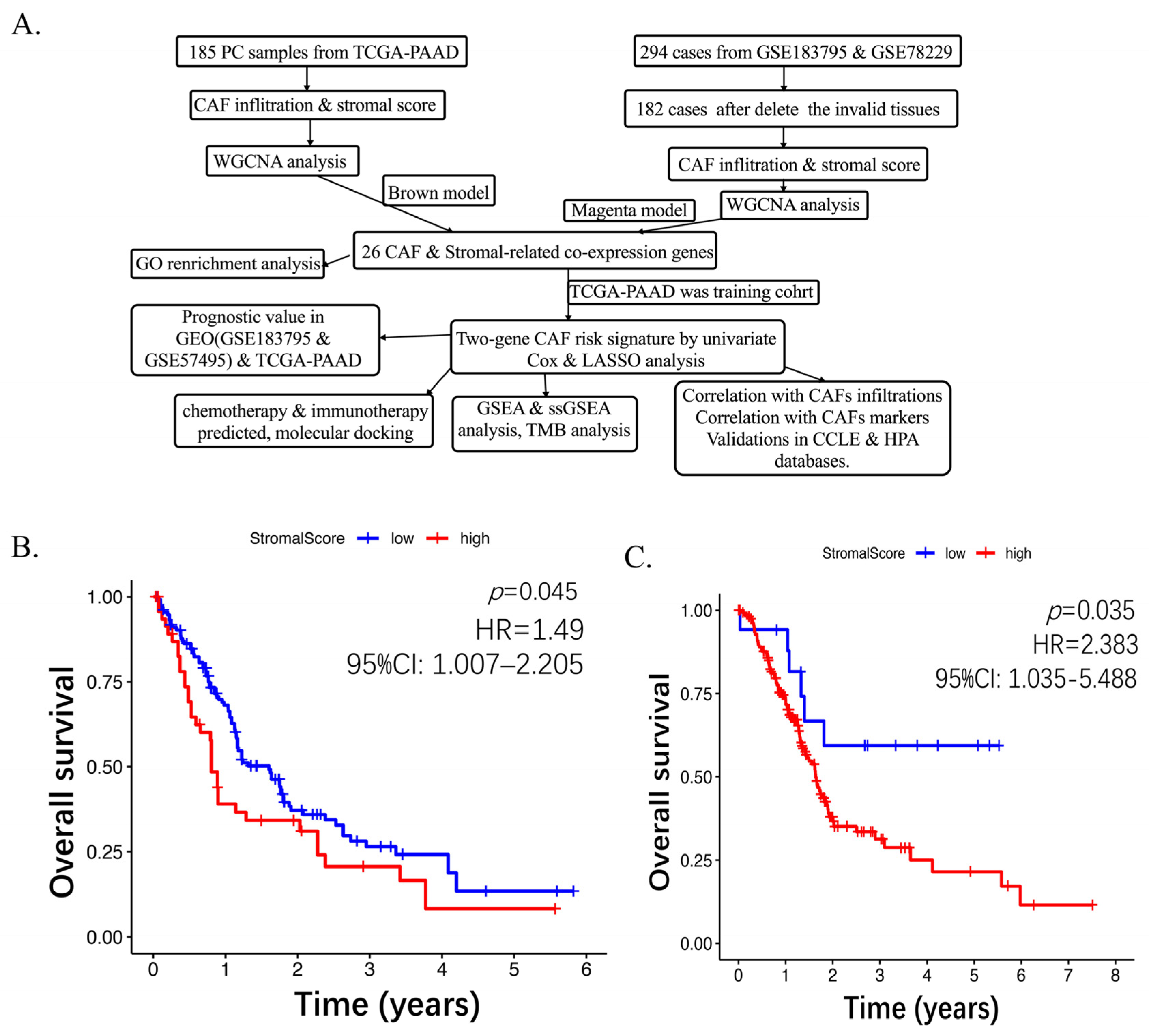
2.1. Higher CAF Infiltrations Are Related to Worse Overall Survival in Pancreatic Cancer
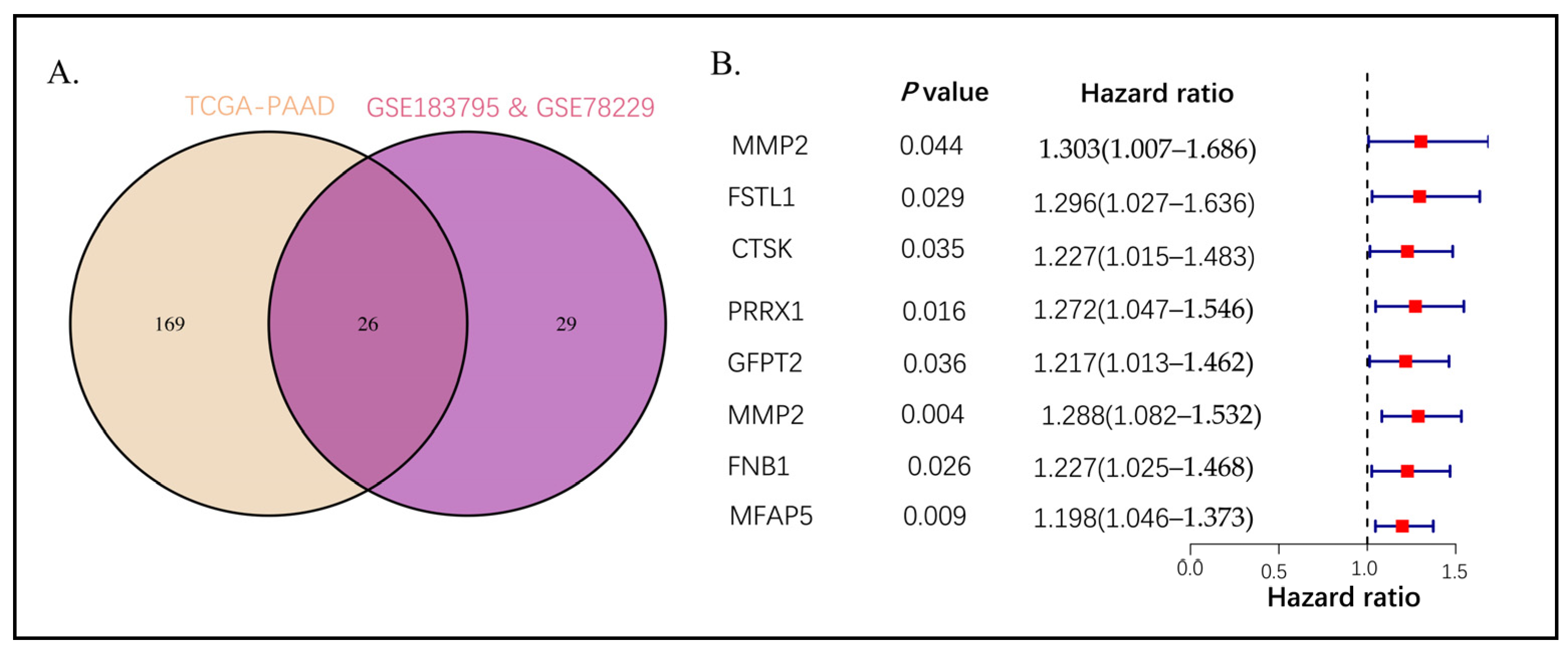
2.2. Intersection Genes Enrichment Related to ECM
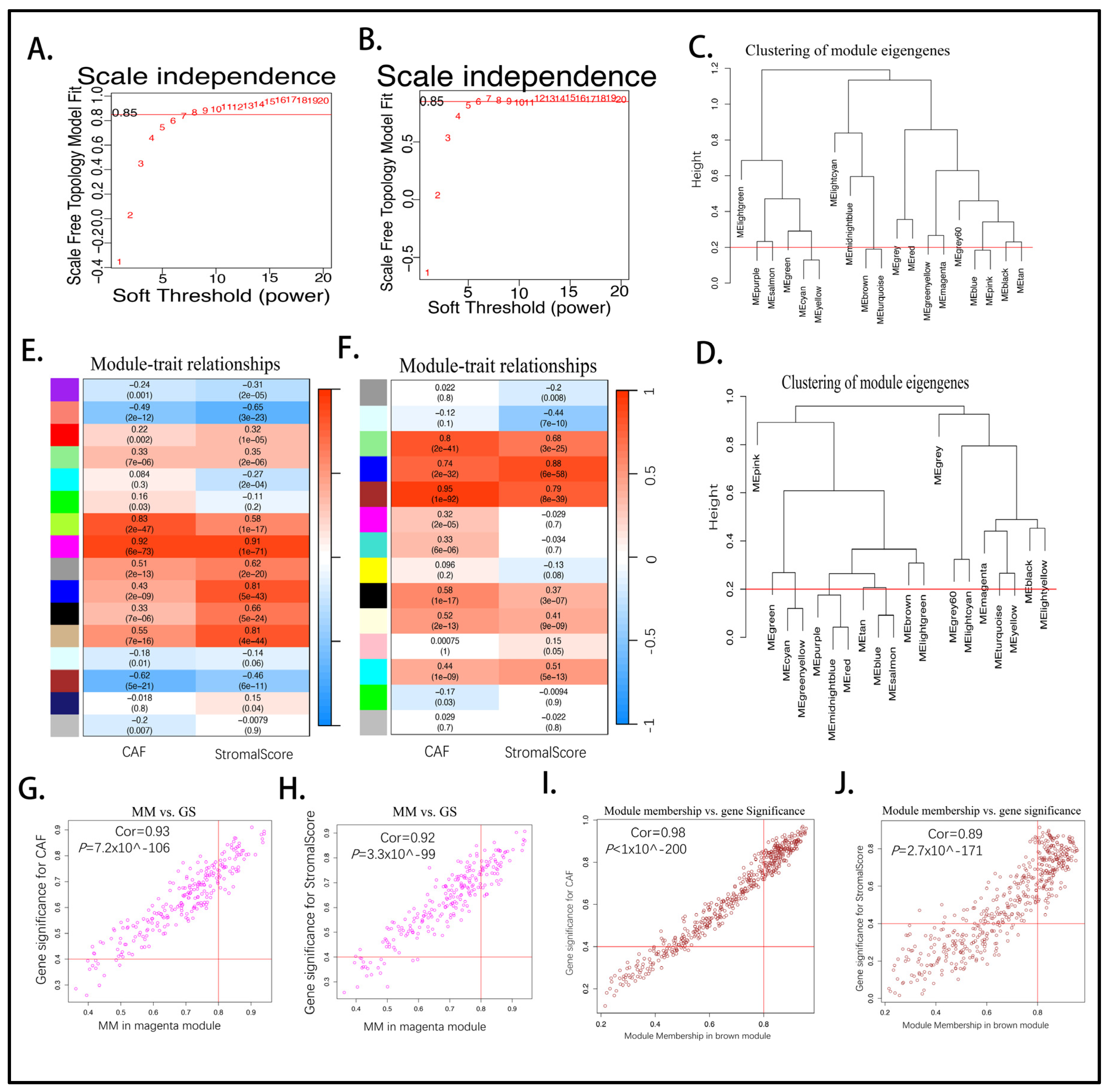
2.3. Establishing the Prognostic Risk Model Genes from the Stromal CAF-Based
2.4. High CAF Infiltrations and Markers Can Be Shown from CAF Signature Genes
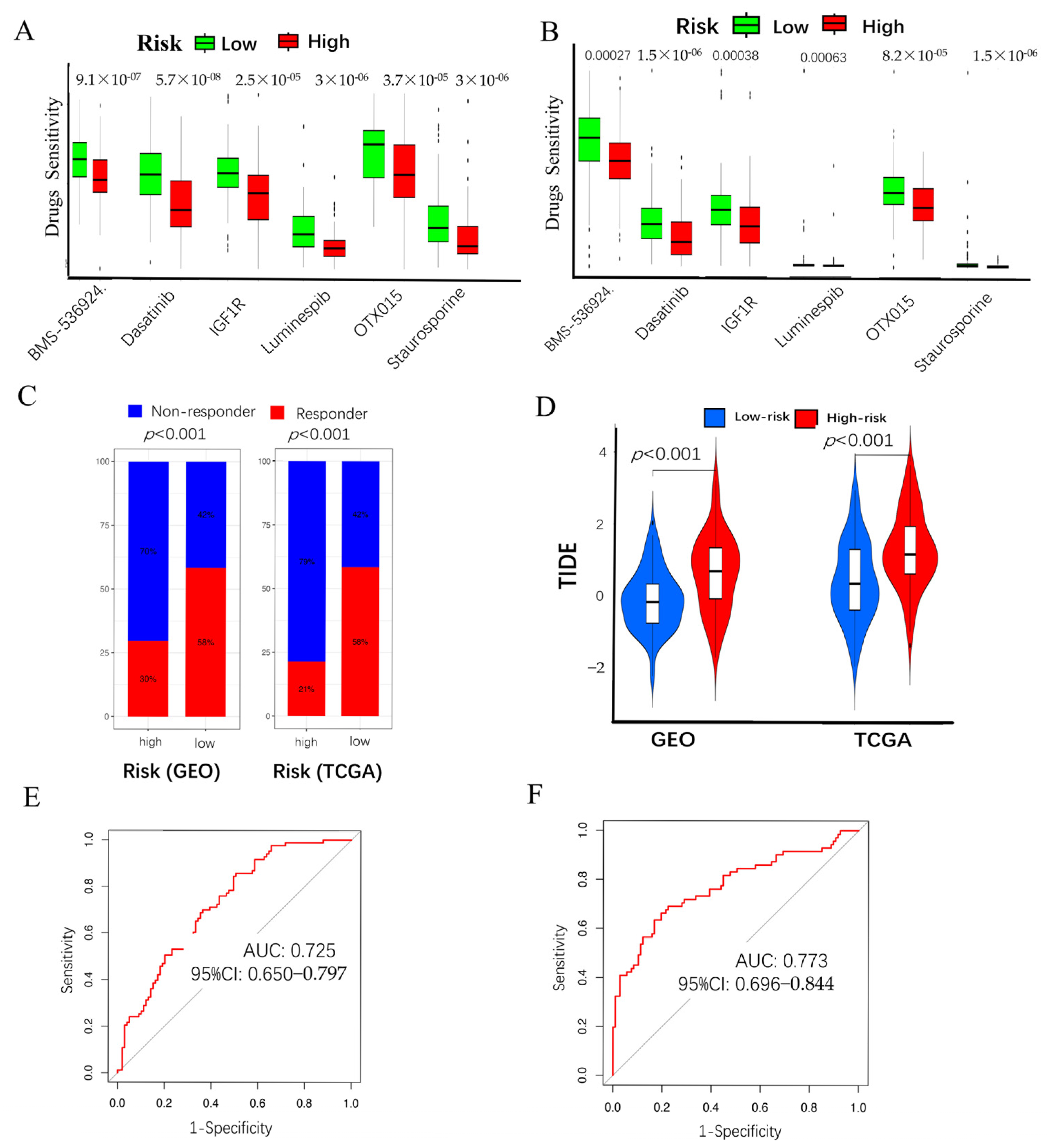
2.5. Chemo-Immuno-Therapy Sensitivity Prediction and Molecular Docking
2.6. High CAF Risk Is Correlated with Tumor Mutation Burden
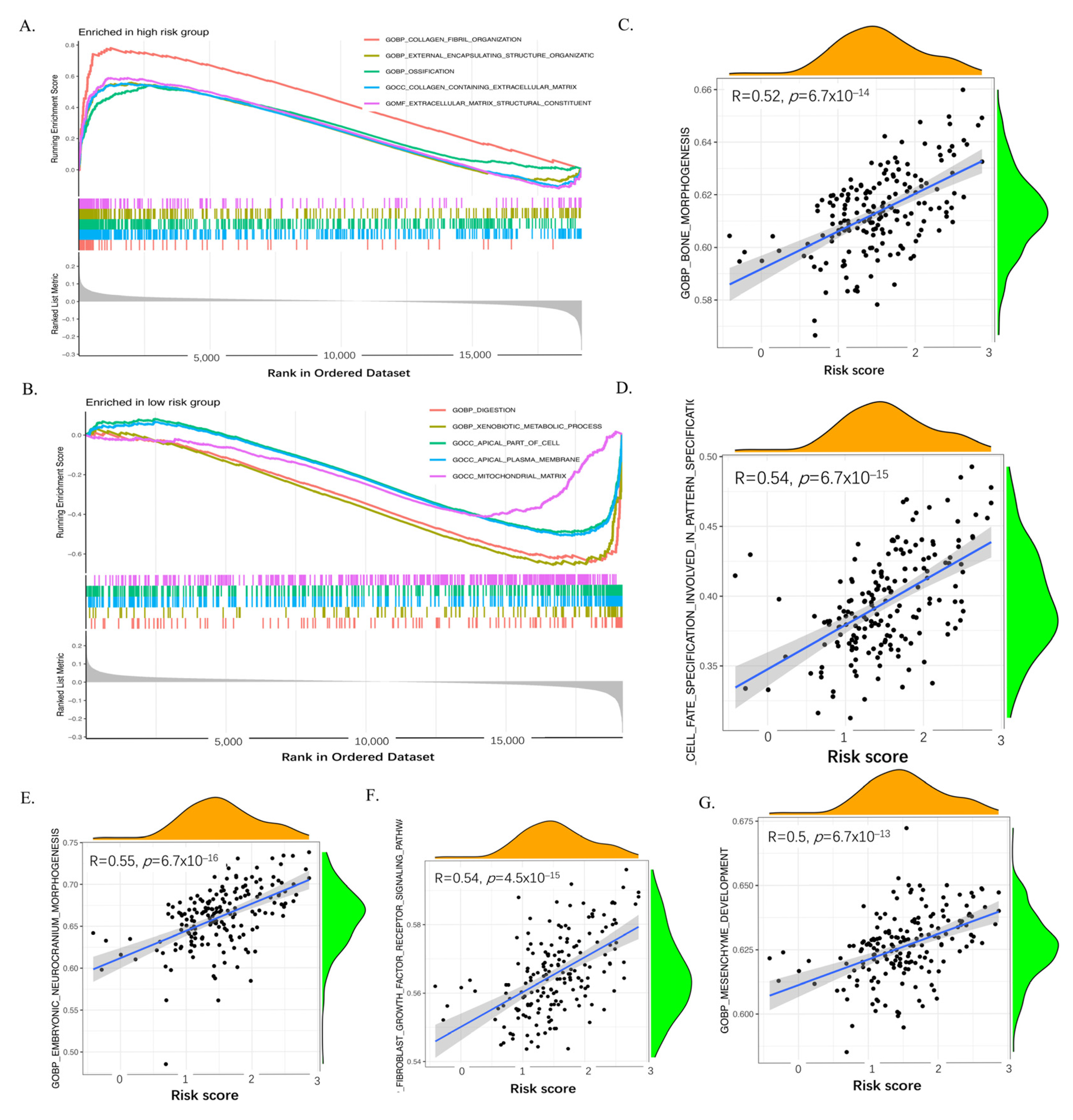
2.7. GSEA and ssGSEA of the Six Genes of the CAF Signature
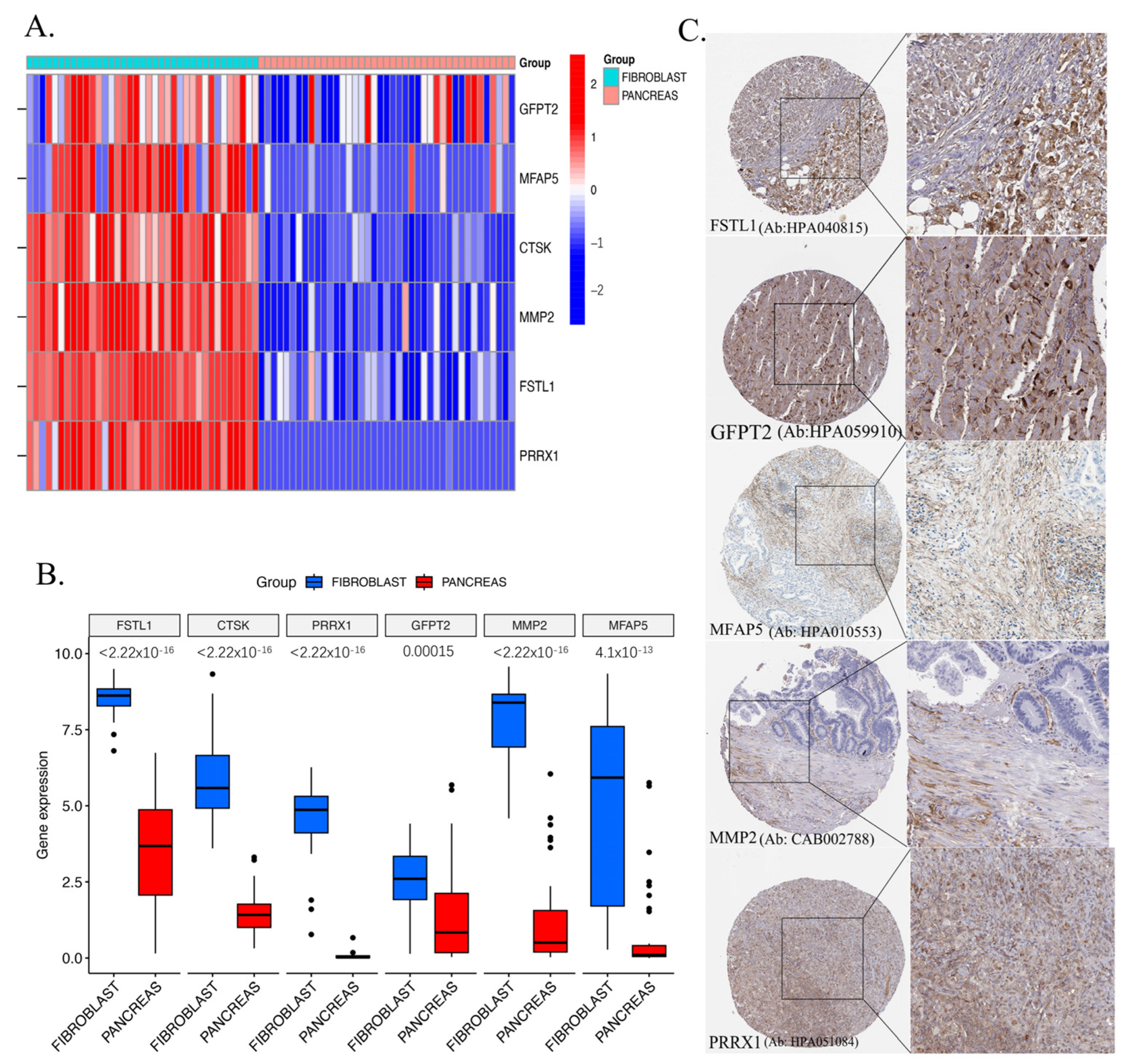
2.8. Validation of Key Genes in CCLE and HPA Databases
3. Discussion
4. Materials and Methods
4.1. Accumulating and Processing Data
4.2. Quantification of Cancer-Associated Fibroblasts (CAFs)
4.3. CAF and Stromal Co-Expression Network Module
4.4. Kaplan–Meier Curve and Log-Rank Analysis
4.5. Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
4.6. CAF-Associated Model Construction and Validation
4.7. Sensitivity Drugs Prediction and Molecular Docking
4.8. Gathering and Analyzing Data on Somatic Changes
4.9. GSEA and ssGSEA Enrichment Analyses
4.10. Validation Via Cancer Cell Line Encyclopedia (CCLE) and Human Protein Atlas (HPA) Databases
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PC | Pancreatic cancer |
| CAFs | Cancer associated fibroblasts |
| GEO | Gene Expression Omnibus |
| TCGA | the Cancer Genome Atlas |
| WGCNA | weighted gene co-expression network analysis |
| TMB | tumor mutation burden |
| GSEA | Gene set enrichment analysis |
| OS | overall survival |
| TME | tumor microenvironment |
| PSCs | pancreatic stellate cells |
| ECM | extracellular matrix |
| TGF-β | transforming growth factor-β |
| IL-6 | interleukin-6 |
| CCL2 | CC-chemokine ligand 2 |
| ROS | reactive oxygen species |
| EBV | Epstein–Barr virus |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| DEGs | differentially expressed genes |
| GS | gene significance |
| MM | module membership |
| GO | gene ontology |
| BP | biological process |
| CC | cell component |
| MF | molecular function |
| KEGG | kyoto encyclopedia of genes and genomes |
| GSEA | Gene Set Enrichment Analysis |
| ssGSEA | single-sample gene set enrichment analysis |
| ADM | acinar-ductal metaplasia |
| FPKM | per million mapped reads |
| MAF | Mutation Annotation Format |
| TPM | transcripts per million |
| EPIC | Estimating the Proportion of Immune and Cancer cells |
| MCP-counter | Microenvironment Cell Populations-counter |
| TIDE | Tumor Immune Dysfunction and Exclusion |
| MAD | median absolute deviation |
| TOM | topological overlap matrix |
| MEs | Module eigengenes |
| GDSC | the Genomics of Drug Sensitivity in Cancer |
| ICB | immune checkpoint blockade |
| AUC | area under the curve |
| CCLE | Cancer Cell Line Encyclopedias |
| HPA | Human Protein Atlas |
