Preliminary Evaluation of Plasma circ_0009910, circ_0027478, and miR-1236-3p as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinicopathological Characteristics and Laboratory Results of the Studied Groups
2.2. The Plasma Expression Distribution of has_circ_0009910, hsa_circ_0027478, and miR-1236-3p
2.3. Plasma Levels of AFP in the Studied Groups
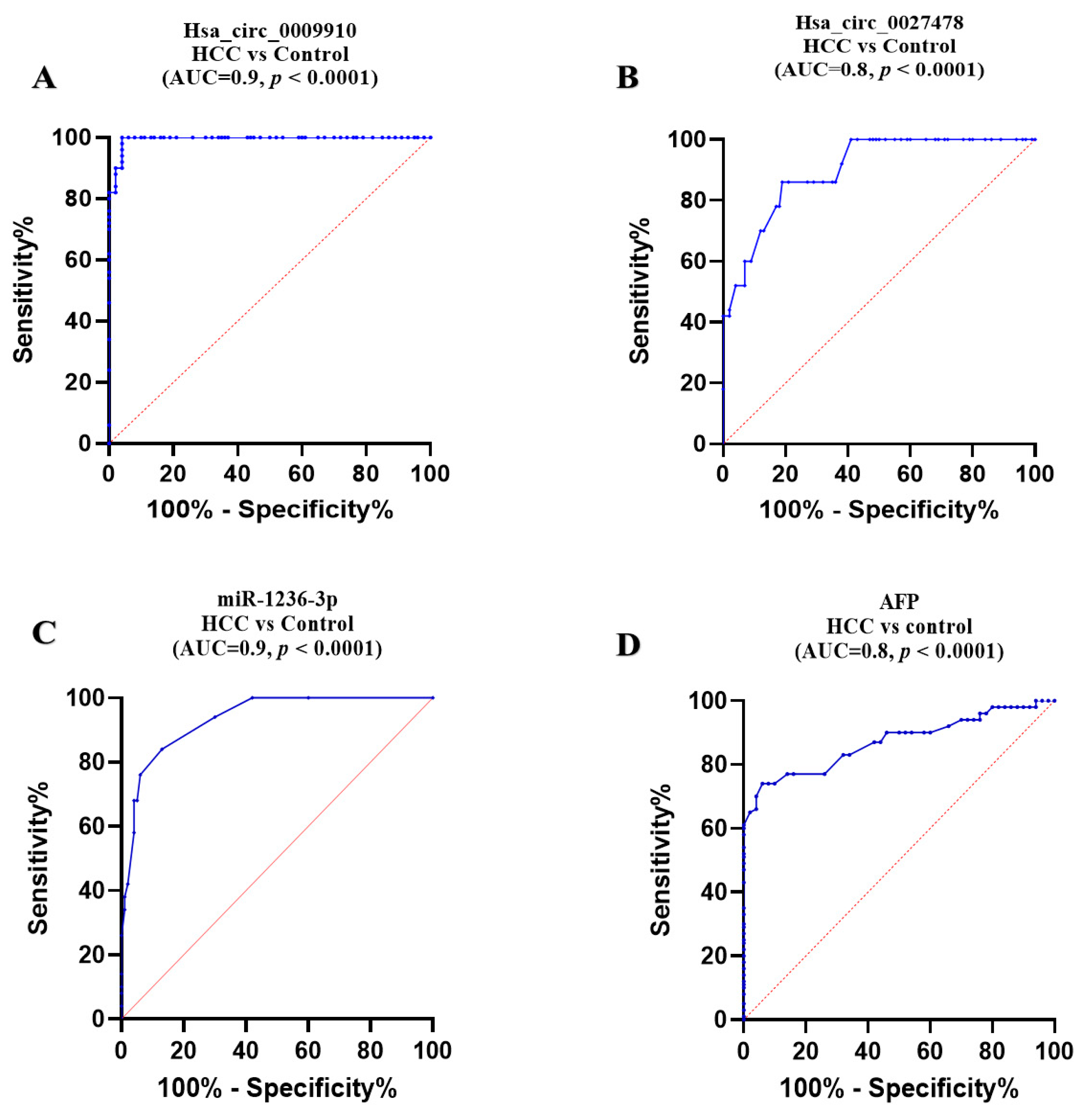
2.4. Comparative Diagnostic Accuracy of the Studied Parameters
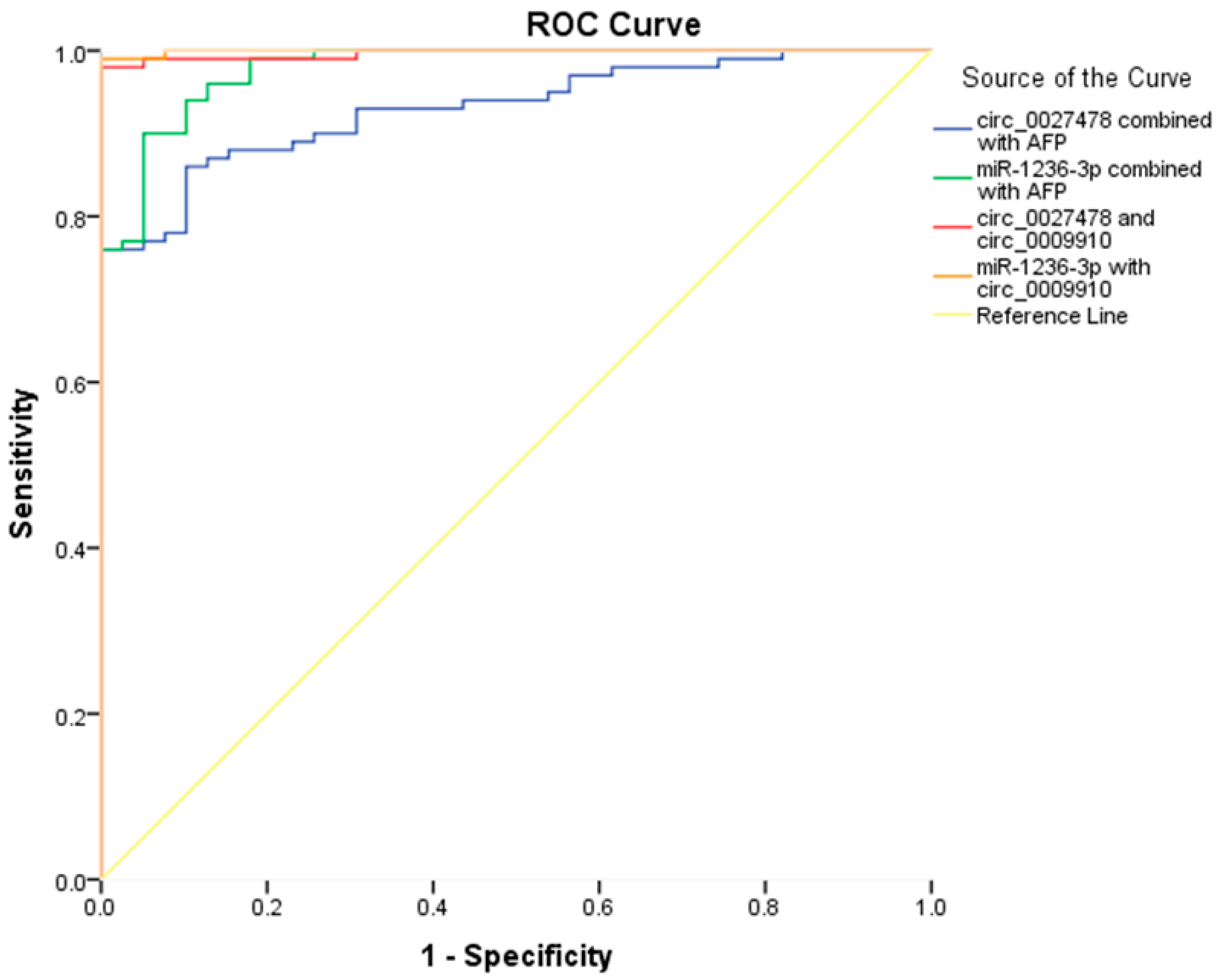
2.5. Enhancing the Diagnostic Performance of AFP Through Integration with the Studied circRNAs and miR-1236
2.6. Prognostic Significance of the Studied Biomarkers
2.7. Analysis Using Logistic Regression
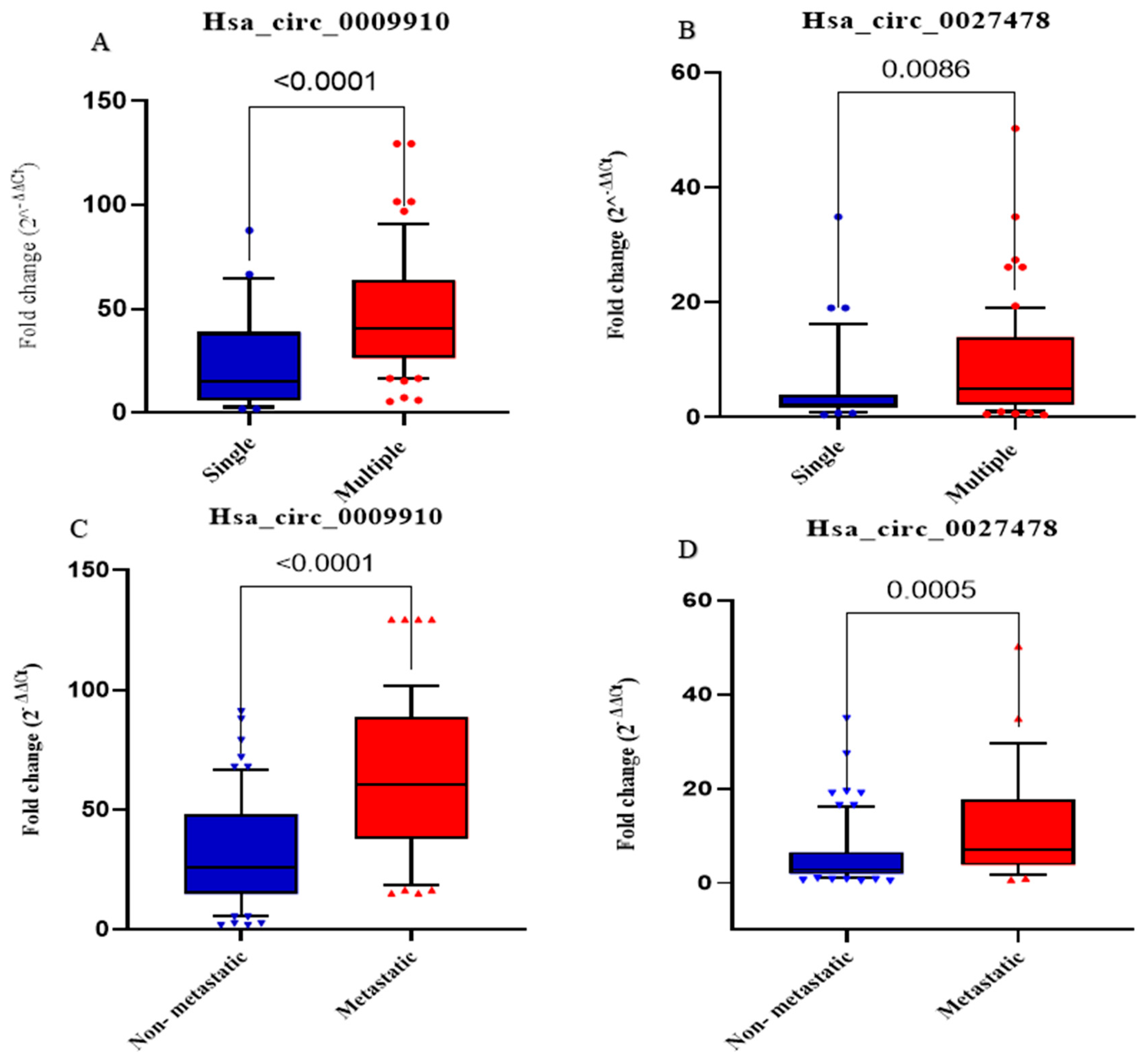
2.8. Association Between Expression Levels of Analyzed Parameters and Clinicopathological Features in Patients with HCC
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Blood Specimens
4.3. Selection of circRNAs and Prediction of circRNA–miRNA Interactions
4.4. Sample Size Calculation
4.5. Assessment of Plasma circ_0009910, circ_0027478, and miR-1236 Using RT-qPCR
4.6. Assessment of Serum AFP, Using ELISA
4.7. The Difference Between Staging Systems in the Current Study
4.8. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chidambaranathan-Reghupaty, S.; Fisher, P.B.; Sarkar, D. Chapter One—Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. In Advances in Cancer Research; Sarkar, D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2021; Volume 149, pp. 1–61. [Google Scholar]
- Yang, J.D.; Heimbach, J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ 2020, 371, m3544. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, H.; Zhang, L.; Zhu, A.X.; Bernards, R.; Qin, W.; Wang, C. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Rumgay, H.; Ferlay, J.; de Martel, C.; Georges, D.; Ibrahim, A.S.; Zheng, R.; Wei, W.; Lemmens, V.E.; Soerjomataram, I. Global, regional and national burden of primary liver cancer by subtype. Eur. J. Cancer. 2022, 161, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Wong, G.; Anstee, Q.M.; Henry, L. The global burden of liver disease. Clin. Gastroenterol. Hepatol. 2023, 21, 1978–1991. [Google Scholar] [CrossRef]
- Sharma, K.K.; Mohsin, M.; Mittal, P.; Ali, Z.; Fatma, N.; Upadhyay, P.; Gupta, R.; Verma, A.; Kumar, G. Diagnosis of the initial stage of hepatocellular carcinoma: A review. Curr. Pharm. Des. 2024, 30, 1708–1724. [Google Scholar] [CrossRef]
- Wang, W.; Wei, C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020, 7, 308–319. [Google Scholar] [CrossRef]
- Sparchez, Z.; Craciun, R.; Caraiani, C.; Horhat, A.; Nenu, I.; Procopet, B.; Sparchez, M.; Stefanescu, H.; Mocan, T. Ultrasound or sectional imaging techniques as screening tools for hepatocellular carcinoma: Fall forward or move forward? J. Clin. Med. 2021, 10, 903. [Google Scholar] [CrossRef]
- Tsuchiya, N.; Sawada, Y.; Endo, I.; Saito, K.; Uemura, Y.; Nakatsura, T. Biomarkers for the early diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 2015, 21, 10573–10583. [Google Scholar] [CrossRef]
- Manea, I.; Iacob, R.; Iacob, S.; Cerban, R.; Dima, S.; Oniscu, G.; Popescu, I.; Gheorghe, L. Liquid biopsy for early detection of hepatocellular carcinoma. Front. Med. 2023, 10, 1218705. [Google Scholar] [CrossRef]
- Nahon, P.; Najean, M.; Layese, R.; Zarca, K.; Segar, L.B.; Cagnot, C.; Ganne-Carrié, N.; N’kontchou, G.; Pol, S.; Chaffaut, C.; et al. Early hepatocellular carcinoma detection using magnetic resonance imaging is cost-effective in high-risk patients with cirrhosis. JHEP Rep. 2022, 4, 100390. [Google Scholar] [CrossRef]
