The Alpha/Beta-Hydrolase Fold Superfamily in Brassica napus: Expression Profiles and Functional Implications of Clade-3 BnABH Proteins in Response to Abiotic Stress
Abstract
:1. Introduction
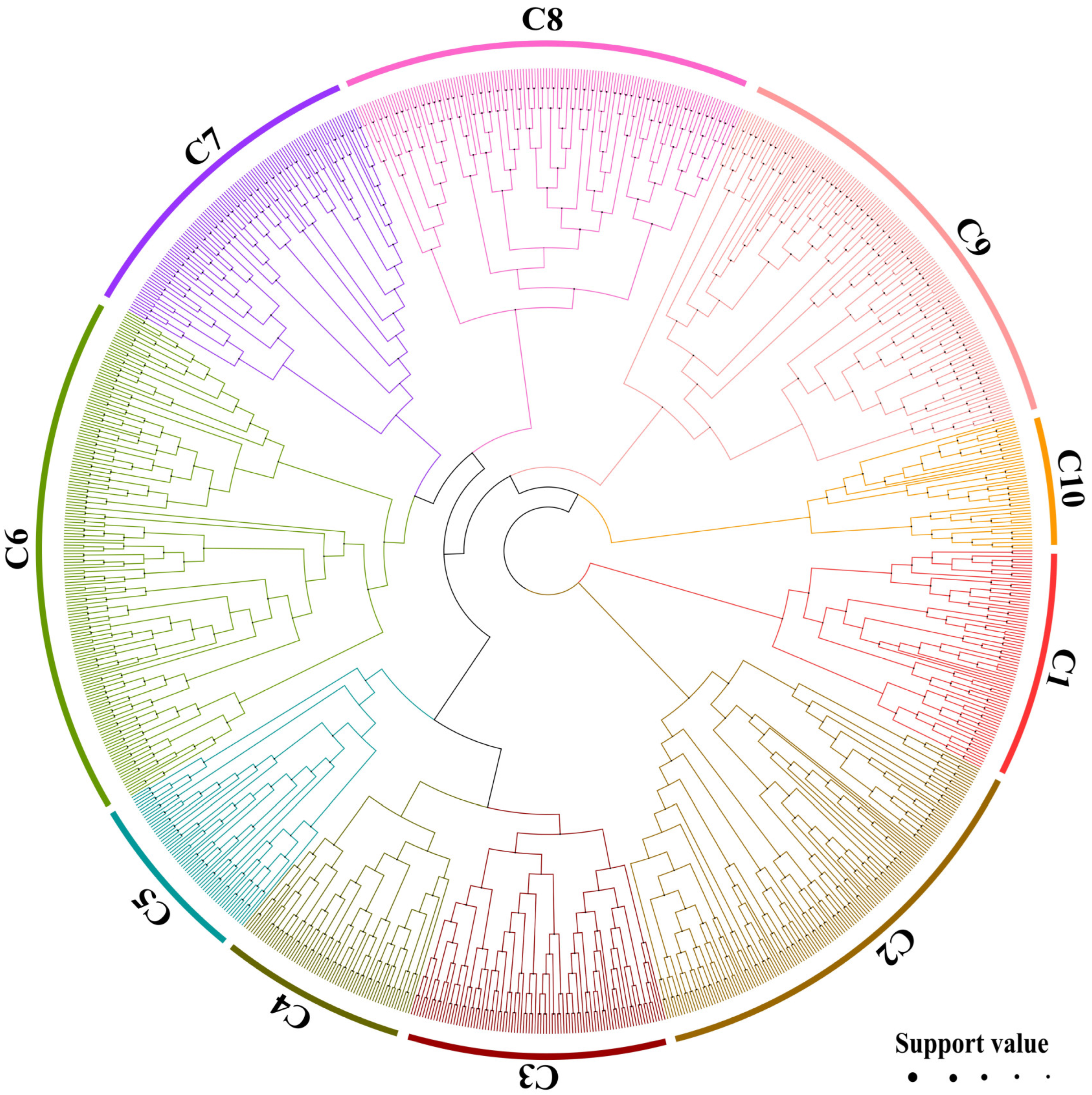
2. Results
2.1. Genome-Wide Identification and Characterization of BnABH Genes
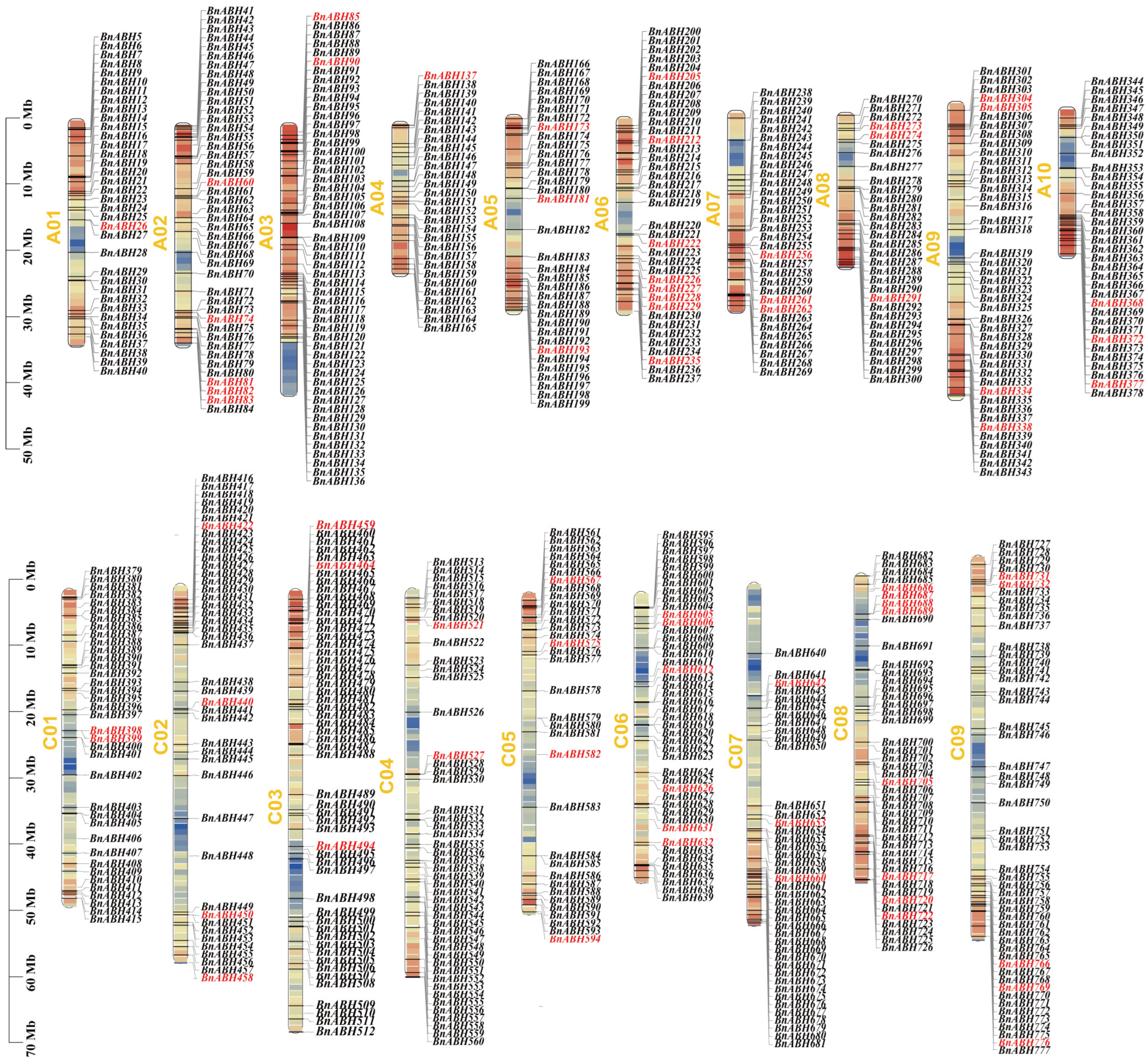
2.2. Chromosome Distribution of BnABH Genes
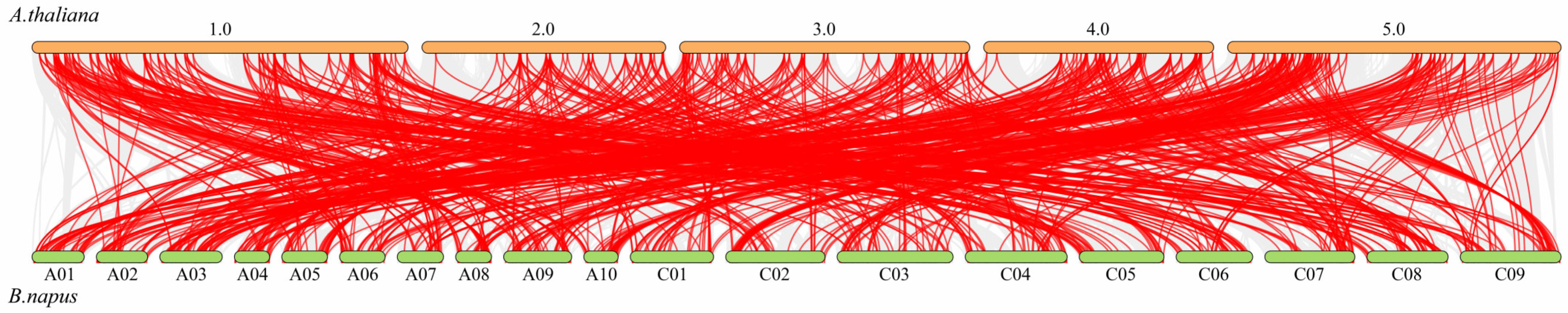
2.3. Gene Duplication and Collinearity Survey of the BnABH Genes
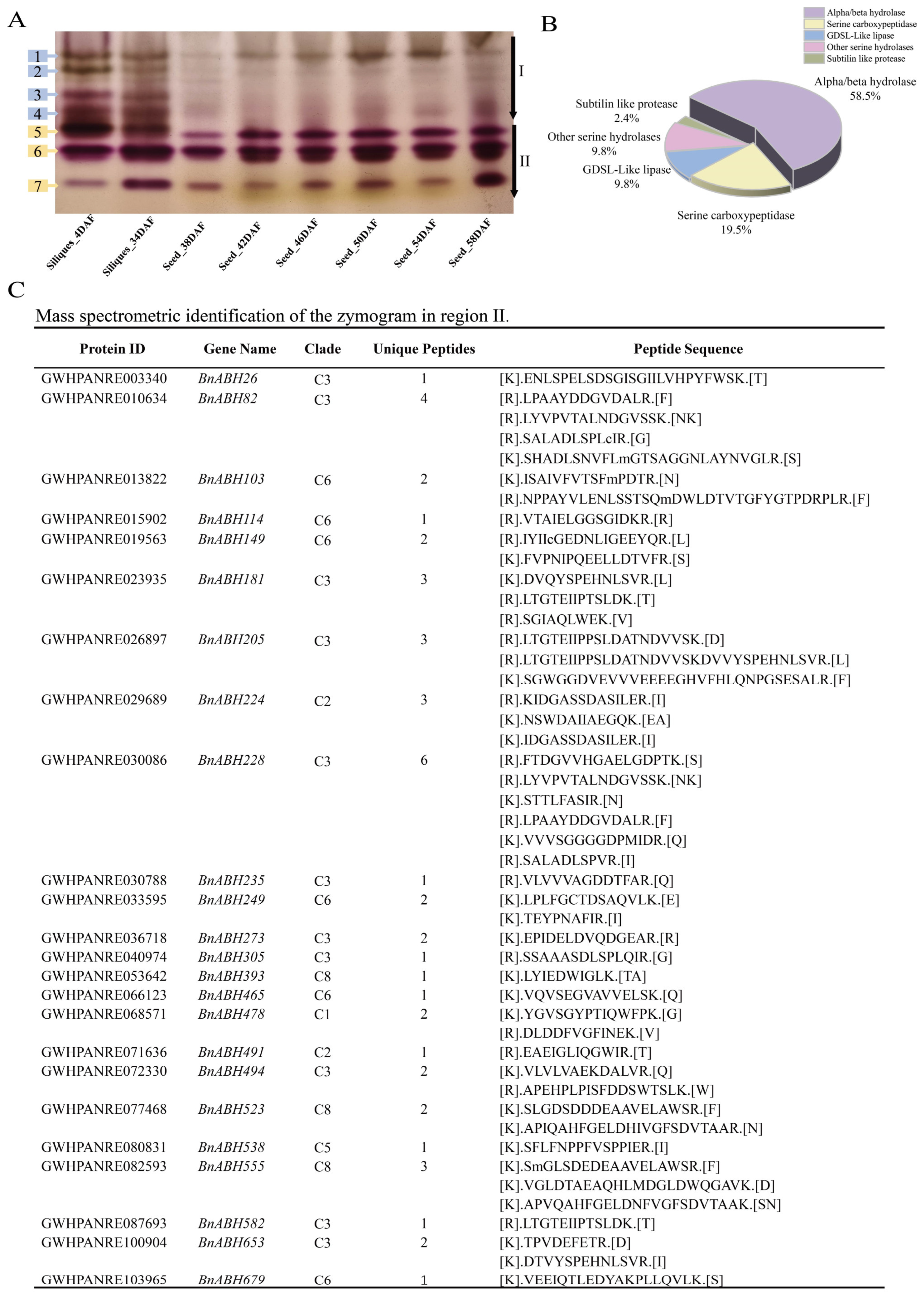
2.4. Eleven Members of the Clade-C3 BnABH Protein Group Are Mainly Related to Esterase Isozyme Activity During Seed Maturation
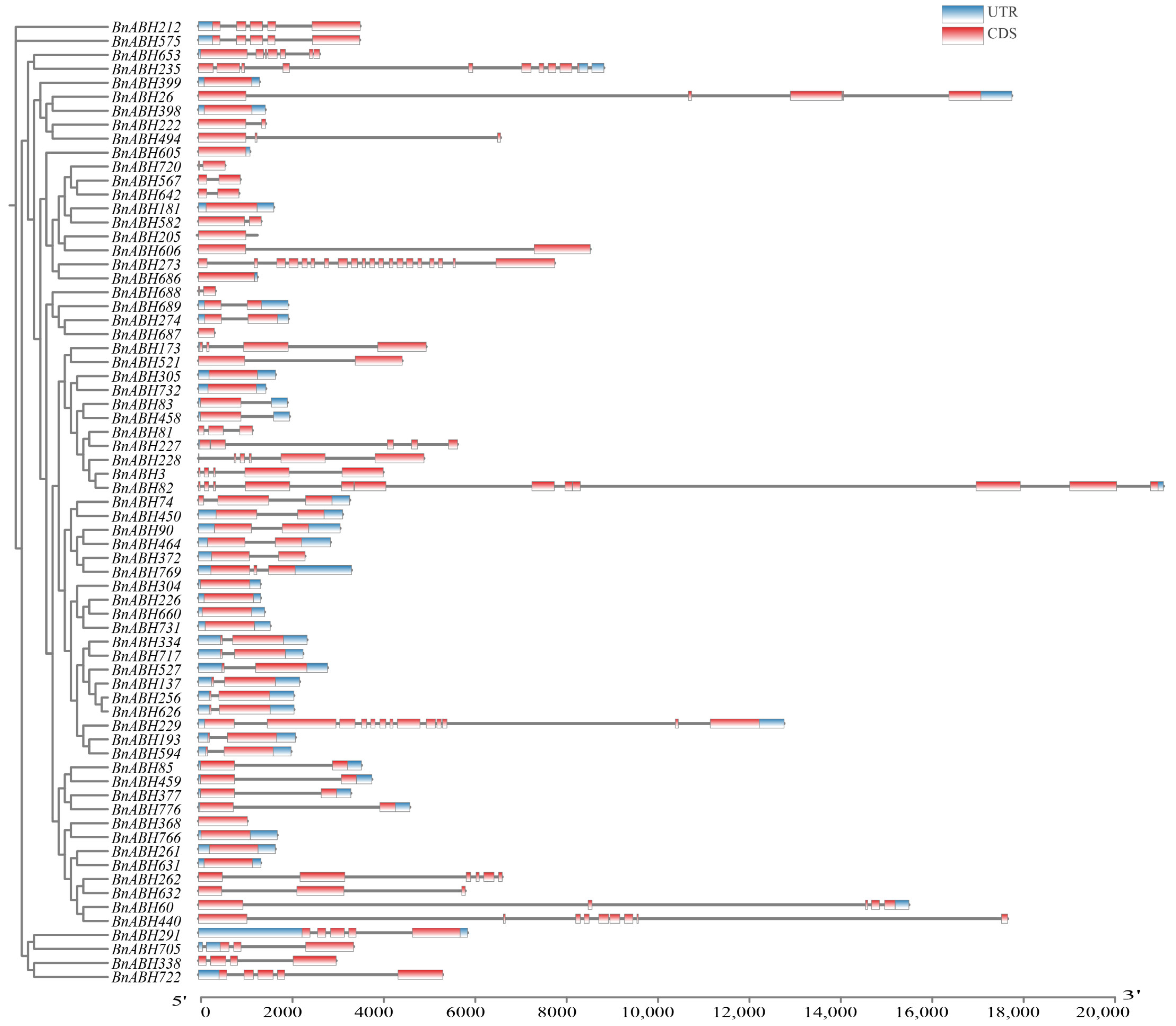
2.5. Gene Structure and Conserved Motif Analysis of BnABHs in Clade C3
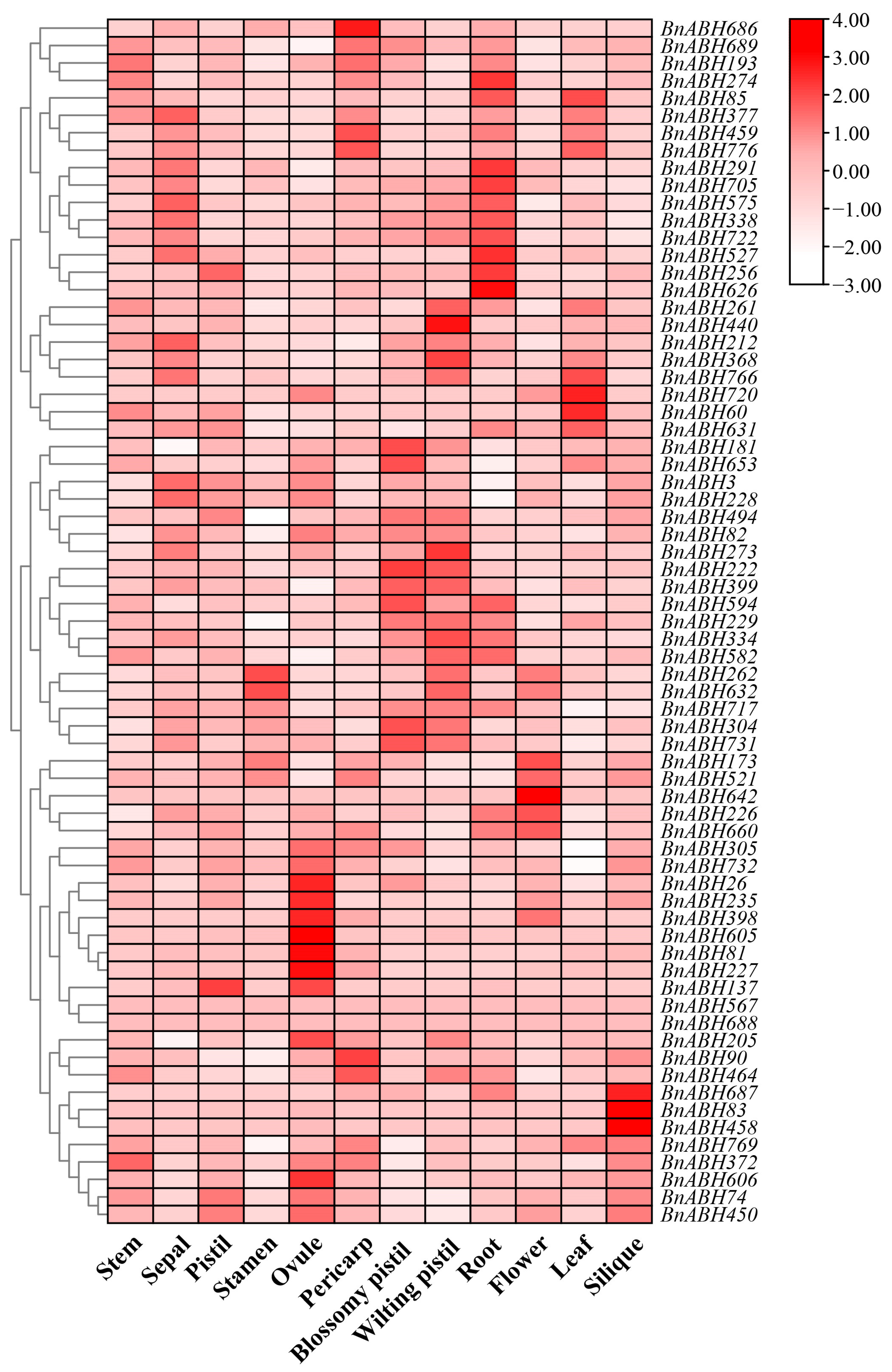
2.6. Expression Patterns of BnABH Genes in Clade C3
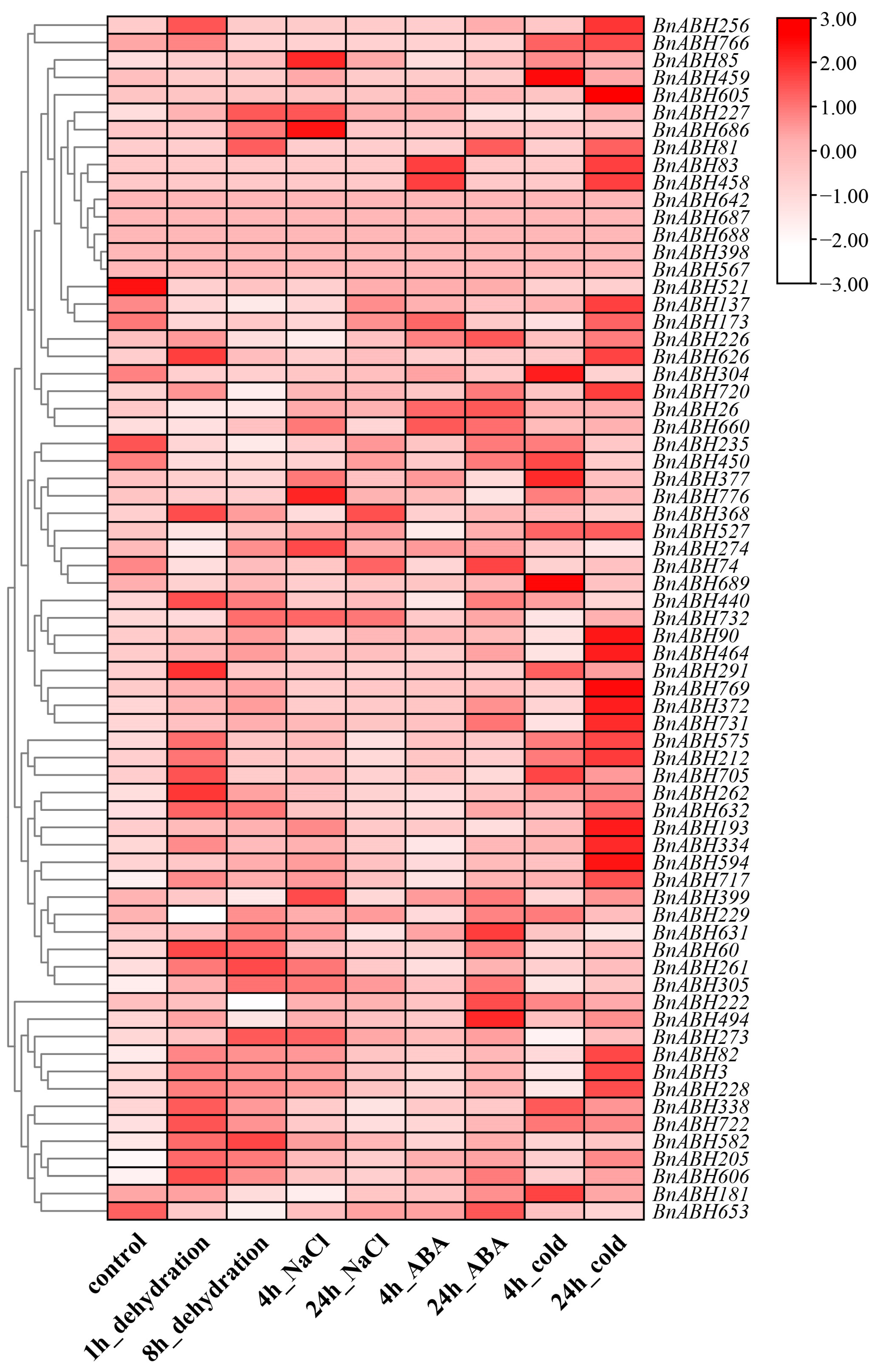
2.7. Expression Profiles of the Clade-C3 BnABH Genes in Response to Abiotic Stresses
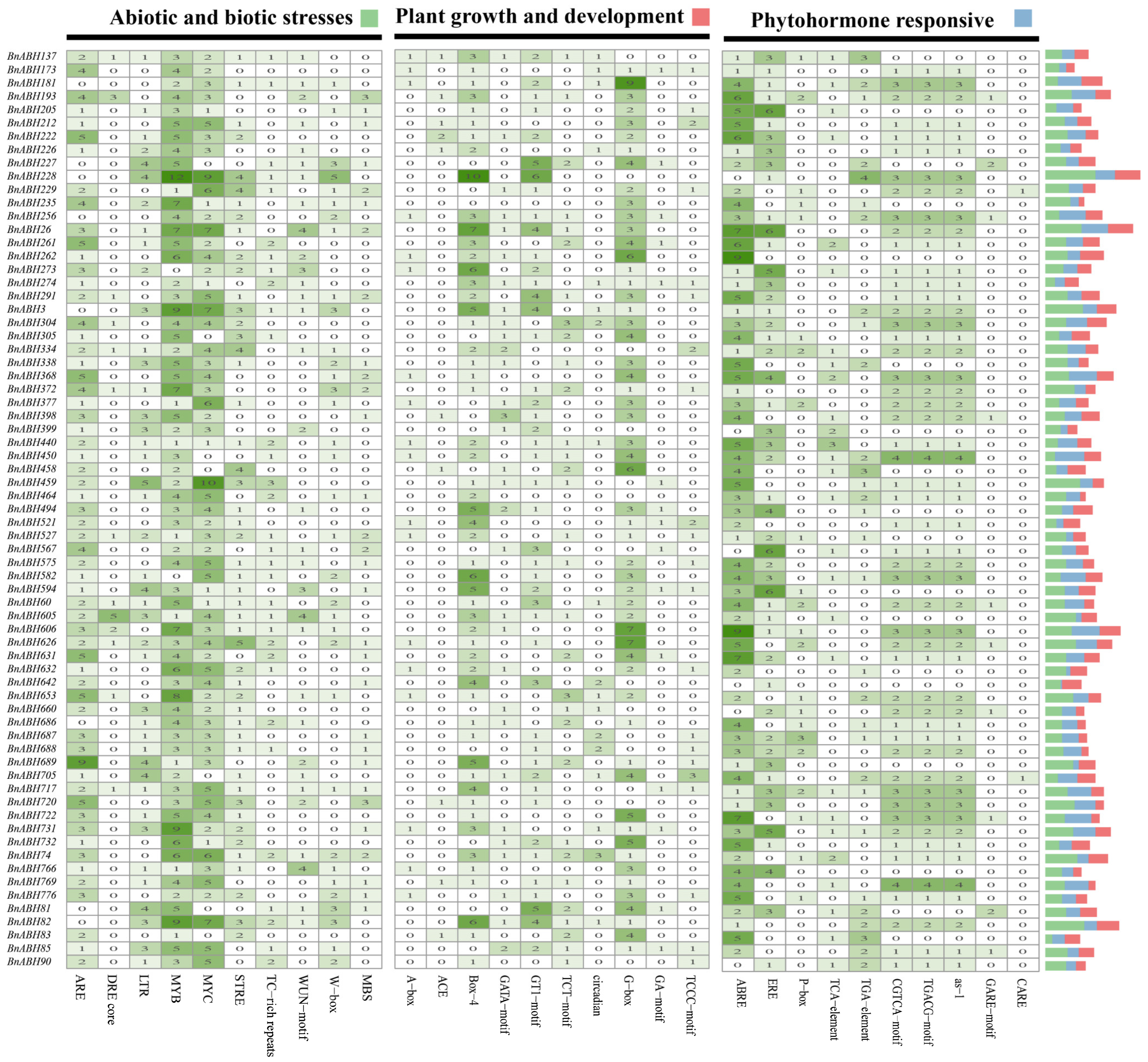
2.8. Analysis of Cis-Elements in Promoters of the BnABH Genes in Clade C3
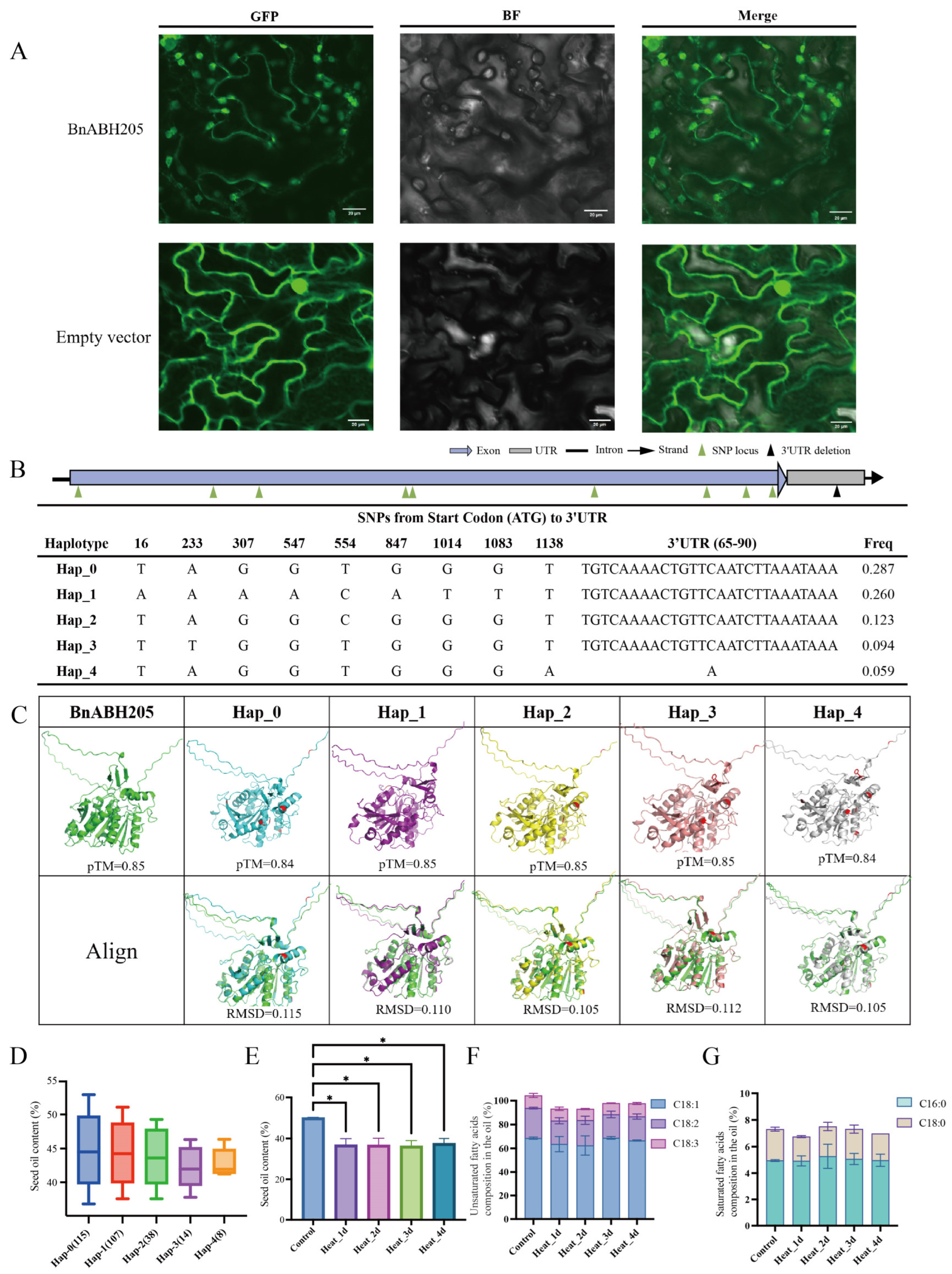
2.9. Subcellular Localization of BnABH205
2.10. Five Haplotypes of BnABH205 Exhibit Correlation with SOC
3. Discussion
4. Materials and Methods
4.1. Identification of the ABH Gene Family in Brassica napus
4.2. Analysis of Predicted ABH Proteins
4.3. Genome-Scale Synteny Analysis of ABH Genes
4.4. Chromosome Localization and Gene Structure Analyses
4.5. Expression Profiles of BnABH Genes Determined by Transcriptome Data
4.6. Identification of Cis-Elements in the Promoters of the Clade-C3 BnABH Genes
