Prognostic Value of a Multivariate Gut Microbiome Model for Progression from Normal Cognition to Mild Cognitive Impairment Within 4 Years
Abstract
1. Introduction
2. Results
2.1. Patients Demographics
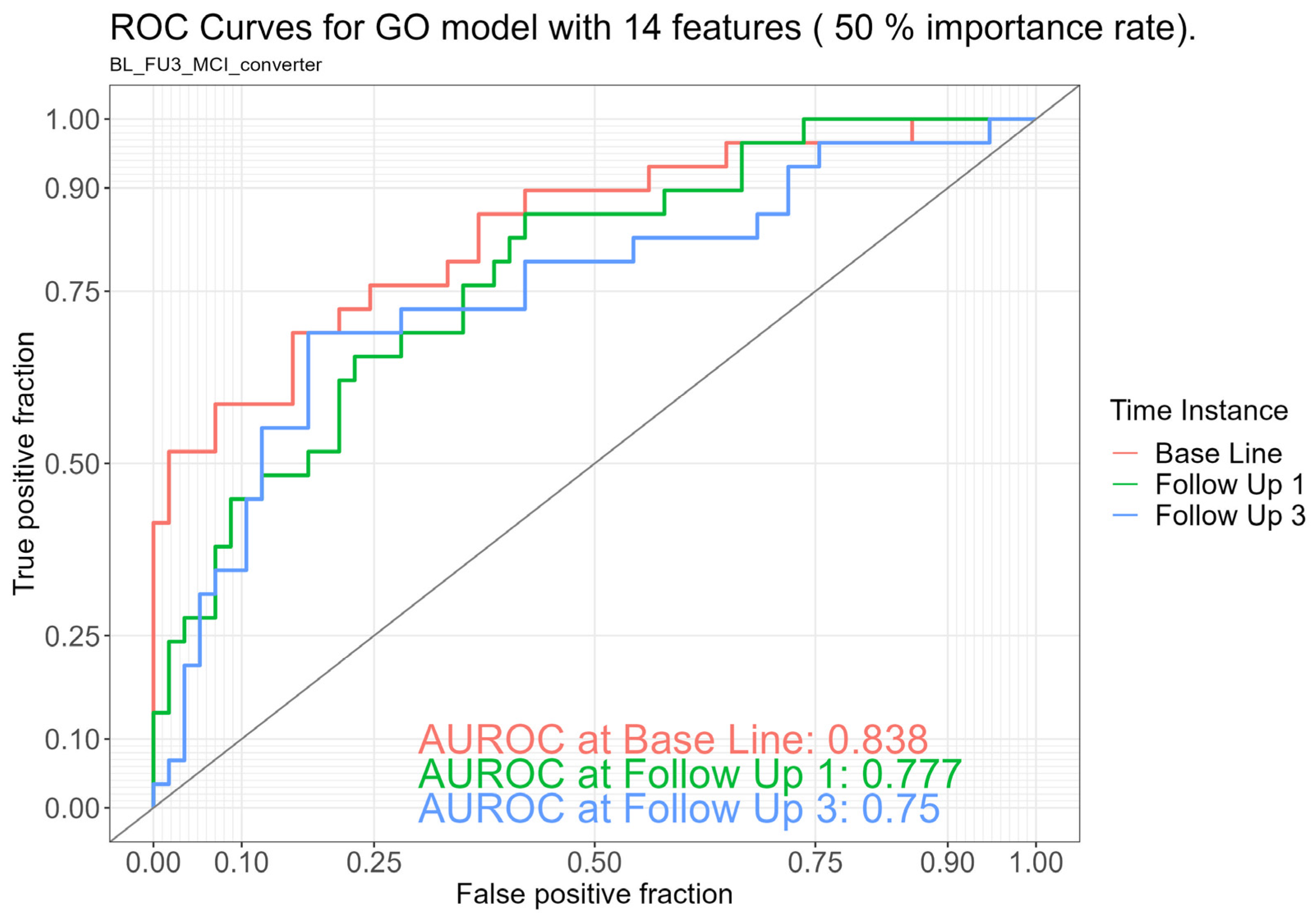
2.2. Discriminatory Ability of the Gut Microbiome Between Stable Healthy Controls and MCI Converters
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Determination of the ApoE4 Genotype
4.3. Stool Collection, DNA Extraction, and Shotgun Metagenome Sequencing
4.4. Metagenomic Assembly
4.5. Taxonomic Classification
4.6. Functional Classification
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeCarli, C. Mild cognitive impairment: Prevalence, prognosis, aetiology, and treatment. Lancet Neurol. 2003, 2, 15–21. [Google Scholar] [CrossRef]
- Chen, Y.X.; Liang, N.; Li, X.L.; Yang, S.H.; Wang, Y.P.; Shi, N.N. Diagnosis and Treatment for Mild Cognitive Impairment: A Systematic Review of Clinical Practice Guidelines and Consensus Statements. Front. Neurol. 2021, 12, 719849. [Google Scholar] [CrossRef]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.; Sandhu, K.V.; Bastiaanssen, T.F.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Stolzer, I.; Scherer, E.; Suss, P.; Rothhammer, V.; Winner, B.; Neurath, M.F.; Günther, C. Impact of Microbiome-Brain Communication on Neuroinflammation and Neurodegeneration. Int. J. Mol. Sci. 2023, 24, 14925. [Google Scholar] [CrossRef]
- Jemimah, S.; Chabib, C.M.M.; Hadjileontiadis, L.; AlShehhi, A. Gut microbiome dysbiosis in Alzheimer’s disease and mild cognitive impairment: A systematic review and meta-analysis. PLoS ONE 2023, 18, e0285346. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Laske, C.; Müller, S.; Preische, O.; Ruschil, V.; Munk, M.H.J.; Honold, I.; Peter, S.; Schoppmeier, U.; Willmann, M. Signature of Alzheimer’s Disease in Intestinal Microbiome: Results From the AlzBiom Study. Front. Neurosci. 2022, 16, 792996. [Google Scholar] [CrossRef]
- Li, B.; He, Y.; Ma, J.; Huang, P.; Du, J.; Cao, L.; Wang, Y.; Xiao, Q.; Tang, H.; Chen, S. Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement 2019, 15, 1357–1366. [Google Scholar] [CrossRef]
- Liu, P.; Wu, L.; Peng, G.; Han, Y.; Tang, R.; Ge, J.; Zhang, L.; Jia, L.; Yue, S.; Zhou, K.; et al. Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 2019, 80, 633–643. [Google Scholar] [CrossRef]