- Koustas, E.; Trifylli, E.-M.; Sarantis, P.; Papadopoulos, N.; Papanikolopoulos, K.; Aloizos, G.; Damaskos, C.; Garmpis, N.; Garmpi, A.; Matthaios, D.; et al. An insight into the arising role of MicroRNAs in hepatocellular carcinoma: Future diagnostic and therapeutic approaches. Int. J. Mol. Sci. 2023, 24, 7168. [Google Scholar] [CrossRef] [PubMed]
- Caputo, V.; De Falco, V.; Ventriglia, A.; Famiglietti, V.; Martinelli, E.; Morgillo, F.; Martini, G.; Della Corte, C.M.; Ciardiello, D.; Poliero, L.; et al. Comprehensive genome profiling by next generation sequencing of circulating tumor DNA in solid tumors: A single academic institution experience. Ther. Adv. Med Oncol. 2022, 14, 17588359221096878. [Google Scholar] [CrossRef]
- Saleem, A.; Khan, M.U.; Zahid, T.; Khurram, I.; Ghani, M.U.; Ullah, I.; Munir, R.; Calina, D.; Sharifi-Rad, J. Biological role and regulation of circular RNA as an emerging biomarker and potential therapeutic target for cancer. Mol. Biol. Rep. 2024, 51, 296. [Google Scholar] [CrossRef] [PubMed]
- Vakili, O.; Asili, P.; Babaei, Z.; Mirahmad, M.; Keshavarzmotamed, A.; Asemi, Z.; Mafi, A. Circular RNAs in Alzheimer’s disease: A new perspective of diagnostic and therapeutic targets. CNS Neurol. Disord. Drug Targets CNS Neurol. Disord. 2023, 22, 1335–1354. [Google Scholar] [CrossRef]
- Liu, L.; Gu, M.; Ma, J.; Wang, Y.; Li, M.; Wang, H.; Yin, X.; Li, X. CircGPR137B/miR-4739/FTO feedback loop suppresses tumorigenesis and metastasis of hepatocellular carcinoma. Mol. Cancer 2022, 21, 149. [Google Scholar] [CrossRef] [PubMed]
- Du, A.; Li, S.; Zhou, Y.; Disoma, C.; Liao, Y.; Zhang, Y.; Chen, Z.; Yang, Q.; Liu, P.; Liu, S.; et al. MM6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol. Cancer 2022, 21, 109. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, L.; Zhang, R.; Hou, P.; Wang, J.; Wu, L.; Li, J. Circ-ZEB1 promotes PIK3CA expression by silencing miR-199a-3p and affects the proliferation and apoptosis of hepatocellular carcinoma. Mol. Cancer. 2022, 21, 72. [Google Scholar] [CrossRef]
- Li, Y.; Wu, A.; Chen, L.; Cai, A.; Hu, Y.; Zhou, Z.; Qi, Q.; Wu, Y.; Xia, D.; Dong, P.; et al. Hsa_circ_0000098 is a novel therapeutic target that promotes hepatocellular carcinoma development and resistance to doxorubicin. J. Exp. Clin. Cancer Res. 2022, 41, 267. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, X.; Cong, H. Advances in the protein-encoding functions of circular RNAs associated with cancer (Review). Oncol. Rep. 2023, 50, 160. [Google Scholar] [CrossRef]
- Sun, M.; Yang, Y. Biological functions and applications of circRNAs—Next generation of RNA-based therapy. J. Mol. Cell Biol. 2023, 15, mjad031. [Google Scholar] [CrossRef]
- Fontemaggi, G.; Turco, C.; Esposito, G.; Di Agostino, S. New molecular mechanisms and clinical impact of circRNAs in human cancer. Cancers 2021, 13, 3154. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wang, L.; Hu, G.; Zhang, Y.; Du, J.; Ding, J.; Ji, X.; Shen, H.; Huang, H.; Ye, F.; et al. CircPVT1 promotes ER-positive breast tumorigenesis and drug resistance by targeting ESR1 and MAVS. EMBO J. 2023, 42, e112408. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- An, J.X.; Ma, M.H.; Zhang, C.D.; Shao, S.; Zhou, N.M.; Dai, D.Q. miR-1236-3p inhibits invasion and metastasis in gastric cancer by targeting MTA2. Cancer Cell Int. 2018, 18, 66. [Google Scholar] [CrossRef]
- Sletten, M.; Skogstrom, K.B.; Lind, S.M.; Tinholt, M.; Stavik, B.; Rayner, S.; Iversen, N. Elevated TFPI is a prognostic factor in hepatocellular carcinoma: Putative role of miR-7-5p and miR-1236-3p. Thromb. Res. 2024, 241, 109073. [Google Scholar] [CrossRef]
