Genome-Wide Analysis of bZIP Transcription Factor Family and Its Expression in Graft Healing of Soapberry (Sapindus mukorossi Gaertn.)
Abstract
1. Introduction
2. Results
2.1. Examining Differentially Expressed Genes in Soapberry Associated with Graft Union
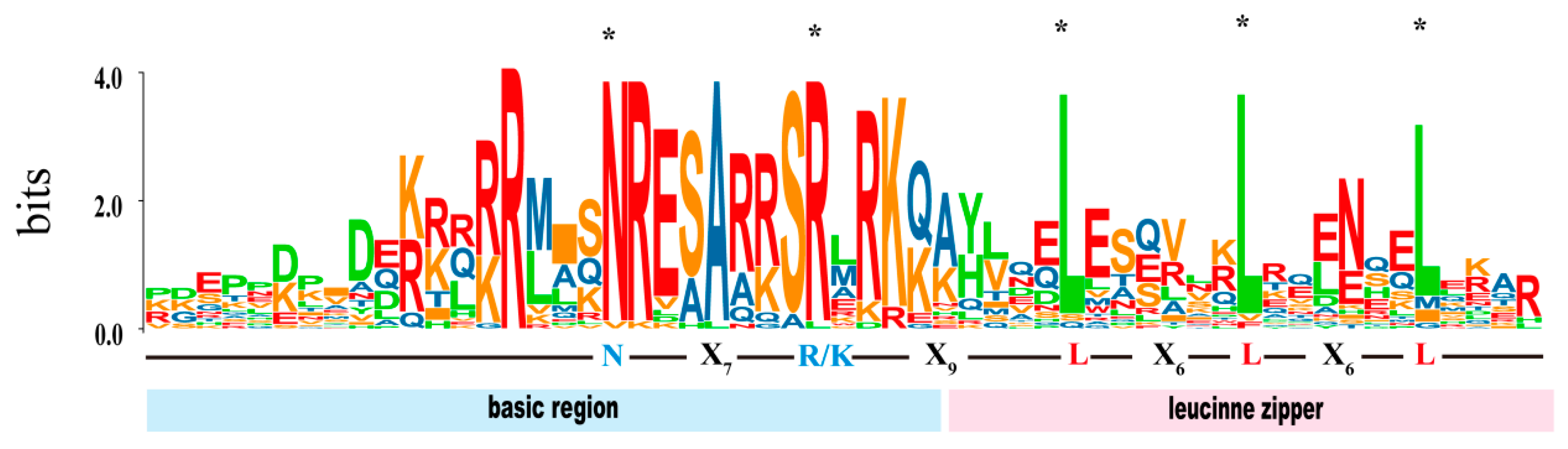
2.2. Identification and Characterization of SmbZIPs in Soapberry
2.3. Chromosomal Location and Analysis of SmbZIPs
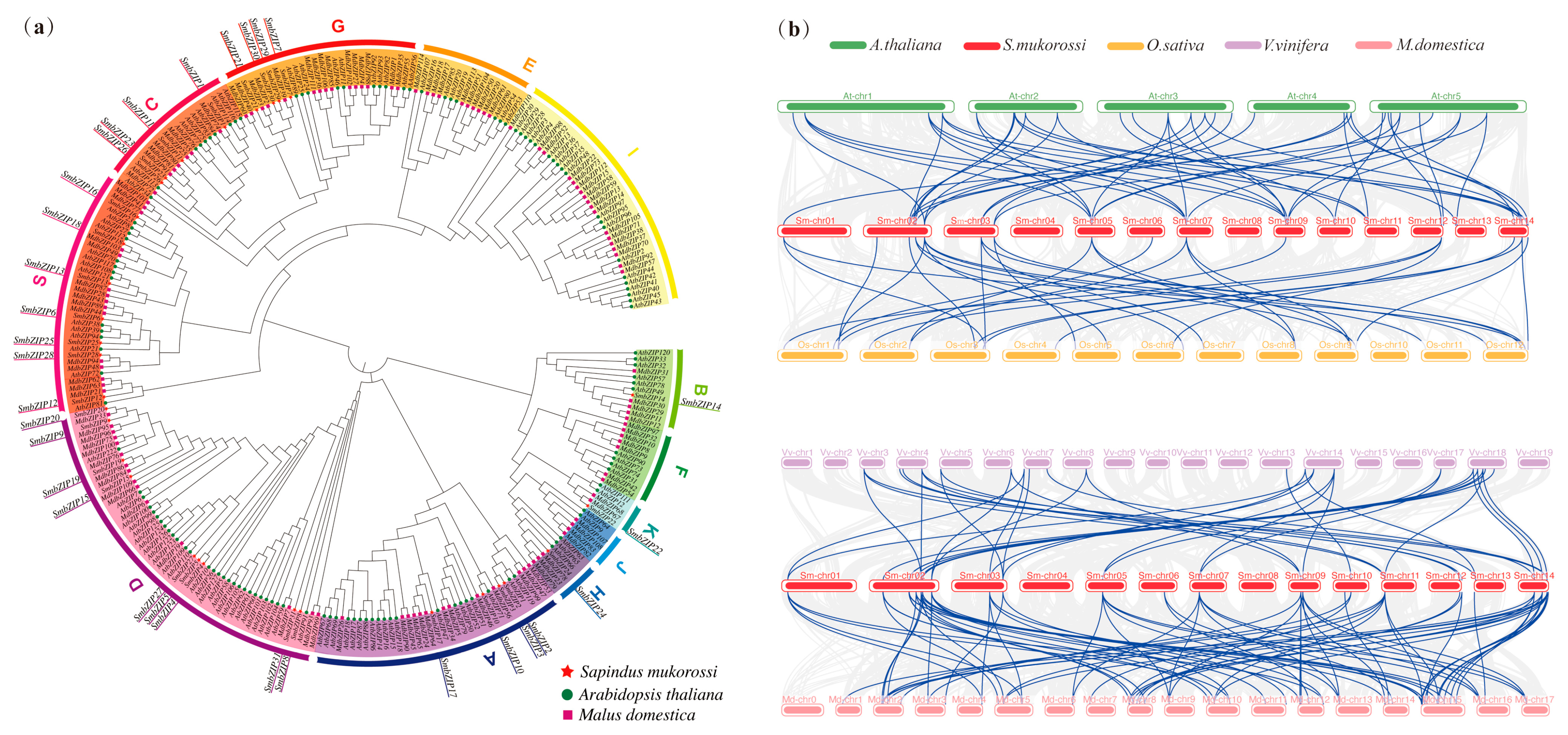
2.4. Phylogenetic Relationship and Collinearity Analysis of SmbZIPs
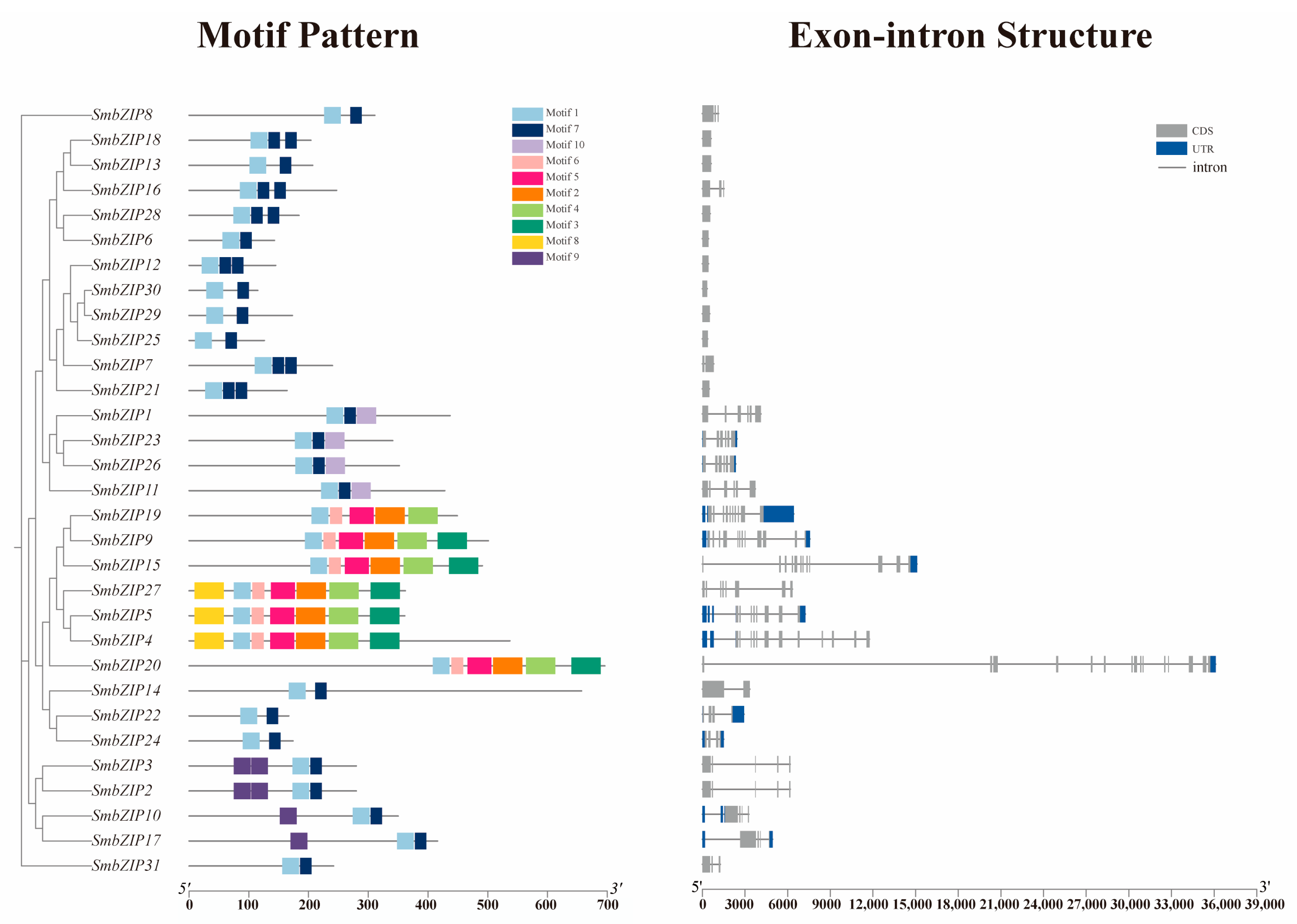
2.5. Gene Structure, Conserved Motif Analysis of SmbZIPs
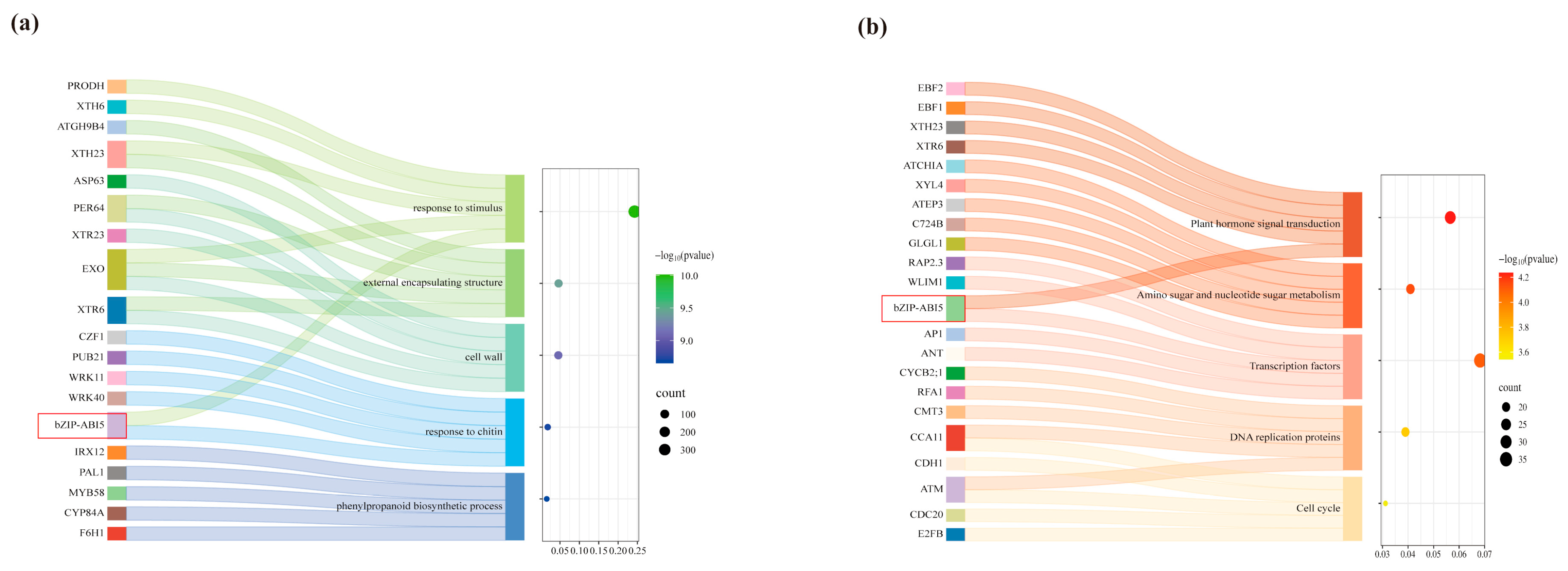
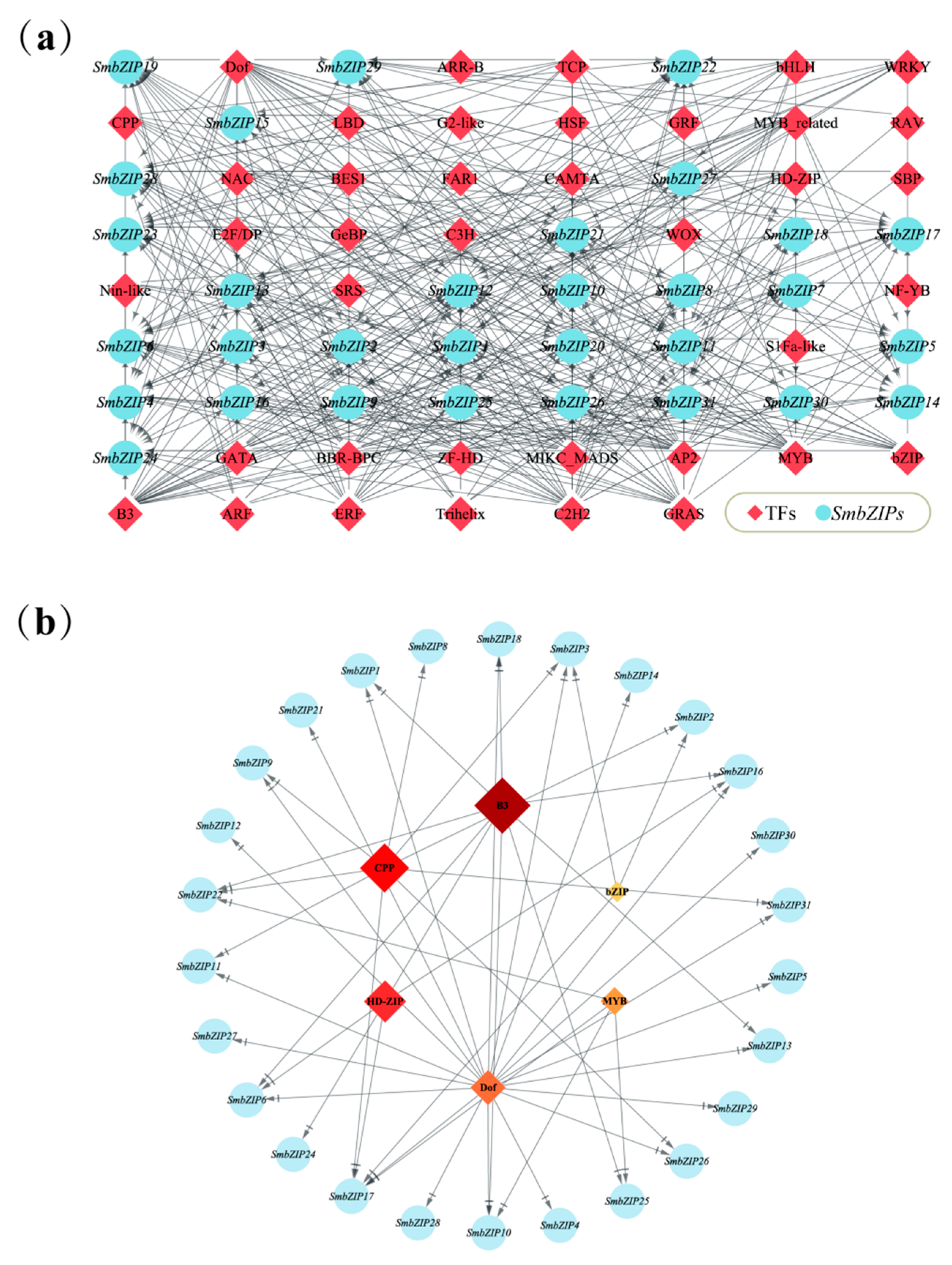
2.6. Prediction Analysis of SmbZIP-Mediated Regulatory Network
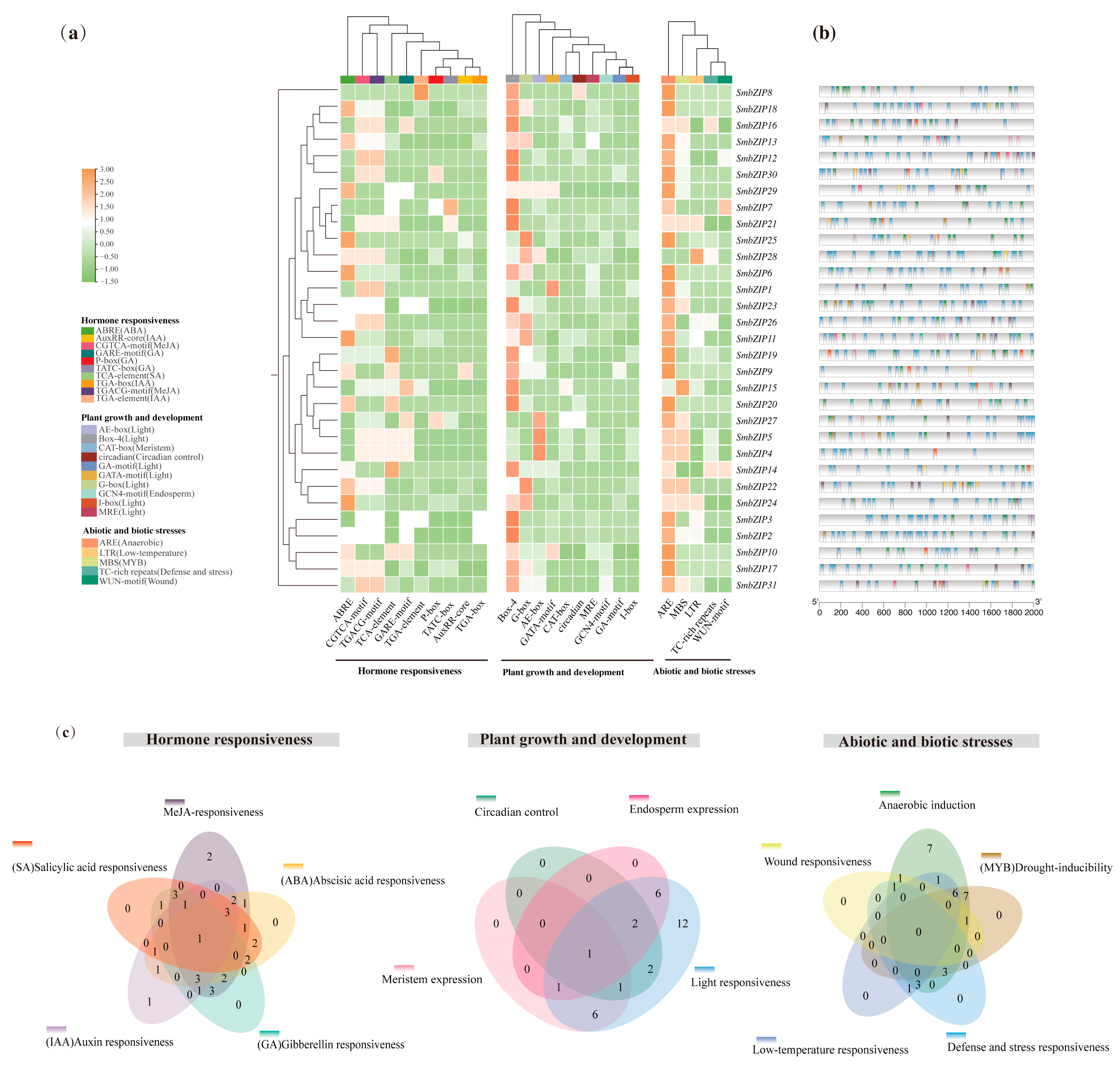
2.7. Analysis of Cis-Regulatory Elements of SmbZIPs
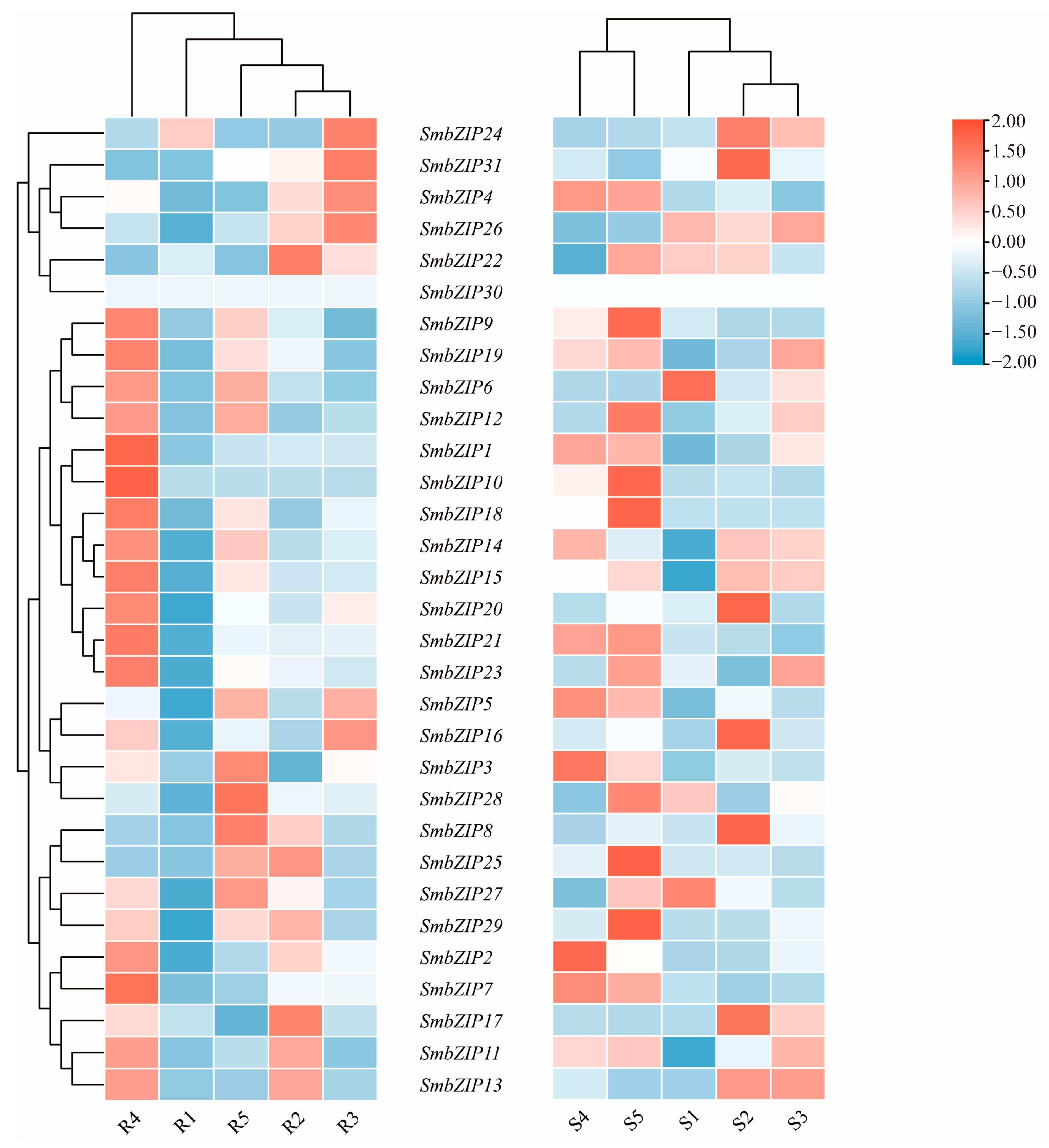
2.8. SmbZIP Gene Expression in Scion and Rootstock Tissues
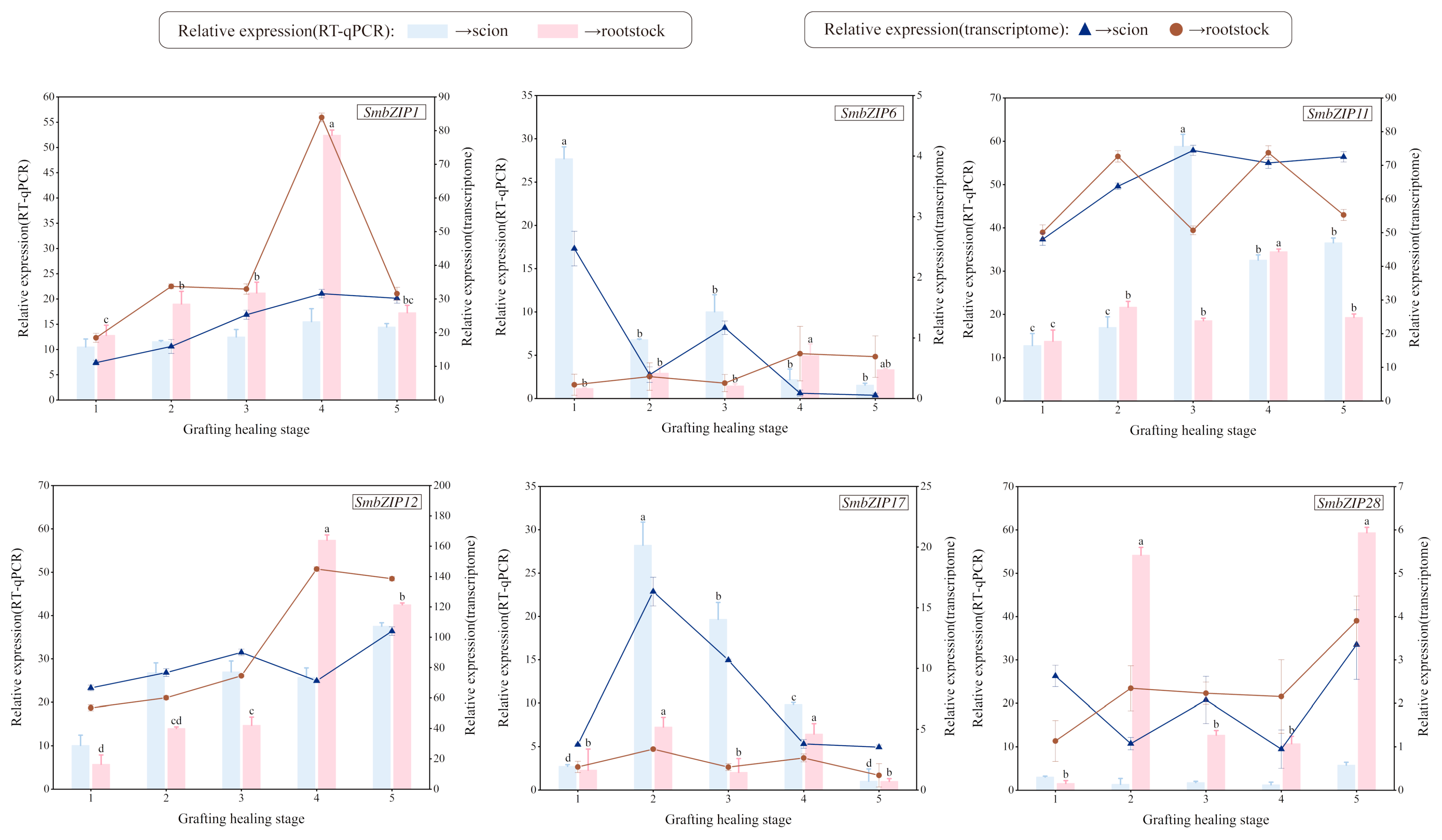
2.9. Analysis of qRT-PCR Gene Expression in SmbZIP Gene Famliy
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Gene Screening for Differential Expression
4.3. Identification of bZIP Genes in Soapberry
4.4. Analysis of Protein Physicochemical Properties and Chromosomal Distribution
4.5. Multiple Sequence Alignment and Phylogenetic Analysis
4.6. Analysis of Conserved Motifs and Gene Structure
4.7. Analysis of Collinearity
4.8. Analysis of Cis-Regulatory Elements
4.9. Analysis of TF Regulatory Network
4.10. Expression Patterns of SmbZIP Genes
4.11. Quantitative Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhao, Y.; Zhu, J.-K. Thriving under Stress: How Plants Balance Growth and the Stress Response. Dev. Cell 2020, 55, 529–543. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, H.; Liu, S.; Wang, X.; Zhang, Y.; Meng, Y.; Luo, D.; Chen, R. The NAC Transcription Factor CaNAC064 Is a Regulator of Cold Stress Tolerance in Peppers. Plant Sci. 2020, 291, 110346. [Google Scholar] [CrossRef]