4.7. Plant Growth, Esterase Isozyme Electrophoresis, and Mass Spectrometry Analysis
4.8. Three-Dimensional Structure Simulation and Subcellular Localization Analysis
4.9. Analysis of Variations in BnABH205 and Assessment of Seed Oil Content
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, Z.; Zhang, C.; Tang, F.; Yang, B.; Zhang, L.; Liu, J.; Huo, Q.; Wang, S.; Li, S.; Wei, L.; et al. Identification of candidate genes controlling oil content by combination of genome-wide association and transcriptome analysis in the oilseed crop Brassica napus. Biotechnol. Biofuels 2019, 12, 216. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Q.; Pak, H.; Yan, T.; Chen, M.; Chen, X.; Wu, D.; Jiang, L. Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus. BMC Plant Biol. 2021, 21, 6. [Google Scholar] [CrossRef]
- Li, L.; Tian, Z.; Chen, J.; Tan, Z.; Zhang, Y.; Zhao, H.; Wu, X.; Yao, X.; Wen, W.; Chen, W.; et al. Characterization of novel loci controlling seed oil content in Brassica napus by marker metabolite-based multi-omics analysis. Genome Biol. 2023, 24, 141. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, M.; Chao, H.; Yin, Y.; Xia, Y.; Cheng, X.; Chen, K.; Yan, S.; Wang, X.; Xiong, Y.; et al. A rare dominant allele DYSOC1 determines seed coat color and improves seed oil content in Brassica napus. Sci. Adv. 2025, 11, eads7620. [Google Scholar] [CrossRef]
- Xiao, Z.; Zhang, C.; Qu, C.; Wei, L.; Zhang, L.; Yang, B.; Lu, K.; Li, J. Identification of candidate genes regulating seed oil content by QTL mapping and transcriptome sequencing in Brassica napus. Front. Plant Sci. 2022, 13, 1067121. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Xing, L.; Xu, J.; Jiang, T.; Tang, X.; Wang, Y.; Huang, S.; Hao, W.; Zhou, X.; Zhang, Y.; et al. Genome-wide exploration of the GDSL-type esterase/lipase gene family in rapeseed reveals several BnGELP proteins active during early seedling development. Front. Plant Sci. 2023, 14, 1139972. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ali, U.; Cao, Z.; Zeng, C.; Xiao, M.; Wei, F.; Yao, X.; Guo, L.; Lu, S. Fatty acid exporter 1 enhances seed oil content in Brassica napus. Mol. Breed. 2022, 42, 75. [Google Scholar] [CrossRef]
- Dolui, A.K.; Vijayaraj, P. Functional omics identifies serine hydrolases that mobilize storage lipids during rice seed germination. Plant Physiol. 2020, 184, 693–708. [Google Scholar] [CrossRef]
- Lai, C.P.; Huang, L.M.; Chen, L.O.; Chan, M.T.; Shaw, J.F. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 2017, 95, 181–197. [Google Scholar] [CrossRef]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef]
- Su, H.G.; Zhang, X.H.; Wang, T.T.; Wei, W.L.; Wang, Y.X.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; Xu, Z.S.; et al. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in Soybean. Front. Plant Sci. 2020, 11, 726. [Google Scholar] [CrossRef] [PubMed]
- Chatonnet, A.; Perochon, M.; Velluet, E.; Marchot, P. The ESTHER database on alpha/beta hydrolase fold proteins—An overview of recent developments. Chem. Biol. Interact. 2023, 383, 110671. [Google Scholar] [CrossRef]