- Guo, L.; Jia, L.; Luo, L.; Xu, X.; Xiang, Y.; Ren, Y.; Ren, D.; Shen, L.; Liang, T. Critical Roles of Circular RNA in Tumor Metastasis via Acting as a Sponge of miRNA/isomiR. Int. J. Mol. Sci. 2022, 23, 7024. [Google Scholar] [CrossRef]
- Gao, R.; Cai, C.; Gan, J.; Yang, X.; Shuang, Z.; Liu, M.; Li, S.; Tang, H. miR-1236 down-regulates alpha-fetoprotein, thus causing PTEN accumulation, which inhibits the PI3K/Akt pathway and malignant phenotype in hepatoma cells. Oncotarget 2015, 6, 6014–6028. [Google Scholar] [CrossRef]
- Liu, M.; Liu, K.D.; Zhang, L.; Cai, J.; Yao, H.W.; Bai, Y.K.; Zhang, Z.T. Circ_0009910 regulates growth and metastasis and is associated with poor prognosis in gastric cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8248–8256. [Google Scholar]
- Li, Y.; Lin, S.; An, N. Hsa_circ_0009910: Oncogenic circular RNA targets microRNA-145 in ovarian cancer cells. Cell Cycle 2020, 19, 1857–1868. [Google Scholar] [CrossRef]
- Deng, N.; Li, L.; Gao, J.; Zhou, J.; Wang, Y.; Wang, C.; Liu, Y. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem. Biophys. Res. Commun. 2018, 495, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Ping, L.; Jian-Jun, C.; Chu-Shu, L.; Guang-Hua, L.; Ming, Z. Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol. Dis. 2019, 75, 41–47. [Google Scholar] [CrossRef]
- Suk, K.T.; Kim, D.J. Staging of liver fibrosis or cirrhosis: The role of hepatic venous pressure gradient measurement. World J Hepatol. 2015, 7, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, R.; Eltabbakh, M.; El Kassas, M. Unique situation of hepatocellular carcinoma in Egypt: A review of epidemiology and control measures. World J. Gastrointest. Oncol. 2021, 13, 1919–1938. [Google Scholar] [CrossRef] [PubMed]
- Kotsari, M.; Dimopoulou, V.; Koskinas, J.; Armakolas, A. Immune System and Hepatocellular Carcinoma (HCC): New Insights into HCC Progression. Int. J. Mol. Sci. 2023, 24, 11471. [Google Scholar] [CrossRef]
- Sukowati, C.H.C.; Cabral, L.K.D.; Tiribelli, C.; Pascut, D. Circulating Long and Circular Noncoding RNA as Non-Invasive Diagnostic Tools of Hepatocellular Carcinoma. Biomedicines 2021, 9, 90. [Google Scholar] [CrossRef]
- Song, R.; Ma, S.; Xu, J.; Ren, X.; Guo, P.; Liu, H.; Li, P.; Yin, F.; Liu, M.; Wang, Q.; et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol. Cancer 2023, 22, 16. [Google Scholar] [CrossRef]
- Li, J.; Hu, Z.Q.; Yu, S.Y.; Mao, L.; Zhou, Z.J.; Wang, P.C.; Gong, Y.; Su, S.; Zhou, J.; Fan, J.; et al. CircRPN2 inhibits aerobic glycolysis and metastasis in hepatocellular carcinoma. Cancer Res. 2022, 82, 1055–1069. [Google Scholar] [CrossRef]
- Zhou, W.-Y.; Cai, Z.-R.; Liu, J.; Wang, D.-S.; Ju, H.-Q.; Xu, R.-H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Su, X.; Wang, P.; Lin, W. The value of circulating circular RNA in cancer diagnosis, monitoring, prognosis, and guiding treatment. Front. Oncol. 2021, 11, 736546. [Google Scholar] [CrossRef]
- Qiu, L.; Wang, T.; Ge, Q.; Xu, H.; Wu, Y.; Tang, Q.; Chen, K. Circular RNA signature in hepatocellular carcinoma. J. Cancer 2019, 10, 3361. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Ding, W.B.; Wang, M.C.; Guo, X.G.; Xu, J.; Xu, Q.G.; Yang, Y.; Sun, S.H.; Liu, J.F.; Qin, L.X.; et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: A large-scale, multicenter study. Int. J. Cancer 2020, 146, 1754–1763. [Google Scholar] [CrossRef]
- Li, M.; Li, H.; Li, C.; Wang, S.; Jiang, W.; Liu, Z.; Zhou, S.; Liu, X.; McNutt, M.A.; Li, G. Alpha-fetoprotein: A new member of intracellular signal molecules in regulation of the PI3K/AKT signaling in human hepatoma cell lines. Int. J. Cancer 2011, 128, 524–532. [Google Scholar] [CrossRef]