- Ling, Z.; Zhu, M.; Yan, X.; Cheng, Y.; Shao, L.; Liu, X.; Jiang, R.; Wu, S. Structural and Functional Dysbiosis of Fecal Microbiota in Chinese Patients With Alzheimer’s Disease. Front. Cell Dev. Biol. 2020, 8, 634069. [Google Scholar] [CrossRef]
- Ferreiro, A.L.; Choi, J.; Ryou, J.; Newcomer, E.P.; Thompson, R.; Bollinger, R.M.; Hall-Moore, C.; Ndao, I.M.; Sax, L.; Benzinger, T.L.S.; et al. Gut microbiome composition may be an indicator of preclinical Alzheimer’s disease. Sci. Transl. Med. 2023, 15, eabo2984. [Google Scholar] [CrossRef]
- Laske, C.; Müller, S.; Munk, M.H.J.; Honold, I.; Willmann, M.; Peter, S.; Schoppmeier, U. Prognostic Value of Gut Microbiome for Conversion from Mild Cognitive Impairment to Alzheimer’s Disease Dementia within 4 Years: Results from the AlzBiom Study. Int. J. Mol. Sci. 2024, 25, 1906. [Google Scholar] [CrossRef]
- Riedel, B.C.; Thompson, P.M.; Brinton, R.D. Age, APOE and sex: Triad of risk of Alzheimer’s disease. J. Steroid Biochem. Mol. Biol. 2016, 160, 134–147. [Google Scholar] [CrossRef]
- Kang, S.Y.; Kim, Y.J.; Jang, W.; Son, K.Y.; Park, H.S.; Kim, Y.S. Body mass index trajectories and the risk for Alzheimer’s disease among older adults. Sci. Rep. 2021, 11, 3087. [Google Scholar] [CrossRef]
- de la Cuesta-Zuluaga, J.; Kelley, S.T.; Chen, Y.; Escobar, J.S.; Mueller, N.T.; Ley, R.E.; McDonald, D.; Huang, S.; Swafford, A.D.; Knight, R.; et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems 2019, 4, 10-1128. [Google Scholar] [CrossRef]
- Haro, C.; Rangel-Zúñiga, O.A.; Alcalá-Díaz, J.F.; Gómez-Delgado, F.; Pérez-Martínez, P.; Delgado-Lista, J.; Quintana-Navarro, G.M.; Landa, B.B.; Navas-Cortés, J.A.; Tena-Sempere, M.; et al. Intestinal Microbiota Is Influenced by Gender and Body Mass Index. PLoS ONE 2016, 11, e0154090. [Google Scholar] [CrossRef]
- Tran, T.T.T.; Corsini, S.; Kellingray, L.; Hegarty, C.; Le Gall, G.; Narbad, A.; Müller, M.; Tejera, N.; O’Toole, P.W.; Minihane, A.M.; et al. APOE genotype influences the gut microbiome structure and function in humans and mice: Relevance for Alzheimer’s disease pathophysiology. FASEB J. 2019, 33, 8221–8231. [Google Scholar] [CrossRef]
- Cerroni, R.; Pietrucci, D.; Teofani, A.; Chillemi, G.; Liguori, C.; Pierantozzi, M.; Unida, V.; Selmani, S.; Mercuri, N.B.; Stefani, A. Not just a Snapshot: An Italian Longitudinal Evaluation of Stability of Gut Microbiota Findings in Parkinson’s Disease. Brain Sci. 2022, 12, 739. [Google Scholar] [CrossRef]
- Anderson, N.D. State of the science on mild cognitive impairment (MCI). CNS Spectr. 2019, 24, 78–87. [Google Scholar] [CrossRef]