- Kollist, H.; Zandalinas, S.I.; Sengupta, S.; Nuhkat, M.; Kangasjärvi, J.; Mittler, R. Rapid Responses to Abiotic Stress: Priming the Landscape for the Signal Transduction Network. Trends Plant Sci. 2019, 24, 25–37. [Google Scholar] [CrossRef]
- Shao, H.; Wang, H.; Tang, X. NAC Transcription Factors in Plant Multiple Abiotic Stress Responses: Progress and Prospects. Front. Plant Sci. 2015, 6, 902. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. IJMS 2021, 22, 6125. [Google Scholar] [CrossRef]
- Guo, Z.; Dzinyela, R.; Yang, L.; Hwarari, D. bZIP Transcription Factors: Structure, Modification, Abiotic Stress Responses and Application in Plant Improvement. Plants 2024, 13, 2058. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP Transcription Factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Nijhawan, A.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Genomic Survey and Gene Expression Analysis of the Basic Leucine Zipper Transcription Factor Family in Rice. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP Transcription Factor Family—An Update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Van Leene, J.; Blomme, J.; Kulkarni, S.R.; Cannoot, B.; De Winne, N.; Eeckhout, D.; Persiau, G.; Van De Slijke, E.; Vercruysse, L.; Vanden Bossche, R.; et al. Functional Characterization of the Arabidopsis Transcription Factor bZIP29 Reveals Its Role in Leaf and Root Development. J. Exp. Bot. 2016, 67, 5825–5840. [Google Scholar] [CrossRef]
- Wang, Z.; Cheng, K.; Wan, L.; Yan, L.; Jiang, H.; Liu, S.; Lei, Y.; Liao, B. Genome-Wide Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Six Legume Genomes. BMC Genom. 2015, 16, 1053. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Y.; Shi, H.; Guo, M.; Chai, M.; He, Q.; Yan, M.; Cao, D.; Zhao, L.; Cai, H.; et al. Evolutionary and Expression Analyses of Soybean Basic Leucine Zipper Transcription Factor Family. BMC Genom. 2018, 19, 159. [Google Scholar] [CrossRef] [PubMed]