- Chevalier, F.; Nieminen, K.; Sanchez-Ferrero, J.C.; Rodriguez, M.L.; Chagoyen, M.; Hardtke, C.S.; Cubas, P. Strigolactone promotes degradation of DWARF14, an alpha/beta hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 2014, 26, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zheng, Z.; La Clair, J.J.; Chory, J.; Noel, J.P. Smoke-derived karrikin perception by the alpha/beta-hydrolase KAI2 from Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 8284–8289. [Google Scholar] [CrossRef]
- Hamiaux, C.; Drummond, R.S.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef]
- Melville, K.T.; Kamran, M.; Yao, J.; Costa, M.; Holland, M.; Taylor, N.L.; Fritz, G.; Flematti, G.R.; Waters, M.T. Perception of butenolides by Bacillus subtilis via the alpha/beta hydrolase RsbQ. Curr. Biol. 2024, 34, 623–631.e626. [Google Scholar] [CrossRef]
- Waters, M.T.; Scaffidi, A.; Flematti, G.; Smith, S.M. Substrate-induced degradation of the alpha/beta-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Mol. Plant 2015, 8, 814–817. [Google Scholar] [CrossRef]
- Gawarecka, K.; Siwinska, J.; Poznanski, J.; Onysk, A.; Surowiecki, P.; Sztompka, K.; Surmacz, L.; Ahn, J.H.; Korte, A.; Swiezewska, E.; et al. Cis-prenyltransferase 3 and alpha/beta-hydrolase are new determinants of dolichol accumulation in Arabidopsis. Plant Cell Environ. 2022, 45, 479–495. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Guo, Y.; Austin, M.B.; Ramsey, J.; Fridman, E.; Pichersky, E.; Noel, J.P. Emergent decarboxylase activity and attenuation of alpha/beta-hydrolase activity during the evolution of methylketone biosynthesis in tomato. Plant Cell 2012, 24, 1596–1607. [Google Scholar] [CrossRef]
- Bui, S.; Gil-Guerrero, S.; van der Linden, P.; Carpentier, P.; Ceccarelli, M.; Jambrina, P.G.; Steiner, R.A. Evolutionary adaptation from hydrolytic to oxygenolytic catalysis at the alpha/beta-hydrolase fold. Chem. Sci. 2023, 14, 10547–10560. [Google Scholar] [CrossRef]