- Jiang, L.; Huang, Y.; Yang, R.; Jiang, X.; Yan, H. Hsa_circ_0009910 Regulates Cisplatin Sensitivity of Ovarian Cancer Cells by Targeting miR-455-5p/PAX2. Pak. J. Zool. 2023, 55, 1285. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, L.; Hou, J.; Meng, J.; Lin, K.; Wu, X.; Chen, X. Three novel circRNAs upregulated in tissue and plasma from hepatocellular carcinoma patients and their regulatory network. Cancer Cell Int. 2021, 21, 72. [Google Scholar] [CrossRef]
- Wang, D.; Ming, X.; Xu, J.; Xiao, Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol. Oncol. 2021, 39, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Lyu, L.H.; Zhang, C.Y.; Yang, W.J.; Jin, A.L.; Zhu, J.; Wang, H.; Liu, T.; Wang, B.L.; Cheng, J.W.; Yang, X.R.; et al. Hsa_circ_0003945 promotes progression of hepatocellular carcinoma by mediating miR-34c-5p/LGR4/beta-catenin axis activity. J. Cell Mol. Med. 2022, 26, 2218–2229. [Google Scholar] [CrossRef]
- Li, H.W.; Liu, J. Circ_0009910 promotes proliferation and metastasis of hepatocellular carcinoma cells through miR-335-5p/ROCK1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1725–1735. [Google Scholar]
- Hao, Q.; Han, Y.; Xia, W.; Wang, Q.; Qian, H. Systematic Review and Meta-Analysis of the Utility of Circular RNAs as Biomarkers of Hepatocellular Carcinoma. Can. J. Gastroenterol. Hepatol. 2019, 2019, 1684039. [Google Scholar] [CrossRef]
- Zhang, C.; Ding, R.; Sun, Y.; Huo, S.T.; He, A.; Wen, C.; Chen, H.; Du, W.W.; Lai, W.; Wang, H. Circular RNA in tumor metastasis. Mol. Ther. Nucleic Acids 2021, 23, 1243–1257. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Cui, Q.; Hu, P.; Hu, S.; Qian, Y. Circular RNA hsa_circ_0006091 as a novel biomarker for hepatocellular carcinoma. Bioengineered 2022, 13, 1988–2003. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, F.; Li, P. Circular RNAs: Characteristics, Function and Clinical Significance in Hepatocellular Carcinoma. Cancers 2018, 10, 258. [Google Scholar] [CrossRef]
- Terrinoni, A.; Calabrese, C.; Basso, D.; Aita, A.; Caporali, S.; Plebani, M.; Bernardini, S. The circulating miRNAs as diagnostic and prognostic markers. Clin. Chem. Lab. Med. CCLM 2019, 57, 932–953. [Google Scholar] [CrossRef]
- Turshudzhyan, A.; Wu, G.Y. Persistently Rising Alpha-fetoprotein in the Diagnosis of Hepatocellular Carcinoma: A Review. J. Clin. Transl. Hepatol. 2022, 10, 159–163. [Google Scholar] [CrossRef]
- Gopal, P.; Yopp, A.C.; Waljee, A.K.; Chiang, J.; Nehra, M.; Kandunoori, P.; Singal, A.G. Factors that affect accuracy of alpha-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2014, 12, 870–877. [Google Scholar] [CrossRef] [PubMed]
- Parikh, N.D.; Mehta, A.S.; Singal, A.G.; Block, T.; Marrero, J.A.; Lok, A.S. Biomarkers for the Early Detection of Hepatocellular Carcinoma. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Jiang, Z.; Li, T.; Hu, Y.; Guo, J. Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 2018, 7, 3101–3109. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, F.; Ma, C.; Cheng, Q. Involvement of microRNAs and their potential diagnostic, therapeutic, and prognostic role in hepatocellular carcinoma. J. Clin. Lab. Anal. 2022, 36, e24673. [Google Scholar] [CrossRef]
- Meng, H.; Niu, R.; Huang, C.; Li, J. Circular RNA as a novel biomarker and therapeutic target for HCC. Cells 2022, 11, 1948. [Google Scholar] [CrossRef]
- Bai, D.S.; Zhang, C.; Chen, P.; Jin, S.J.; Jiang, G.Q. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci. Rep. 2017, 7, 12870. [Google Scholar] [CrossRef]