- Collin, A.; Daszkowska-Golec, A.; Kurowska, M.; Szarejko, I. Barley ABI5 (Abscisic Acid INSENSITIVE 5) Is Involved in Abscisic Acid-Dependent Drought Response. Front. Plant Sci. 2020, 11, 1138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, J.; Jia, N.; Wang, M.; Lu, Y.; Wang, D.; Zhang, J.; Zhang, H.; Wang, X. Genome-Wide Identification and Characterization of the bZIP Gene Family and Their Function in Starch Accumulation in Chinese Chestnut (Castanea mollissima Blume). Front. Plant Sci. 2023, 14, 1166717. [Google Scholar] [CrossRef]
- Liu, J.; Chen, N.; Chen, F.; Cai, B.; Dal Santo, S.; Tornielli, G.B.; Pezzotti, M.; Cheng, Z.-M. Genome-Wide Analysis and Expression Profile of the bZIP Transcription Factor Gene Family in Grapevine (Vitis vinifera). BMC Genom. 2014, 15, 281. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Fu, F.; Zhang, H.; Song, F. Genome-Wide Systematic Characterization of the bZIP Transcriptional Factor Family in Tomato (Solanum lycopersicum L.). BMC Genom. 2015, 16, 771. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Feng, X.; Li, Y.; Guzmán, C.; Lin, N.; Xu, Q.; Zhang, Y.; Tang, H.; Qi, P.; Deng, M.; et al. Genome-Wide Identification of bZIP Transcription Factor Genes Related to Starch Synthesis in Barley ( Hordeum vulgare L.). Genome 2021, 64, 1067–1080. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Meng, D.; Li, M.; Cheng, L. Genome-Wide Identification and Expression Analysis of the bZIP Gene Family in Apple (Malus domestica). Tree Genet. Genomes 2016, 12, 82. [Google Scholar] [CrossRef]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-Wide Analysis of bZIP-Encoding Genes in Maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, D.; Jia, L.; Huang, X.; Ma, G.; Wang, S.; Zhu, M.; Zhang, A.; Guan, M.; Lu, K.; et al. Genome-Wide Identification and Structural Analysis of bZIP Transcription Factor Genes in Brassica napus. Genes 2017, 8, 288. [Google Scholar] [CrossRef]
- Sun, S.; Ke, X.; Cui, L.; Yang, G.; Bi, Y.; Song, F.; Xu, X. Enzymatic Epoxidation of Sapindus Mukorossi Seed Oil by Perstearic Acid Optimized Using Response Surface Methodology. Ind. Crops Prod. 2011, 33, 676–682. [Google Scholar] [CrossRef]