- Dubois, B.; Albert, M.L. Amnestic MCI or prodromal Alzheimer’s disease? Lancet Neurol. 2004, 3, 246–248. [Google Scholar] [CrossRef]
- Katzeff, J.S.; Kim, W.S. ATP-binding cassette transporters and neurodegenerative diseases. Essays Biochem. 2021, 65, 1013–1024. [Google Scholar]
- Kumar, V.; Kim, S.H.; Bishayee, K. Dysfunctional Glucose Metabolism in Alzheimer’s Disease Onset and Potential Pharmacological Interventions. Int. J. Mol. Sci. 2022, 23, 9540. [Google Scholar] [CrossRef]
- Maity, S.; Farrell, K.; Navabpour, S.; Narayanan, S.N.; Jarome, T.J. Epigenetic Mechanisms in Memory and Cognitive Decline Associated with Aging and Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 12280. [Google Scholar] [CrossRef]
- Prajjwal, P.; Asharaf, S.; Makhanasa, D.; Yamparala, A.; Tariq, H.; Aleti, S.; Gadam, S.; Vora, N. Association of Alzheimer’s dementia with oral bacteria, vitamin B12, folate, homocysteine levels, and insulin resistance along with its pathophysiology, genetics, imaging, and biomarkers. Dis. Mon. 2023, 69, 101546. [Google Scholar] [CrossRef] [PubMed]
- Younesian, S.; Yousefi, A.M.; Momeny, M.; Ghaffari, S.H.; Bashash, D. The DNA Methylation in Neurological Diseases. Cells 2022, 11, 3439. [Google Scholar] [CrossRef]
- Zott, B.; Simon, M.M.; Hong, W.; Unger, F.; Chen-Engerer, H.J.; Frosch, M.P.; Sakmann, B.; Walsh, D.M.; Konnerth, A. A vicious cycle of ß amyloid-dependent neuronal hyperactivation. Neurodegeneration 2019, 365, 559–565. [Google Scholar]
- Reiss, A.B.; Voloshyna, I. Regulation of cerebral cholesterol metabolism in alzheimer disease. J. Investig. Med. 2012, 60, 576–582. [Google Scholar] [CrossRef]
- Nanjundaiah, S.; Chidambaram, H.; Chandrashekar, M.; Chinnathambi, S. Role of Microglia in Regulating Cholesterol and Tau Pathology in Alzheimer’s Disease. Cell. Mol. Neurobiol. 2021, 41, 651–668. [Google Scholar] [CrossRef]
- Zhang, Q.; Huang, S.; Cao, Y.; Dong, G.; Chen, Y.; Zhu, X.; Yun, W.; Zhang, M. Remnant cholesterol and mild cognitive impairment: A cross-sectional study. Front. Aging Neurosci. 2023, 15, 1069076. [Google Scholar] [CrossRef]
- Qian, X.-H.; Liu, X.-L.; Zhang, B.; Lin, Y.; Xu, J.-H.; Ding, G.-Y.; Tang, H.-D. Investigating the causal association between branched-chain amino acids and Alzheimer’s disease: A bidirectional Mendelian randomized study. Front. Nutr. 2023, 10, 1103303. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, S.; Shin, S.J.; Park, Y.H.; Nam, Y.; Kim, C.W.; Lee, K.W.; Kim, S.-M.; Jung, I.D.; Yang, H.D.; et al. Gram-negative bacteria and their lipopolysaccharides in Alzheimer’s disease: Pathologic roles and therapeutic implications. Transl. Neurodegener. 2021, 10, 49. [Google Scholar] [CrossRef]