- Liang, W.-C.; Wong, C.-W.; Liang, P.-P.; Shi, M.; Cao, Y.; Rao, S.-T.; Tsui, S.K.-W.; Waye, M.M.-Y.; Zhang, Q.; Fu, W.-M.; et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019, 20, 84. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Sun, X.; Huang, Q. Circ_0000105 promotes liver cancer by regulating miR-498/PIK3R1. J. Gene Med. 2020, 22, e3256. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.-Y.; Chen, L.; Zhang, P.; Zang, H.; Zhu, B.; Shao, W.-B. Circ_0091579 promotes proliferative ability and metastasis of liver cancer cells by regulating microRNA-490-3p. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 10264–10273. [Google Scholar] [PubMed]
- Babaei, Z.; Keyvanloo Shahrestanaki, M.; Aghaei, M. MiR-1236: Key controller of tumor development and progression: Focus on the biological functions and molecular mechanisms. Pathol. Res. Pract. 2023, 248, 154671. [Google Scholar] [CrossRef]
- Bian, T.; Jiang, D.; Liu, J.; Yuan, X.; Feng, J.; Li, Q.; Zhang, Q.; Li, X.; Liu, Y.; Zhang, J. miR-1236-3p suppresses the migration and invasion by targeting KLF8 in lung adenocarcinoma A549 cells. Biochem. Biophys. Res. Commun. 2017, 492, 461–467. [Google Scholar] [CrossRef]
- Li, C.; Ge, Q.; Liu, J.; Zhang, Q.; Wang, C.; Cui, K.; Chen, Z. Effects of miR-1236-3p and miR-370-5p on activation of p21 in various tumors and its inhibition on the growth of lung cancer cells. Tumor Biol. 2017, 39, 1010428317710824. [Google Scholar] [CrossRef]
- Li, Q.-h.; Liu, Y.; Chen, S.; Zong, Z.-h.; Du, Y.-p.; Sheng, X.-j.; Zhao, Y. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed. Pharmacother. 2019, 114, 108832. [Google Scholar] [CrossRef]
- Duan, X.; Liu, D.; Wang, Y.; Chen, Z. Circular RNA hsa_circ_0074362 Promotes Glioma Cell Proliferation, Migration, and Invasion by Attenuating the Inhibition of miR-1236-3p on HOXB7 Expression. DNA Cell Biol. 2018, 37, 917–924. [Google Scholar] [CrossRef]
- Elimam, H.; Mageed, S.S.A.; Hatawsh, A.; Moussa, R.; Radwan, A.F.; Elfar, N.; Alhamshry, N.A.A.; Abd-Elmawla, M.A.; Mohammed, O.A.; Zaki, M.B.; et al. Unraveling the influence of LncRNA in gastric cancer pathogenesis: A comprehensive review focus on signaling pathways interplay. Med. Oncol. 2024, 41, 218. [Google Scholar] [CrossRef]
- Zhou, H.; Huang, X.; Yang, X.; Jiang, F.; Shao, F.; Shi, W.; Huang, K.; Pan, J.; Zhang, Y.; Chen, J.; et al. CircRAPGEF5 promotes the proliferation and metastasis of lung adenocarcinoma through the miR-1236-3p/ZEB1 axis and serves as a potential biomarker. Int. J. Biol. Sci. 2022, 18, 2116. [Google Scholar] [CrossRef]
- Kang, H. Sample size determination and power analysis using the G*Power software. J. Educ. Eval Heal. Prof. 2021, 18, 1–12. [Google Scholar] [CrossRef]
- Moro, R. The alpha-fetoprotein receptor (RECAF): Characterization and potential uses for cancer diagnosis and therapy. Alpha-, 2016. [Google Scholar]
- Pan, Y.; Chen, H.; Yu, J. Biomarkers in hepatocellular carcinoma: Current status and future perspectives. Biomedicines 2020, 8, 576. [Google Scholar] [CrossRef] [PubMed]
- Ishaq, Y.; Ikram, A.; Alzahrani, B.; Khurshid, S. The Role of miRNAs, circRNAs and Their Interactions in Development and Progression of Hepatocellular Carcinoma: An Insilico Approach. Genes 2022, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Radwan, A.F.; Shaker, O.G.; El-Boghdady, N.A.; Senousy, M.A. Association of MALAT1 and PVT1 Variants, Expression Profiles and Target miRNA-101 and miRNA-186 with Colorectal Cancer: Correlation with Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2021, 22, 6147. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2014, 43, D146–D152. [Google Scholar] [CrossRef]