- Melnyk, C.W.; Schuster, C.; Leyser, O.; Meyerowitz, E.M. A Developmental Framework for Graft Formation and Vascular Reconnection in Arabidopsis thaliana. Curr. Biol. 2015, 25, 1306–1318. [Google Scholar] [CrossRef]
- Padilla, Y.G.; Gisbert-Mullor, R.; López-Galarza, S.; Albacete, A.; Martínez-Melgarejo, P.A.; Calatayud, Á. Short-Term Water Stress Responses of Grafted Pepper Plants Are Associated with Changes in the Hormonal Balance. Front. Plant Sci. 2023, 14, 1170021. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liao, Y.; Liu, J.; Zhao, T.; Jia, L.; Chen, Z. Advances in Understanding the Graft Healing Mechanism: A Review of Factors and Regulatory Pathways. Hortic. Res. 2024, 11, uhae175. [Google Scholar] [CrossRef]
- Wu, Y.; Li, X.; Zhang, J.; Zhao, H.; Tan, S.; Xu, W.; Pan, J.; Yang, F.; Pi, E. ERF Subfamily Transcription Factors and Their Function in Plant Responses to Abiotic Stresses. Front. Plant Sci. 2022, 13, 1042084. [Google Scholar] [CrossRef] [PubMed]
- Vinson, C.R.; Sigler, P.B.; McKnight, S.L. Scissors-Grip Model for DNA Recognition by a Family of Leucine Zipper Proteins. Science 1989, 246, 911–916. [Google Scholar] [CrossRef]
- Zhao, K.; Chen, S.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-Wide Analysis and Expression Profile of the bZIP Gene Family in Poplar. BMC Plant Biol. 2021, 21, 122. [Google Scholar] [CrossRef]
- He, G.-H.; Xu, J.-Y.; Wang, Y.-X.; Liu, J.-M.; Li, P.-S.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S. Drought-Responsive WRKY Transcription Factor Genes TaWRKY1 and TaWRKY33 from Wheat Confer Drought and/or Heat Resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, J.; Han, X.; Zhang, Y.; Yang, Q.; Tong, Z. Identification and expression analysis of bZIP gene family under ABA treatment in Phoebe bournei. J. Zhejiang AF Univ. 2024, 41, 275–285. [Google Scholar]