- Doifode, T.; Giridharan, V.V.; Generoso, J.S.; Bhatti, G.; Collodel, A.; Schulz, P.E.; Forlenza, O.V.; Barichello, T. The impact of the microbiota-gut-brain axis on Alzheimer’s disease pathophysiology. Pharmacol Res. 2021, 164, 105314. [Google Scholar] [CrossRef]
- Qian, X.H.; Xie, R.Y.; Liu, X.L.; Chen, S.D.; Tang, H.D. Mechanisms of Short-Chain Fatty Acids Derived from Gut Microbiota in Alzheimer’s Disease. Aging Dis. 2022, 13, 1252–1266. [Google Scholar] [CrossRef]
- Hatayama, K.; Ebara, A.; Okuma, K.; Tokuno, H.; Hasuko, K.; Masuyama, H.; Ashikari, I.; Shirasawa, T. Characteristics of Intestinal Microbiota in Japanese Patients with Mild Cognitive Impairment and a Risk-Estimating Method for the Disorder. Biomedicines 2023, 11, 1789. [Google Scholar] [CrossRef]
- Buck, S.S.; Gordon, J.I. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA 2006, 103, 10011–10016. [Google Scholar]
- Ng, Q.X.; Lim, Y.L.; Yaow, C.; Ng, W.K.; Thumboo, J.; Liew, T. Effect of Probiotic Supplementation on Gut Microbiota in Patients with Major Depressive Disorders: A Systematic Review. Nutrients 2023, 15, 1351. [Google Scholar] [CrossRef]
- Folstein, M.F.; Folstein, S.E.; McHugh, P.R. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J. Psychiatr. Res. 1975, 12, 189–198. [Google Scholar] [CrossRef]
- Morris, J.C. Clinical dementia rating: A reliable and valid diagnostic and staging measure for dementia of the Alzheimer type. Int. Psychogeriatr. 1997, 9 (Suppl. 1), 173–176. [Google Scholar] [CrossRef]
- Morris, J.C. The Clinical Dementia Rating (CDR): Current versino and scoring rules. Neurology 1993, 43, 2412–2414. [Google Scholar] [CrossRef]
- Petersen, R.C. Mild cognitive impairment as a diagnostic entity. J. Intern. Med. 2004, 256, 183–194. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignment. Genome Biol. 2014, 15, 1–12. [Google Scholar] [CrossRef]
- Truong, D.T.; Franzosa, E.A.; Tickle, T.L.; Scholz, M.; Weingart, G.; Pasolli, E.; Tett, A.; Huttenhower, C.; Segata, N. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Method 2015, 12, 902–903. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. Available online: https://github.com/vegandevs/vegan (accessed on 30 January 2024).