- Bzowka, M.; Mitusinska, K.; Raczynska, A.; Skalski, T.; Samol, A.; Bagrowska, W.; Magdziarz, T.; Gora, A. Evolution of tunnels in alpha/beta-hydrolase fold proteins-What can we learn from studying epoxide hydrolases? PLoS Comput. Biol. 2022, 18, e1010119. [Google Scholar] [CrossRef] [PubMed]
- Lenfant, N.; Hotelier, T.; Bourne, Y.; Marchot, P.; Chatonnet, A. Proteins with an alpha/beta hydrolase fold: Relationships between subfamilies in an ever-growing superfamily. Chem. Biol. Interact. 2013, 203, 266–268. [Google Scholar] [CrossRef]
- Liu, F.; Xia, Y.; Wu, L.; Fu, D.; Hayward, A.; Luo, J.; Yan, X.; Xiong, X.; Fu, P.; Wu, G.; et al. Enhanced seed oil content by overexpressing genes related to triacylglyceride synthesis. Gene 2015, 557, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Kourani, M.; Mohareb, F.; Rezwan, F.I.; Anastasiadi, M.; Hammond, J.P. Genetic and physiological responses to heat stress in Brassica napus. Front. Plant Sci. 2022, 13, 832147. [Google Scholar] [CrossRef] [PubMed]
- Brunel-Muguet, S.; D’Hooghe, P.; Bataille, M.P.; Larre, C.; Kim, T.H.; Trouverie, J.; Avice, J.C.; Etienne, P.; Durr, C. Heat stress during seed filling interferes with sulfur restriction on grain composition and seed germination in oilseed rape (Brassica napus L.). Front. Plant Sci. 2015, 6, 213. [Google Scholar] [CrossRef]
- Huang, R.; Liu, Z.; Xing, M.; Yang, Y.; Wu, X.; Liu, H.; Liang, W. Heat stress suppresses Brassica napus seed oil accumulation by inhibition of photosynthesis and BnWRI1 pathway. Plant Cell Physiol. 2019, 60, 1457–1470. [Google Scholar] [CrossRef]