- Hu, X.; Liang, J.; Wang, W.; Cai, C.; Ye, S.; Wang, N.; Han, F.; Wu, Y.; Zhu, Q. Comprehensive Genome-wide Analysis of the DREB Gene Family in Moso Bamboo (Phyllostachys edulis): Evidence for the Role of PeDREB28 in Plant Abiotic Stress Response. Plant J. 2023, 116, 1248–1270. [Google Scholar] [CrossRef]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N.; et al. Genome-Wide Identification and Expression Profiling of the TCP Family Genes in Spike and Grain Development of Wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M.; Corpas, F.J. Class III Peroxidases (POD) in Pepper (Capsicum annuum L.): Genome-Wide Identification and Regulation during Nitric Oxide (NO)-Influenced Fruit Ripening. Antioxidants 2023, 12, 1013. [Google Scholar] [CrossRef]
- Yang, Z. Bayes Empirical Bayes Inference of Amino Acid Sites Under Positive Selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.-R.; Yu, Y.-H.; Ni, P.-Y.; Zhang, G.-H.; Guo, D.-L. Genome-Wide Identification of Small Heat-Shock Protein (HSP20) Gene Family in Grape and Expression Profile during Berry Development. BMC Plant Biol. 2019, 19, 433. [Google Scholar] [CrossRef] [PubMed]
- Bishopp, A.; Help, H.; El-Showk, S.; Weijers, D.; Scheres, B.; Friml, J.; Benková, E.; Mähönen, A.P.; Helariutta, Y. A Mutually Inhibitory Interaction between Auxin and Cytokinin Specifies Vascular Pattern in Roots. Curr. Biol. 2011, 21, 917–926. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Sánchez Rodríguez, F.; Wu, M.-F.; Solé-Gil, A.; Leal Gavarrón, M.; Siligato, R.; Miyashima, S.; et al. High Levels of Auxin Signalling Define the Stem-Cell Organizer of the Vascular Cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Thomas, H.; Van Den Broeck, L.; Spurney, R.; Sozzani, R.; Frank, M. Gene Regulatory Networks for Compatible versus Incompatible Grafts Identify a Role for SlWOX4 during Junction Formation. Plant Cell 2022, 34, 535–556. [Google Scholar] [CrossRef]
- Zhang, A.; Matsuoka, K.; Kareem, A.; Robert, M.; Roszak, P.; Blob, B.; Bisht, A.; De Veylder, L.; Voiniciuc, C.; Asahina, M.; et al. Cell-Wall Damage Activates DOF Transcription Factors to Promote Wound Healing and Tissue Regeneration in Arabidopsis thaliana. Curr. Biol. 2022, 32, 1883–1894.e7. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Lund, H. Renewable Energy Strategies for Sustainable Development. Energy 2007, 32, 912–919. [Google Scholar] [CrossRef]