| ROC | receiver operating characteristic |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, T.; Sun, L.; Yuan, Y.; Zhu, Y. Potential mechanisms of cancer-associated fibroblasts in therapeutic resistance. Biomed. Pharmacother. 2023, 166, 115425. [Google Scholar] [CrossRef] [PubMed]
- Liot, S.; Balas, J.; Aubert, A.; Prigent, L.; Mercier-Gouy, P.; Verrier, B.; Bertolino, P.; Hennino, A.; Valcourt, U.; Lambert, E. Stroma Involvement in Pancreatic Ductal Adenocarcinoma: An Overview Focusing on Extracellular Matrix Proteins. Front. Immunol. 2021, 12, 612271. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef]
- Glaviano, A.; Lau, H.S.-H.; Carter, L.M.; Lee, E.H.C.; Lam, H.Y.; Okina, E.; Tan, D.J.J.; Tan, W.; Ang, H.L.; Carbone, D.; et al. Harnessing the tumor microenvironment: Targeted cancer therapies through modulation of epithelial-mesenchymal transition. J. Hematol. Oncol. 2025, 18, 6. [Google Scholar]
- Verloy, R.; Privat-Maldonado, A.; Smits, E.; Bogaerts, A. Cold Atmospheric Plasma Treatment for Pancreatic Cancer-The Importance of Pancreatic Stellate Cells. Cancers 2020, 12, 2782. [Google Scholar] [CrossRef]
- Peng, J.; Sun, B.-F.; Chen, C.-Y.; Zhou, J.-Y.; Chen, Y.-S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.-S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, Y.; Xu, H.; Zhang, X.; Mao, T.; Cui, J.; Yao, J.; Wang, Y.; Jiao, F.; Xiao, X.; et al. Single-cell analysis of pancreatic ductal adenocarcinoma identifies a novel fibroblast subtype associated with poor prognosis but better immunotherapy response. Cell Discov. 2021, 7, 36. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, G.; Huang, H.; Fu, Z.; Cao, Z.; Zheng, L.; You, L.; Zhang, T. Preclinical models of pancreatic ductal adenocarcinoma: Challenges and opportunities in the era of precision medicine. J. Exp. Clin. Cancer Res. 2021, 40, 8. [Google Scholar] [CrossRef]
- Werba, G.; Weissinger, D.; Kawaler, E.A.; Zhao, E.; Kalfakakou, D.; Dhara, S.; Wang, L.; Lim, H.B.; Oh, G.; Jing, X.; et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat. Commun. 2023, 14, 797. [Google Scholar] [CrossRef]
- Ye, B.; Wang, Q.; Zhu, X.; Zeng, L.; Luo, H.; Xiong, Y.; Li, Q.; Zhu, Q.; Zhao, S.; Chen, T.; et al. Single-cell RNA sequencing identifies a novel proliferation cell type affecting clinical outcome of pancreatic ductal adenocarcinoma. Front. Oncol. 2023, 13, 1236435. [Google Scholar] [CrossRef] [PubMed]
- Cros, J.; Raffenne, J.; Couvelard, A.; Poté, N. Tumour Heterogeneity in Pancreatic Adenocarcinoma. Pathobiology 2018, 85, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef]
- Su, S.; Chen, J.; Yao, H.; Liu, J.; Yu, S.; Lao, L.; Wang, M.; Luo, M.; Xing, Y.; Chen, F.; et al. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell 2018, 172, 841–856.e16. [Google Scholar] [CrossRef]
- Flavell, R.A.; Sanjabi, S.; Wrzesinski, S.H.; Licona-Limón, P. The polarization of immune cells in the tumour environment by TGFbeta. Nat. Rev. Immunol. 2010, 10, 554–567. [Google Scholar] [CrossRef]