- Magno Massuia de Almeida, L.; Coquemont-Guyot, M.; Elie, N.; Morvan-Bertrand, A.; Avice, J.C.; Mollier, A.; Brunel-Muguet, S. Repeated heat stress events during the reproductive phase impact the dynamic development of seeds in Brassica napus L. Plant Sci. Int. J. Exp. Plant Biol. 2023, 327, 111559. [Google Scholar] [CrossRef]
- Sun, F.; Fan, G.; Hu, Q.; Zhou, Y.; Guan, M.; Tong, C.; Li, J.; Du, D.; Qi, C.; Jiang, L.; et al. The high-quality genome of Brassica napus cultivar ’ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 2017, 92, 452–468. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Song, M.; Tang, X.; Huang, S.; Linhu, B.; Jin, P.; Guo, W.; Li, F.; Xing, L.; et al. Genome-wide identification of the cytochrome P450 superfamily genes and targeted editing of BnCYP704B1 confers male sterility in rapeseed. Plants 2023, 12, 365. [Google Scholar] [CrossRef]
- Song, M.; Linghu, B.; Huang, S.; Li, F.; An, R.; Xie, C.; Zhu, Y.; Hu, S.; Mu, J.; Zhang, Y. Genome-wide survey of leucine-rich repeat receptor-like protein kinase genes and CRISPR/Cas9-targeted mutagenesis BnBRI1 in Brassica napus. Front. Plant Sci. 2022, 13, 865132. [Google Scholar] [CrossRef]
- Han, X.; Wu, X.; Zhang, Y.; Tang, Q.; Zeng, L.; Liu, Y.; Xiang, Y.; Hou, K.; Fang, S.; Lei, W.; et al. Genetic and transcriptome analyses of the effect of genotype-by environment interactions on Brassica napus seed oil content. Plant Cell 2025, 37, koaf062. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zhao, H.; Lu, S.; Yu, L.; Zhang, G.; Zhang, Y.; Yang, Q.Y.; Zhou, Y.; Wang, X.; Ma, W.; et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol. Plant 2021, 14, 470–487. [Google Scholar] [CrossRef]