- Wang, X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics 2016, 32, 1316–1322. [Google Scholar] [CrossRef]
- Dudekula, D.B.; Panda, A.C.; Grammatikakis, I.; De, S.; Abdelmohsen, K.; Gorospe, M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016, 13, 34–42. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih I-h Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Chou, C.-H.; Shrestha, S.; Yang, C.-D.; Chang, N.-W.; Lin, Y.-L.; Liao, K.-W.; Huang, W.-C.; Sun, T.-H.; Tu, S.-J.; Lee, W.-H.; et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, J.; Huang, S.; Li, F.; Huang, J.; Li, X.; Ling, Q.; Ye, W.; Wang, Y.; Yu, W.; et al. Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 2021, 19, 28. [Google Scholar] [CrossRef]
- Shang, X.; Li, G.; Liu, H.; Li, T.; Liu, J.; Zhao, Q.; Wang, C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine 2016, 95, e3811. [Google Scholar] [CrossRef] [PubMed]
- Sperveslage, J.; Hoffmeister, M.; Henopp, T.; Klöppel, G.; Sipos, B. Establishment of robust controls for the normalization of miRNA expression in neuroendocrine tumors of the ileum and pancreas. Endocrine 2014, 46, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, O.M.; Abdelazim, S.A.; Darwish, H.A.; Shaker, O.G.; Senousy, M.A. Association of LncRNA-PAX8-AS1 and LAIR-2 polymorphisms along with their expression with clinical and subclinical hypothyroidism. Sci. Rep. 2023, 13, 6. [Google Scholar] [CrossRef]
- Senousy, M.A.; Shaker, O.G.; Ayeldeen, G.; Radwan, A.F. Association of lncRNA MEG3 rs941576 polymorphism, expression profile, and its related targets with the risk of obesity-related colorectal cancer: Potential clinical insights. Sci. Rep. 2024, 14, 10271. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shaker, O.G.; El-Maraghy, S.A.; Senousy, M.A. Serum interferon-related microRNAs as biomarkers to predict the response to interferon therapy in chronic hepatitis C genotype 4. PLoS ONE 2015, 10, e0120794. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Taha, H.; Elfar, N.; Haffez, H.; Hassan, Z.A. Raptinal silver nanoparticles: New therapeutic advances in hepatocellular carcinoma mouse model. Naunyn Schmiedebergs Arch. Pharmacol. 2021, 394, 279–289. [Google Scholar] [CrossRef]
- Llovet, J.M.; Bru, C.; Bruix, J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin. Liver Dis. 1999, 19, 329–338. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Pugh, R.N.; Murray-Lyon, I.M.; Dawson, J.L.; Pietroni, M.C.; Williams, R. Transection of the oesophagus for bleeding oesophageal varices. Br. J. Surg. 1973, 60, 646–649. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Ohtsuki, T.; Obata, H.; Tomimatsu, M.; Okazaki, N.; Hasegawa, H.; Nakajima, Y.; Ohnishi, K. Natural history of hepatocellular carcinoma and prognosis in relation to treatment. Study of 850 patients. Cancer 1985, 56, 918–928. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Höner zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]





| Healthy Controls (n = 50) | HCC (n = 100) | p-Value | |||
|---|---|---|---|---|---|
| Age in years Age range | 55.4 ± 0.75 (45–65) | 59.2 ± 0.8 (34–89) | 0.22 | ||
| Sex, n (%) | Male | 27 (54%) | 58 (58%) | 0.64 | |
| Female | 23 (46%) | 42 (42%) | |||
| ALT (IU/L) | 16.6 ± 0.8 | 62.3 ± 5.8 | <0.001 | ||
| AST (IU/L) | 18 ± 0.6 | 102.2 ± 12.1 | <0.001 | ||
| AFP (ng/mL) | 3.7 ± 0.2 | 139.2 ± 20 | <0.001 | ||
| Albumin (gm/dL) | 4.6 ± 0.12 | 2.6 ± 0.06 | <0.001 | ||
| Bilirubin (mg/dL) | 0.5 ± 0.03 | 6.8 ± 1.4 | <0.001 | ||