- Yang, X.; Lin, Y.; Shi, Y.; Li, B.; Liu, W.; Yin, W.; Dang, Y.; Chu, Y.; Fan, J.; He, R. FAP Promotes Immunosuppression by Cancer-Associated Fibroblasts in the Tumor Microenvironment via STAT3-CCL2 Signaling. Cancer Res. 2016, 76, 4124–4135. [Google Scholar] [CrossRef]
- Cheteh, E.H.; Augsten, M.; Rundqvist, H.; Bianchi, J.; Sarne, V.; Egevad, L.; Bykov, V.J.; Östman, A.; Wiman, K.G. Human cancer-associated fibroblasts enhance glutathione levels and antagonize drug-induced prostate cancer cell death. Cell Death Dis. 2017, 8, e2848. [Google Scholar] [CrossRef]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhu, Y.; Yang, J.; Liu, X.; Sun, T.; Zhao, Y.; Xiang, F.; Chen, F.; He, H. Transcriptome Analysis Reveals Coexpression Networks and Hub Genes Involved in Papillae Development in Lilium auratum. Int. J. Mol. Sci. 2024, 25, 2436. [Google Scholar] [CrossRef]
- Zhou, H.; Jing, S.; Liu, Y.; Wang, X.; Duan, X.; Xiong, W.; Li, R.; Peng, Y.; Ai, Y.; Fu, D.; et al. Identifying the key genes of Epstein-Barr virus-regulated tumour immune microenvironment of gastric carcinomas. Cell Prolif. 2023, 56, e13373. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Zhang, Y.; Liu, C.; Li, Y.; Lin, C.; Chen, H.; Hou, J.; Gao, G.; Liu, Z.; Yan, Q.; et al. Identification of CENPM as a key gene driving adrenocortical carcinoma metastasis via physical interaction with immune checkpoint ligand FGL1. Clin. Transl. Med. 2025, 15, e70182. [Google Scholar] [CrossRef]
- Mahajan, M.; Dhabalia, S.; Dash, T.; Sarkar, A.; Mondal, S. A comprehensive multi-omics study reveals potential prognostic and diagnostic biomarkers for colorectal cancer. Int. J. Biol. Macromol. 2025, 303, 140443. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, J.; Wang, G. Weighted gene co-expression network analysis for hub genes in colorectal cancer. Pharmacol. Rep. 2024, 76, 140–153. [Google Scholar] [CrossRef]
- Pengping, L.; Kexin, Y.; Yuwei, X.; Ke, S.; Rongguo, L.; Zhenyu, W.; Haigang, J.; Shaowen, W.; Yuqing, H. Construction of an anaplastic thyroid cancer stratification signature to guide immune therapy selection and validation of the pivotal gene HLF through in vitro experiments. Front. Immunol. 2024, 15, 1478904. [Google Scholar] [CrossRef]
- Lin, S.; Zhou, M.; Cheng, L.; Shuai, Z.; Zhao, M.; Jie, R.; Wan, Q.; Peng, F.; Ding, S. Exploring the association of POSTN(+) cancer-associated fibroblasts with triple-negative breast cancer. Int. J. Biol. Macromol. 2024, 268 Pt 1, 131560. [Google Scholar] [CrossRef]
- Zheng, H.; Liu, H.; Li, H.; Dou, W.; Wang, X. Weighted Gene Co-expression Network Analysis Identifies a Cancer-Associated Fibroblast Signature for Predicting Prognosis and Therapeutic Responses in Gastric Cancer. Front. Mol. Biosci. 2021, 8, 744677. [Google Scholar] [CrossRef]