- Nikolaidis, P. A Comparative Overview of Hydrogen Production Processes. Renew. Sustain. Energy Rev. 2017, 67, 597–611. [Google Scholar] [CrossRef]
- Zhao, C.; Meng, G.; Wu, S.; Zhang, C.; Liu, Y. Analysis on Policy Evolution and Industry Development of Forestry Biomass Energy in China. For. Resour. Manag. 2022, 1–7. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Ren, C.; Wang, H.; Zhou, Z.; Jia, J.; Zhang, Q.; Liang, C.; Li, W.; Zhang, Y.; Yu, G. Genome-Wide Identification of the B3 Gene Family in Soybean and the Response to Melatonin under Cold Stress. Front. Plant Sci. 2023, 13, 1091907. [Google Scholar] [CrossRef] [PubMed]
- Flagel, L.E.; Wendel, J.F. Gene Duplication and Evolutionary Novelty in Plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Abscisic-Acid-Dependent Basic Leucine Zipper (bZIP) Transcription Factors in Plant Abiotic Stress. Protoplasma 2017, 254, 3–16. [Google Scholar] [CrossRef]
- Li, X.-Y.; Liu, X.; Yao, Y.; Li, Y.-H.; Liu, S.; He, C.-Y.; Li, J.-M.; Lin, Y.-Y.; Li, L. Overexpression of Arachis Hypogaea AREB1 Gene Enhances Drought Tolerance by Modulating ROS Scavenging and Maintaining Endogenous ABA Content. IJMS 2013, 14, 12827–12842. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 Is a Transcription Activator of Novel ABRE-Dependent ABA Signaling That Enhances Drought Stress Tolerance in Arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Guo, T.; Guo, B.; Chen, Z.; An, X. Comprehensive Analysis of the R2R3-MYB Transcription Factor Gene Family in Populus Trichocarpa. Ind. Crops Prod. 2021, 168, 113614. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Weiste, C. The C/S1 bZIP Network: A Regulatory Hub Orchestrating Plant Energy Homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Li, Q.; Sun, T.; Chai, S.; Wang, C.; Bai, L.; Sun, M.; Li, Y.; Qin, X.; Zhang, Z.; et al. Sugars Promote Graft Union Development in the Heterograft of Cucumber onto Pumpkin. Hortic. Res. 2021, 8, 146. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, Y.