| PT (seconds) | 12.1 ± 0.1 | 18.2 ± 0.91 | <0.001 | ||
| INR | 0.9 ± 0.01 | 1.5 ± 0.07 | <0.001 | ||
| HCV Ab, n (%) | Positive | - | 79, (79%) | ||
| Negative | 21, (21%) | ||||
| Tumor anatomical site, n (%) | |||||
| Left lobe (LT) | - | 44, (44%) | - | ||
| Right lobe (RL) | - | 35, (35%) | - | ||
| LT&RL | - | 21, (21%) | - | ||
| Child staging, n (%) | |||||
| A | - | 19, (19%) | - | ||
| B | - | 27, (27%) | - | ||
| C | - | 54, (54%) | - | ||
| TNM staging system,n (%) | |||||
| Stage I, II (Early) | - | 53, (53%) | - | ||
| Stage III, IV (Late) | - | 47, (47%) | - | ||
| BCLC staging system, n (%) | |||||
| A | - | 28, (28%) | - | ||
| B | - | 21, (21%) | - | ||
| C | - | 34, (34%) | - | ||
| D | - | 17, (17%) | - | ||
| Okuda staging,n (%) | |||||
| 1 | - | 18, (18%) | - | ||
| 2 | - | 34, (34%) | - | ||
| 3 | - | 48, (48%) | - | ||
| Number of lesion, n (%) | |||||
| Single | - | 32, (32%) | - | ||
| Multiple | - | 68, (68%) | - | ||
| Lymph node metastasis,n (%) | |||||
| Present | - | 42, (42%) | - | ||
| Absent | - | 58, (58%) | - | ||
| Distant metastasis, n (%) | |||||
| Present | - | 25, (25%) | - | ||
| Absent | - | 75, (75%) | - | ||
| Cirrhosis, n (%) | |||||
| 0 | - | 16, (16%) | - | ||
| 1 | - | 39, (39%) | - | ||
| 2 | - | 28, (28%) | - | ||
| 3 | - | 17, (17%) | - | ||
| Tumor size,n (%) | |||||
| <3 | - | 40, (40%) | |||
| ≥3 | - | 60, (60%) | |||
| HCC (n = 100) | Control (n = 50) | p-Value | |
|---|---|---|---|
| hsa_circ_0009910 | 35.6 (16.7–54.2) | 0.57 (0.18–1.2 | <0.001 |
| hsa_circ_0027478 | 3.8 (1.9–10.5) | 0.66 (0.1–1.3) | <0.001 |
| hsa_miR_1236-3p | 0.13 (0.05–0.3) | 0.8 (0.5–1.2) | <0.001 |
| Parameter | Coefficient | SE | p-Value | OR | OR (95% CI) |
|---|---|---|---|---|---|
| Univariate | |||||
| Hsa_circ_0009910 | 1.185 | 0.3 | 0.0001 | 3.27 | 1.804–5.936 |
| Hsa_circ_0027478 | 1.3530 | 0.262 | <0.0001 | 3.86 | 2.311–6.475 |
| miRNA-1236 | −6.4 | 1.05 | <0.0001 | 0.001 | 0.0002–0.012 |
| AFP | 0.0992 | 0.02 | <0.0001 | 1.10 | 1.052–1.158 |
| Multivariate | |||||
| Hsa_circ_0009910 | 1.072 | 0.66 | 0.109 | 2.93 | 0.786–10.86 |
| Hsa_circ_0027478 | 0.928 | 1.04 | 0.374 | 2.52 | 0.325–19.65 |
| miRNA-1236 | −14.682 | 11.033 | 0.183 | 0.00 | 0.001–1035.8152 |
| AFP | 0.0703 | 0.203 | 0.729 | 1.07 | 0.719–1.59 |
| Constant | −1.08 | ||||
| circ_0009910 | circ_0027478 | miR-1236-3p | AFP | Albumin | Bilirubin | ALT | AST | Size | TNM | BCLC | Metastasis | Cirrhosis | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| circ_0009910 (fold change) | r | 1.000 | 0.325 | −0.165 | 0.455 | 0.076 | 0.010 | 0.112 | 0.164 | 0.608 | 0.728 | 0.507 | 0.415 | 0.616 |
| p | 0.001 | 0.101 | <0.001 | 0.450 | 0.925 | 0.276 | 0.103 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| circ_0027478 (fold change) | r | 0.325 | 1.000 | −0.252 | 0.341 | 0.119 | −0.105 | −0.020 | 0.065 | 0.363 | 0.412 | 0.338 | 0.342 | 0.322 |
| p | 0.001 | 0.011 | 0.001 | 0.237 | 0.289 | 0.843 | 0.518 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| miR-1236-3p (fold change) | r | −0.165 | −0.252 | 1.000 | −0.132 | −0.031 | 0.130 | 0.015 | 0.018 | −0.083 | −0.015 | 0.047 | −0.044 | −0.083 |
| p | 0.101 | 0.011 | 0.189 | 0.762 | 0.197 | 0.880 | 0.859 | 0.414 | 0.881 | 0.644 | 0.664 | 0.413 | ||
| AFP(ng/mL) | r | 0.455 | 0.341 | −0.132 | 1.000 | 0.190 | 0.152 | 0.060 | 0.283 | 0.366 | 0.494 | 0.544 | 0.306 | 581 |
| p | <0.001 | 0.001 | 0.189 | 0.058 | 0.131 | 0.553 | 0.004 | <0.001 | <0.001 | <0.001 | 0.002 | <0.001 | ||
| Albumin (g/dl) | r | 0.076 | 0.119 | −0.031 | 0.190 | 1.000 | −0.272 | −0.110 | −0.097 | 0.176 | 0.049 | 0.065 | −0.084 | −0.070 |