- Zhong, Y.-J.; Luo, Z.-M.; Liu, F.; He, Z.-Q.; Yang, S.-Q.; Ma, W.-J.; Wang, J.-K.; Dai, Y.-S.; Zou, R.-Q.; Hu, Y.-F.; et al. Integrative analyses of bulk and single-cell transcriptomics reveals the infiltration and crosstalk of cancer-associated fibroblasts as a novel predictor for prognosis and microenvironment remodeling in intrahepatic cholangiocarcinoma. J. Transl. Med. 2024, 22, 422. [Google Scholar] [CrossRef]
- Li, Y.; Tian, R.; Liu, J.; Li, J.; Tan, H.; Wu, Q.; Fu, X. Deciphering the immune landscape dominated by cancer-associated fibroblasts to investigate their potential in indicating prognosis and guiding therapeutic regimens in high grade serous ovarian carcinoma. Front. Immunol. 2022, 13, 940801. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, S.; Wang, Z.; Wang, S.; Jiang, L.; Wang, X.; Li, J.; Shen, L. Influential upregulation of KCNE4: Propelling cancer associated fibroblasts-driven colorectal cancer progression. Cancer Cell Int. 2024, 24, 103. [Google Scholar] [CrossRef]
- Du, Y.; Jiang, X.; Wang, B.; Cao, J.; Wang, Y.; Yu, J.; Wang, X.; Liu, H. The cancer-associated fibroblasts related gene CALD1 is a prognostic biomarker and correlated with immune infiltration in bladder cancer. Cancer Cell Int. 2021, 21, 283. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Shen, X.; Wang, Z.; Chen, Y.; Weng, Y.; Yu, F.; Tang, Y.; Lu, P.; Liu, M.; Wang, L.; et al. Tumor cell-intrinsic epigenetic dysregulation shapes cancer-associated fibroblasts heterogeneity to metabolically support pancreatic cancer. Cancer Cell 2024, 42, 869–884.e9. [Google Scholar] [CrossRef] [PubMed]
- Arpinati, L.; Carradori, G.; Scherz-Shouval, R. CAF-induced physical constraints controlling T cell state and localization in solid tumours. Nat. Rev. Cancer 2024, 24, 676–693. [Google Scholar] [CrossRef] [PubMed]
- Levi-Galibov, O.; Lavon, H.; Wassermann-Dozorets, R.; Pevsner-Fischer, M.; Mayer, S.; Wershof, E.; Stein, Y.; Brown, L.E.; Zhang, W.; Friedman, G.; et al. Heat Shock Factor 1-dependent extracellular matrix remodeling mediates the transition from chronic intestinal inflammation to colon cancer. Nat. Commun. 2020, 11, 6245. [Google Scholar] [CrossRef]
- Berestjuk, I.; Lecacheur, M.; Carminati, A.; Diazzi, S.; Rovera, C.; Prod’homme, V.; Ohanna, M.; Popovic, A.; Mallavialle, A.; Larbret, F.; et al. Targeting Discoidin Domain Receptors DDR1 and DDR2 overcomes matrix-mediated tumor cell adaptation and tolerance to BRAF-targeted therapy in melanoma. EMBO Mol. Med. 2022, 14, e11814. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Zhu, W.-W.; Wang, M.-Y.; Zhai, R.-D.; Wang, Q.; Shen, W.-L.; Liu, L.-K. Cancer-associated fibroblasts promote oral squamous cell carcinoma progression through LOX-mediated matrix stiffness. J. Transl. Med. 2021, 19, 513. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, C.-C.; Ni, B.; Zhang, Z.-Z.; Jiang, S.-H.; Hu, L.-P.; Wang, X.; Zhang, X.-X.; Huang, P.-Q.; Yang, Q.; et al. Lysyl oxidase promotes liver metastasis of gastric cancer via facilitating the reciprocal interactions between tumor cells and cancer associated fibroblasts. EBioMedicine 2019, 49, 157–171. [Google Scholar] [CrossRef]