- Zhao, C.; Xie, M.; Liang, L.; Yang, L.; Han, H.; Qin, X.; Zhao, J.; Hou, Y.; Dai, W.; Du, C.; et al. Genome-wide association analysis combined with quantitative trait loci mapping and dynamic transcriptome unveil the genetic control of seed oil content in Brassica napus L. Front. Plant Sci. 2022, 13, 929197. [Google Scholar] [CrossRef] [PubMed]
- Karunarathna, N.L.; Wang, H.; Harloff, H.J.; Jiang, L.; Jung, C. Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 2020, 18, 2251–2266. [Google Scholar] [CrossRef]
- Chen, X.; Tong, C.; Zhang, X.; Song, A.; Hu, M.; Dong, W.; Chen, F.; Wang, Y.; Tu, J.; Liu, S.; et al. A high-quality Brassica napus genome reveals expansion of transposable elements, subgenome evolution and disease resistance. Plant Biotechnol. J. 2021, 19, 615–630. [Google Scholar] [CrossRef]
- Liu, D.; Yu, L.; Wei, L.; Yu, P.; Wang, J.; Zhao, H.; Zhang, Y.; Zhang, S.; Yang, Z.; Chen, G.; et al. BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol. J. 2021, 19, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Fan, C.; Li, J.; Cai, G.; Yang, Q.; Wu, J.; Yi, X.; Zhang, C.; Zhou, Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016, 129, 1203–1215. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0, an upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, H.; Zhang, Y.; Bai, Y.; Zhu, W.; Qin, Y.; Yuan, F.; Zhao, F.; Wang, M.; Hu, J.; et al. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 2015, 168, 659–676. [Google Scholar] [CrossRef]