| p | 0.450 | 0.237 | 0.762 | 0.058 | 0.006 | 0.275 | 0.339 | 0.081 | 0.629 | 0.523 | 0.408 | 0.491 | ||
| Bilirubin (mg/dl) | r | 0.010 | −0.105 | 0.130 | 0.152 | −0.272 | 1.000 | 0.408 | 0.572 | −0.022 | 0.013 | 0.067 | −0.037 | 0.024 |
| p | 0.925 | 0.298 | 0.197 | 0.131 | 0.006 | <0.001 | <0.001 | 0.831 | 0.899 | 0.511 | 0.298 | 0.814 | ||
| ALT(U/L) | r | 0.112 | −0.020 | 0.015 | 0.060 | −0.110 | 0.408 | 1.000 | 0.645 | 0.132 | 0.122 | 0.124 | −0.058 | 0.146 |
| p | 0.267 | 0.843 | 0.880 | 0.553 | 0.275 | <0.001 | <0.001 | 0.189 | 0.228 | 0.220 | 0.566 | 0.148 | ||
| AST(U/L) | r | 0.164 | 0.065 | 0.018 | 0.283 | −0.097 | 0.572 | 0.645 | 1.000 | 0.233 | 0.205 | 0.012 | 0.092 | 0.103 |
| p | 0.103 | 0.518 | 0.859 | 0.004 | 0.339 | <0.001 | <0.001 | 0.020 | 0.041 | 0.909 | 0.360 | 0.308 | ||
| Size(cm) | r | 0.608 | 0.363 | −0.083 | 0.366 | 0.176 | −0.022 | 0.132 | 0.233 | 1.000 | 0.765 | 0.482 | 0.484 | 0.532 |
| p | <0.001 | <0.001 | 0.414 | <0.001 | 0.081 | 0.831 | 0.189 | 0.020 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| TNM | r | 0.728 | 0.412 | −0.015 | 0.494 | 0.049 | 0.013 | 0.122 | 0.205 | 0.765 | 1.000 | 0.698 | 0.778 | 0.793 |
| p | <0.001 | <0.001 | 0.881 | <0.001 | 0.629 | 0.899 | 0.228 | 0.041 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| BCLC | r | 0.507 | 0.338 | 0.047 | 0.554 | 0.065 | 0.067 | 0.124 | 0.012 | 0.482 | 0.698 | 1.000 | 0.496 | 0.833 |
| p | <0.001 | 0.001 | 0.644 | <0.001 | 0.523 | 0.511 | 0.220 | 0.909 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| Metastasis | r | 0.415 | 0.342 | −0.044 | 0.306 | −0.084 | −0.037 | −0.058 | 0.092 | 0.484 | 0.778 | 0.496 | 1.000 | 0.606 |
| p | <0.001 | 0.001 | 0.664 | 0.002 | 0.408 | 0.716 | 0.566 | 0.360 | <0.001 | <0.001 | <0.001 | 0.001 | <0.001 | |
| Cirrhosis | r | 0.616 | 0.322 | −0.083 | 0.581 | −0.070 | 0.024 | 0.146 | 0.103 | 0.532 | 0.793 | 0.833 | 0.606 | 1.000 |
| p | <0.001 | 0.001 | 0.413 | <0.001 | 0.491 | 0.814 | 0.148 | 0.308 | <0.001 | <0.001 | <0.001 | <0.001 | ||
| Gene | Primer Sequence | |
|---|---|---|
| has_circ_0009910 | F. | 5′-GCAGAACTGGACCCCGTTACC-3′ |
| R. | 5′-CAGGGACATTGCGCGGCCAA-3′ | |
| Hsa_circ_0027478 | F. | 5′-CCATTGCCTGGAGTTGGCT-3′ |
| R. | 5′-CCACAGCGTTTACAGAGTCG-3′ | |
| GAPDH | F. | 5′-CCCTTCATTGACCTCAACTA-3′ |
| R. | 5′-TGGAAGATGGTGATGGGATT-3′ | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Awed, M.S.; Ibrahim, A.; Ezzat, O.; Fawzy, A.; Sabir, D.K.; Radwan, A.F. Preliminary Evaluation of Plasma circ_0009910, circ_0027478, and miR-1236-3p as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2025, 26, 4842. https://doi.org/10.3390/ijms26104842
Awed MS, Ibrahim A, Ezzat O, Fawzy A, Sabir DK, Radwan AF. Preliminary Evaluation of Plasma circ_0009910, circ_0027478, and miR-1236-3p as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2025; 26(10):4842. https://doi.org/10.3390/ijms26104842
Chicago/Turabian StyleAwed, Mona Samy, Abeer Ibrahim, Omnia Ezzat, Amal Fawzy, Deema Kamal Sabir, and Abdullah F. Radwan. 2025. "Preliminary Evaluation of Plasma circ_0009910, circ_0027478, and miR-1236-3p as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma" International Journal of Molecular Sciences 26, no. 10: 4842. https://doi.org/10.3390/ijms26104842
APA StyleAwed, M. S., Ibrahim, A., Ezzat, O., Fawzy, A., Sabir, D. K., & Radwan, A. F. (2025). Preliminary Evaluation of Plasma circ_0009910, circ_0027478, and miR-1236-3p as Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. International Journal of Molecular Sciences, 26(10), 4842. https://doi.org/10.3390/ijms26104842