- Li, C.; Qiu, S.; Liu, X.; Guo, F.; Zhai, J.; Li, Z.; Deng, L.; Ge, L.; Qian, H.; Yang, L.; et al. Extracellular matrix-derived mechanical force governs breast cancer cell stemness and quiescence transition through integrin-DDR signaling. Signal Transduct. Target Ther. 2023, 8, 247. [Google Scholar] [CrossRef]
- Hupfer, A.; Brichkina, A.; Koeniger, A.; Keber, C.; Denkert, C.; Pfefferle, P.; Helmprobst, F.; Pagenstecher, A.; Visekruna, A.; Lauth, M. Matrix stiffness drives stromal autophagy and promotes formation of a protumourigenic niche. Proc. Natl. Acad. Sci. USA 2021, 118, e2105367118. [Google Scholar] [CrossRef]
- Malik, R.; Luong, T.; Cao, X.; Han, B.; Shah, N.; Franco-Barraza, J.; Han, L.; Shenoy, V.; Lelkes, P.; Cukierman, E. Rigidity controls human desmoplastic matrix anisotropy to enable pancreatic cancer cell spread via extracellular signal-regulated kinase 2. Matrix Biol. 2019, 81, 50–69. [Google Scholar] [CrossRef] [PubMed]
- Bera, K.; Kiepas, A.; Godet, I.; Li, Y.; Mehta, P.; Ifemembi, B.; Paul, C.D.; Sen, A.; Serra, S.A.; Stoletov, K.; et al. Extracellular fluid viscosity enhances cell migration and cancer dissemination. Nature 2022, 611, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef]
- Puttock, E.H.; Tyler, E.J.; Manni, M.; Maniati, E.; Butterworth, C.; Ramos, M.B.; Peerani, E.; Hirani, P.; Gauthier, V.; Liu, Y.; et al. Extracellular matrix educates an immunoregulatory tumor macrophage phenotype found in ovarian cancer metastasis. Nat. Commun. 2023, 14, 2514. [Google Scholar] [CrossRef]
- Rafaeva, M.; Jensen, A.R.D.; Horton, E.R.; Zornhagen, K.W.; Strøbech, J.E.; Fleischhauer, L.; Mayorca-Guiliani, A.E.; Nielsen, S.R.; Grønseth, D.S.; Kuś, F.; et al. Fibroblast-derived matrix models desmoplastic properties and forms a prognostic signature in cancer progression. Front. Immunol. 2023, 14, 1154528. [Google Scholar] [CrossRef]
- Huang, T.X.; Tan, X.-Y.; Huang, H.-S.; Li, Y.-T.; Liu, B.-L.; Liu, K.-S.; Chen, X.; Chen, Z.; Guan, X.-Y.; Zou, C.; et al. Targeting cancer-associated fibroblast-secreted WNT2 restores dendritic cell-mediated antitumour immunity. Gut 2022, 71, 333–344. [Google Scholar] [CrossRef]
- Cheng, J.T.; Deng, Y.-N.; Yi, H.-M.; Wang, G.-Y.; Fu, B.-S.; Chen, W.-J.; Liu, W.; Tai, Y.; Peng, Y.-W.; Zhang, Q. Hepatic carcinoma-associated fibroblasts induce IDO-producing regulatory dendritic cells through IL-6-mediated STAT3 activation. Oncogenesis 2016, 5, e198. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Yu, C.; You, M.; Zhang, P.; Zhang, S.; Yin, Y.; Zhang, X. A five-gene signature is a prognostic biomarker in pan-cancer and related with immunologically associated extracellular matrix. Cancer Med. 2021, 10, 4629–4643. [Google Scholar] [CrossRef]
- Gross, J.; Lapiere, C.M. Collagenolytic activity in amphibian tissues: A tissue culture assay. Proc. Natl. Acad. Sci. USA 1962, 48, 1014–1022. [Google Scholar] [CrossRef]