- Bai, Y.; Zhu, W.; Hu, X.; Sun, C.; Li, Y.; Wang, D.; Wang, Q.; Pei, G.; Zhang, Y.; Guo, A.; et al. Genome-wide analysis of the bZIP gene family identifies two ABI5-like bZIP transcription factors, BrABI5a and BrABI5b, as positive modulators of ABA signalling in Chinese cabbage. PLoS ONE 2016, 11, e0158966. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Feng, L.; Li, P.; Yang, X.; Li, M.; Liu, H.; Xu, J.; Zhang, J.; Sun, S.; Zhou, X.; et al. The Alpha/Beta-Hydrolase Fold Superfamily in Brassica napus: Expression Profiles and Functional Implications of Clade-3 BnABH Proteins in Response to Abiotic Stress. Int. J. Mol. Sci. 2025, 26, 4746. https://doi.org/10.3390/ijms26104746
Ding Y, Feng L, Li P, Yang X, Li M, Liu H, Xu J, Zhang J, Sun S, Zhou X, et al. The Alpha/Beta-Hydrolase Fold Superfamily in Brassica napus: Expression Profiles and Functional Implications of Clade-3 BnABH Proteins in Response to Abiotic Stress. International Journal of Molecular Sciences. 2025; 26(10):4746. https://doi.org/10.3390/ijms26104746
Chicago/Turabian StyleDing, Yahui, Lianqiang Feng, Pu Li, Xindeng Yang, Muzi Li, Hanxuan Liu, Jiamin Xu, Jitong Zhang, Shouwu Sun, Xiaona Zhou, and et al. 2025. "The Alpha/Beta-Hydrolase Fold Superfamily in Brassica napus: Expression Profiles and Functional Implications of Clade-3 BnABH Proteins in Response to Abiotic Stress" International Journal of Molecular Sciences 26, no. 10: 4746. https://doi.org/10.3390/ijms26104746
APA StyleDing, Y., Feng, L., Li, P., Yang, X., Li, M., Liu, H., Xu, J., Zhang, J., Sun, S., Zhou, X., Hao, W., Zhang, Y., & Xie, C.-G. (2025). The Alpha/Beta-Hydrolase Fold Superfamily in Brassica napus: Expression Profiles and Functional Implications of Clade-3 BnABH Proteins in Response to Abiotic Stress. International Journal of Molecular Sciences, 26(10), 4746. https://doi.org/10.3390/ijms26104746