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Brunner, E.; Bathke, A.C.; Konietschke, F. Rank and Pseudo-Rank Procedures for Independent Observations in Factorial Designs—Using R and SAS (Springer Series in Statistics); Springer: Berlin/Heidelberg, Germany, 2019; p. 521. ISBN 9783030029128. [Google Scholar]
- Noguchi, K.; Gel, Y.R.; Brunner, E.; Konietschke, F. nparL.D.: An R Software Package for the Nonparametric Analysis of Longitudinal Data in Factorial Experiments. J. Stat. Software 2012, 50, 1–23. [Google Scholar] [CrossRef]
- Bischl, B.; Lang, M.; Kotthoff, L.; Schiffner, J.; Richter, J.; Studerus, E.; Casalicchio, G.; Jones, Z.M. mlr: Machine Learning in R. J. Mach. Learn. Res. 2016, 17, 1–5. [Google Scholar]
| Stable HCs (n = 57) | HC-to-MCI Converters (n = 29) | p-Value | |
|---|---|---|---|
| M (SD) | M (SD) | ||
| Age in years | 71.8 (4.5) | 71.6 (3.9) | 0.79 |
| MMSE—baseline | 28.9 (2.3) | 28.9 (1.1) | 0.24 |
| MMSE—1yFU | 28.6 (1.0) | 27.9 (1.5) | 0.04 * |
| MMSE—4yFU | 28.2 (1.6) | 26.5 (1.1) | <0.001 ** |
| GDS | 1.8 (1.9) | 2.2 (2.9) | 0.49 |
| BMI | 25.6 (3.6) | 26.6 (5.9) | 0.44 |
| Ratio (n:n) | Ratio (n:n) | ||
| Gender (male:female) | 28:29 | 12:17 | 0.65 |
| ApoE4 (e4 carriers:single e4 carriers:non-e4-carriers) | 2:11:22 | 1:4:11 | 0.90 |
| Arterial hypertension (yes:no) | 25:32 | 12:17 | 0.83 |
| Diabetes mellitus (yes:no) | 3:54 | 1:28 | 0.71 |
| Rheumatoid arthritis (yes:no) | 2:55 | 2:27 | 0.60 |
| NSAIDs (yes:no) | 13:44 | 8:21 | 0.79 |
| Anticoagulants (yes:no) | 2:55 | 2:27 | 0.60 |
| Antihypertensives (yes:no) | 24:33 | 12:17 | 0.95 |
| Antidiabetics (yes:no) | 3:54 | 1:28 | 0.71 |
| Statins (yes:no) | 7:50 | 2:27 | 0.71 |
| Antidepressants (yes:no) | 1:56 | 3:26 | 0.11 |
| AChE inhibitors (yes:no) | 0:57 | 0:29 | n.a. |
| Genus | Phylum | Genera Levels in MCI Converters vs. Stable HCs (↑ Increased, ↓ Decreased) |
|---|---|---|
| Merdimonas | Bacillota/Firmicutes | ↑ * |
| Butyricicoccus | Bacillota/Firmicutes | ↑ |
| Sharpea | Bacillota/Firmicutes | ↓ |
| Peptoanaerobacter | Bacillota/Firmicutes | ↑ |
| Brevundimonas | Pseudomonadota/a-proteobacteria | ↑ |
| Alkalibacter | Bacillota/Firmicutes | ↑ |
| Acetobacter | Pseudomonadota/aproteobacteria | ↑ |
| Phycicoccus | Actinomycetota | ↑ |
| Tepidanaerobacter | Bacillota/Firmicutes | ↑ |
| Natronincola | Bacillota/Firmicutes | ↓ |
| Anoxybacillus | Bacillota/Firmicutes | ↓ |
| Luteimonas | Pseudomonadota | ↑ |
| Azonexus | Pseudomonadota | ↑ |
| Gilvimarinus | Pseudomonadota | ↑ |
| Dehalococcoides | Chloroflexota | ↑ |
| Desulfovermiculus | Pseudomonadota | ↓ |
| Knoellia | Actinomycetota | ↓ |
| Roseisalinus | Pseudomonadota | ↓ |
| Polycyclovorans | Pseudomonadota | ↑ |
| Thiocystis | Pseudomonadota | ↓ |
| Sulfuricella | Pseudomonadota | ↓ |
| Methanococcoides | Euryarchaeota | ↓ |
| Oceaniovalibus | Pseudomonadota | ↓ |
| Numidum | Bacillota/Firmicutes | ↓ |
| Arcticibacter | Bacteroidota | ↓ |