- Montoya Mira, J.L.; Quentel, A.; Patel, R.K.; Keith, D.; Sousa, M.; Minnier, J.; Kingston, B.R.; David, L.; Esener, S.C.; Sears, R.C.; et al. Early detection of pancreatic cancer by a high-throughput protease-activated nanosensor assay. Sci. Transl. Med. 2025, 17, eadq3110. [Google Scholar] [CrossRef] [PubMed]
- Page-McCaw, A.; Ewald, A.J.; Werb, Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007, 8, 221–233. [Google Scholar] [CrossRef] [PubMed]
- McQuibban, G.A.; Gong, J.-H.; Tam, E.M.; McCulloch, C.A.G.; Clark-Lewis, I.; Overall, C.M. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 2000, 289, 1202–1206. [Google Scholar] [CrossRef]
- Parks, W.C.; Wilson, C.L.; López-Boado, Y.S. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 2004, 4, 617–629. [Google Scholar] [CrossRef]
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef]
- Li, X.; Bao, C.; Ma, Z.; Xu, B.; Ying, X.; Liu, X.; Zhang, X. Perfluorooctanoic acid stimulates ovarian cancer cell migration, invasion via ERK/NF-κB/MMP-2/-9 pathway. Toxicol Lett. 2018, 294, 44–50. [Google Scholar] [CrossRef]
- Agrawal, S.; Anderson, P.; Durbeej, M.; van Rooijen, N.; Ivars, F.; Opdenakker, G.; Sorokin, L.M. Dystroglycan is selectively cleaved at the parenchymal basement membrane at sites of leukocyte extravasation in experimental autoimmune encephalomyelitis. J. Exp. Med. 2006, 203, 1007–1019. [Google Scholar] [CrossRef]
- Littlepage, L.E.; Sternlicht, M.D.; Rougier, N.; Phillips, J.; Gallo, E.; Yu, Y.; Williams, K.; Brenot, A.; Gordon, J.I.; Werb, Z. Matrix metalloproteinases contribute distinct roles in neuroendocrine prostate carcinogenesis, metastasis, and angiogenesis progression. Cancer Res. 2010, 70, 2224–2234. [Google Scholar] [CrossRef]
- Langenskiöld, M.; Holmdahl, L.; Falk, P.; Ivarsson, M.-L. Increased plasma MMP-2 protein expression in lymph node-positive patients with colorectal cancer. Int. J. Color. Dis. 2005, 20, 245–252. [Google Scholar] [CrossRef]
- Nakamura, E.S.; Koizumi, K.; Kobayashi, M.; Saiki, I. Inhibition of lymphangiogenesis-related properties of murine lymphatic endothelial cells and lymph node metastasis of lung cancer by the matrix metalloproteinase inhibitor MMI270. Cancer Sci. 2004, 95, 25–31. [Google Scholar] [CrossRef]
- Wong, J.C.; Chan, S.K.; Schaeffer, D.F.; Sagaert, X.; Lim, H.J.; Kennecke, H.; Owen, D.A.; Suh, K.W.; Kim, Y.-B.; Tai, I.T. Absence of MMP2 expression correlates with poor clinical outcomes in rectal cancer, and is distinct from MMP1-related outcomes in colon cancer. Clin. Cancer Res. 2011, 17, 4167–4176. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Reichert, M.; Takano, S.; von Burstin, J.; Kim, S.-B.; Lee, J.-S.; Ihida-Stansbury, K.; Hahn, C.; Heeg, S.; Schneider, G.; Rhim, A.D.; et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev. 2013, 27, 288–300. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, K.; Maurer, C.; Peschke, K.; Teller, S.; Schuck, K.; Steiger, K.; Engleitner, T.; Öllinger, R.; Nomura, A.; Wirges, N.; et al. Mesenchymal Plasticity Regulated by Prrx1 Drives Aggressive Pancreatic Cancer Biology. Gastroenterology 2021, 160, 346–361.e24. [Google Scholar] [CrossRef]