-L. Regulation of Cell Division and Expansion by Sugar and Auxin Signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly Regulated Genes Are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Johansson, H.; Lee, K.P.; Bou-Torrent, J.; Stewart, K.; Steel, G.; Rodríguez-Concepción, M.; Halliday, K.J. The HY5-PIF Regulatory Module Coordinates Light and Temperature Control of Photosynthetic Gene Transcription. PLoS Genet. 2014, 10, e1004416. [Google Scholar] [CrossRef]
- Nie, X.; Ji, X.; Liu, Y.; Zheng, L.; Wang, Y. Elucidation of the Specific Formation of Homo- and Heterodimeric Forms of ThbZIP1 and Its Role in Stress. IJMS 2014, 15, 10005–10017. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four A Rabidopsis AREB / ABF Transcription Factors Function Predominantly in Gene Expression Downstream of SnRK2 Kinases in Abscisic Acid Signalling in Response to Osmotic Stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Zhou, M.; Fan, J.; Gao, Y.; Zheng, C.; Xu, Y.; Jia, L.; An, X.; Chen, Z. Identification and Analysis of UGT Genes Associated with Triterpenoid Saponin in Soapberry (Sapindus Mukorossi Gaertn.). BMC Plant Biol. 2024, 24, 588. [Google Scholar] [CrossRef]
- Notaguchi, M.; Kurotani, K.; Sato, Y.; Tabata, R.; Kawakatsu, Y.; Okayasu, K.; Sawai, Y.; Okada, R.; Asahina, M.; Ichihashi, Y.; et al. Cell-Cell Adhesion in Plant Grafting Is Facilitated by b-1,4-Glucanases. Science 2020, 369, 698–702. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in Homology Search: HMMER3 and Convergent Evolution of Coiled-Coil Regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Molineris, I.; Sales, G.; Bianchi, F.; Di Cunto, F.; Caselle, M. A New Approach for the Identification of Processed Pseudogenes. J. Comput. Biol. 2010, 17, 755–765. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as Designed by Its Users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for Motif Discovery and Searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a Database of Plant cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Jin, J.; Gao, G. PlantRegMap: Charting Functional Regulatory Maps in Plants. Nucleic Acids Res. 2019, 48, D1104–D1113. [Google Scholar] [CrossRef]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for Visualization and Analysis of Biological Networks. In Data Mining in Proteomics; Springer: New York, NY, USA, 2011; Volume 696, pp. 291–303. ISBN 978-1-60761-986-4. [Google Scholar]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer Premier: Program for Design of Degenerate Primers from a Protein Sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Xu, Y.; Zhao, G.; Ji, X.; Liu, J.; Zhao, T.; Gao, Y.; Gao, S.; Hao, Y.; Gao, Y.; Wang, L.; et al. Metabolome and Transcriptome Analysis Reveals the Transcriptional Regulatory Mechanism of Triterpenoid Saponin Biosynthesis in Soapberry (Sapindus Mukorossi Gaertn.). J. Agric. Food Chem. 2022, 70, 7095–7109. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, N.; Wang, L.; Zhong, J.; Jia, L.; Chen, Z. Genome-Wide Analysis of bZIP Transcription Factor Family and Its Expression in Graft Healing of Soapberry (Sapindus mukorossi Gaertn.). Int. J. Mol. Sci. 2025, 26, 4862. https://doi.org/10.3390/ijms26104862
Chen N, Wang L, Zhong J, Jia L, Chen Z. Genome-Wide Analysis of bZIP Transcription Factor Family and Its Expression in Graft Healing of Soapberry (Sapindus mukorossi Gaertn.). International Journal of Molecular Sciences. 2025; 26(10):4862. https://doi.org/10.3390/ijms26104862
Chicago/Turabian StyleChen, Na, Lixian Wang, Jing Zhong, Liming Jia, and Zhong Chen. 2025. "Genome-Wide Analysis of bZIP Transcription Factor Family and Its Expression in Graft Healing of Soapberry (Sapindus mukorossi Gaertn.)" International Journal of Molecular Sciences 26, no. 10: 4862. https://doi.org/10.3390/ijms26104862
APA StyleChen, N., Wang, L., Zhong, J., Jia, L., & Chen, Z. (2025). Genome-Wide Analysis of bZIP Transcription Factor Family and Its Expression in Graft Healing of Soapberry (Sapindus mukorossi Gaertn.). International Journal of Molecular Sciences, 26(10), 4862. https://doi.org/10.3390/ijms26104862