- Takano, S.; Reichert, M.; Bakir, B.; Das, K.K.; Nishida, T.; Miyazaki, M.; Heeg, S.; Collins, M.A.; Marchand, B.; Hicks, P.D.; et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev. 2016, 30, 233–247. [Google Scholar] [CrossRef]
- Leung, C.S.; Yeung, T.-L.; Yip, K.-P.; Pradeep, S.; Balasubramanian, L.; Liu, J.; Wong, K.-K.; Mangala, L.S.; Armaiz-Pena, G.N.; Lopez-Berestein, G.; et al. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 2014, 5, 5092. [Google Scholar] [CrossRef]
- Li, J.H.; Zhu, X.-X.; Li, F.-X.; Huang, C.-S.; Huang, X.-T.; Wang, J.-Q.; Gao, Z.-X.; Li, S.-J.; Xu, Q.-C.; Zhao, W.; et al. MFAP5 facilitates the aggressiveness of intrahepatic Cholangiocarcinoma by activating the Notch1 signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 476. [Google Scholar] [CrossRef]
- Zhu, L.; Chen, X.; Zhu, Y.; Qin, J.; Niu, T.; Ding, Y.; Xiao, Y.; Jiang, Y.; Liu, K.; Lu, J.; et al. Dihydroartemisinin Inhibits the Proliferation of Esophageal Squamous Cell Carcinoma Partially by Targeting AKT1 and p70S6K. Front. Pharmacol. 2020, 11, 587470. [Google Scholar] [CrossRef]
- Lee, S.; Seo, C.H.; Lim, B.; Yang, J.O.; Oh, J.; Kim, M.; Lee, S.; Lee, B.; Kang, C.; Lee, S. Accurate quantification of transcriptome from RNA-Seq data by effective length normalization. Nucleic Acids Res. 2011, 39, e9. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.-C.; Konaté, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar]
- Yang, S.; Tang, W.; Azizian, A.; Gaedcke, J.; Ströbel, P.; Wang, L.; Cawley, H.; Ohara, Y.; Valenzuela, P.; Zhang, L.; et al. Dysregulation of HNF1B/Clusterin axis enhances disease progression in a highly aggressive subset of pancreatic cancer patients. Carcinogenesis 2022, 43, 1198–1210. [Google Scholar] [CrossRef]
- Hilmi, M.; Delecourt, F.; Raffenne, J.; Bourega, T.; Dusetti, N.; Iovanna, J.; Blum, Y.; Richard, M.; Neuzillet, C.; Couvelard, A.; et al. Redefining phenotypic intratumor heterogeneity of pancreatic ductal adenocarcinoma: A bottom-up approach. J. Pathol. 2025, 265, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Racle, J.; De Jonge, K.; Baumgaertner, P.; Speiser, D.E.; Gfeller, D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. eLife 2017, 6, e26476. [Google Scholar] [CrossRef]
- Aran, D. Cell-Type Enrichment Analysis of Bulk Transcriptomes Using xCell. In Bioinformatics for Cancer Immunotherapy: Methods and Protocols; Boegel, S., Ed.; Springer: New York, NY, USA, 2020; pp. 263–276. [Google Scholar]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautes-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Li, X.; Xiang, Y.; Li, F.; Yin, C.; Li, B.; Ke, X. WNT/β-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2293. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Thirteen years of clusterProfiler. Innovation 2024, 5, 100722. [Google Scholar] [CrossRef]
- Simon, N.; Friedman, J.H.; Hastie, T.; Tibshirani, R. Regularization Paths for Cox’s Proportional Hazards Model via Coordinate Descent. J. Stat. Softw. 2011, 39, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.J.; Fernández, X.M. The 2025 Nucleic Acids Research database issue and the online molecular biology database collection. Nucleic Acids Res. 2025, 53, D1–D9. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., III; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar]


| Target | PDB ID | Drug | PubChem ID | Binding Energy (kcal/mol) |
|---|---|---|---|---|
| MMP2 | 1RTG | Staurosporine | 44259 | −7 |
| Dasatinib | 3062316 | −8.2 | ||
| OTX015 | 9936746 | −8.4 | ||
| BMS-536924 | 135440466 | −8 | ||
| Luminespib | 135539077 | −8.1 | ||
| FSTL1 | 6JZA | Staurosporine | 44259 | −7.4 |
| Dasatinib | 3062316 | −7.3 | ||
| OTX015 | 9936746 | −7.6 | ||
| BMS-536924 | 135440466 | −7 | ||
| Luminespib | 135539077 | −7 | ||
| GFPT2 | 7NUT | Staurosporine | 44259 | −8 |
| Dasatinib | 3062316 | −8.5 | ||
| OTX015 | 9936746 | −8.5 | ||
| BMS-536924 | 135440466 | −9 | ||
| Luminespib | 135539077 | −9.1 | ||
| CTSK | 8V57 | Staurosporine | 44259 | −9 |
| Dasatinib | 3062316 | −7.6 | ||
| OTX015 | 9936746 | −8.5 | ||
| BMS-536924 | 135440466 | −7.9 | ||
| Luminespib | 135539077 | −7.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Lu, Y.; Czubayko, F.; Chen, J.; Zheng, S.; Mo, H.; Liu, R.; Weber, G.F.; Grützmann, R.; Pilarsky, C.; et al. Identification of Cancer Associated Fibroblasts Related Genes Signature to Facilitate Improved Prediction of Prognosis and Responses to Therapy in Patients with Pancreatic Cancer. Int. J. Mol. Sci. 2025, 26, 4876. https://doi.org/10.3390/ijms26104876
Zhou Y, Lu Y, Czubayko F, Chen J, Zheng S, Mo H, Liu R, Weber GF, Grützmann R, Pilarsky C, et al. Identification of Cancer Associated Fibroblasts Related Genes Signature to Facilitate Improved Prediction of Prognosis and Responses to Therapy in Patients with Pancreatic Cancer. International Journal of Molecular Sciences. 2025; 26(10):4876. https://doi.org/10.3390/ijms26104876
Chicago/Turabian StyleZhou, Yong, Yanxi Lu, Franziska Czubayko, Jisheng Chen, Shuwen Zheng, Huaqing Mo, Rui Liu, Georg F. Weber, Robert Grützmann, Christian Pilarsky, and et al. 2025. "Identification of Cancer Associated Fibroblasts Related Genes Signature to Facilitate Improved Prediction of Prognosis and Responses to Therapy in Patients with Pancreatic Cancer" International Journal of Molecular Sciences 26, no. 10: 4876. https://doi.org/10.3390/ijms26104876
APA StyleZhou, Y., Lu, Y., Czubayko, F., Chen, J., Zheng, S., Mo, H., Liu, R., Weber, G. F., Grützmann, R., Pilarsky, C., & David, P. (2025). Identification of Cancer Associated Fibroblasts Related Genes Signature to Facilitate Improved Prediction of Prognosis and Responses to Therapy in Patients with Pancreatic Cancer. International Journal of Molecular Sciences, 26(10), 4876. https://doi.org/10.3390/ijms26104876