| Agarivorans | Pseudomonadota | ↓ |
| Thermovenabulum | Bacillota/Firmicutes | ↓ * |
| Herminiimonas | Pseudomonadota | ↓ |
| Natrinema | Euryarchaeota | ↑ |
| Oceanicoccus | Pseudomonadota | ↓ |
| Bergeriella | Pseudomonadota | ↑ |
| Endomicrobium | Elusimicrobiota | ↓ |
| Caviibacter | Fusobacteria | ↑ |
| Sagittula | Pseudomonadota | ↓ |
| Dehalobacter | Bacillota/Firmicutes | ↓ |
| Olegusella | Actinomycetota | ↓ |
| Labrenzia | Pseudomonadota | ↑ |
| Methanobrevibacter | Euryarchaeota | ↓ * |
| GO Label | Name/Term | Definition/Molecular Functions | Genera Levels in MCI Converters vs. Stable HCs (↑ Increased, ↓ Decreased) | |
|---|---|---|---|---|
| GO.0015416 | ABC-type phosphonate transporter activity | Enables the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: ATP + H2O + phosphonate(out) = ADP + phosphate + phosphonate(in). | ↑ ** | |
| GO.0051484 | Isopentenyl diphosphate | Cholesterol pathway; mevalonate-independent pathway involved in terpenoid biosynthetic process. | ↑ ** | |
| GO.0004339 | Glucan 1,4-alphaglucosidase activity | Enzyme in glycogenolysis: catalysis of the hydrolysis of terminal (1->4)-linked alpha-D glucose residues successively from non-reducing ends of the chains with release of beta-D-glucose. | ↑ * | |
| GO.0046777 | Protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). | ↑ | |
| GO.0009018 | Sucrose phosphorylase activity | Catalysis of the reaction: sucrose + phosphate = D-fructose + alpha-D-glucose 1-phosphate. | ↑ | |
| GO.0043802 | Hydrogenobyrinic acid a,c-diamide synthase (glutaminehydrolysing) activity, CopB | Part of the biosynthetic pathway to cobalamin in aerobic bacteria. Catalysis of the reaction: 2 L-glutamine + 2 ATP + 2 H2O + hydrogenobyrinate = 2 L-glutamate + 2 ADP + 4 H+ + hydrogenobyrinate a,c-diamide + 2 phosphate. | ↑ * | |
| GO.0004415 | Hyalurononglucosam inidase activity | Catalysis of the random hydrolysis of (1->4) linkages between N-Acetyl-beta-D-glucosamine and D-Dlucuronate residues in hyaluronate. | ↑ | |
| GO.0004631 | Phosphomevalonate kinase activity | Catalysis of the reaction: (R)-5-phosphomevalonate + ATP = (R)-5-diphosphomevalonate + ADP + H+. | ↑ | |
| GO.0060567 | Negative regulation of termination of DNA-templated transcription | Any process that decreases the rate, frequency, or extent of DNA-dependent transcription termination, the process in which transcription is completed. | ↑ * | |
| GO.0004328 | Formamidase activity | Catalysis of the reaction: formamide + H2O = formate + NH4. | ↓ | |
| GO.0018710 | Acetone carboxylase activity | Catalysis of the reaction: acetone + ATP + CO2 + 2 H2O = acetoacetate + AMP + 4 H+ + 2 phosphate. | ↑ | |
| GO.0004491 | Methylmalonate-semialdehyde dehydrogenase (acylating, NAD) activity | Synthesis of branched-chain amino acids, pyrimidine catabolic pathway catalysis of the reaction: 2-methyl-3-oxopropanoate + CoA + NAD+ = propanoyl-CoA + CO2 + NADH + H+. | ↓ * | |
| GO.0016730 | Oxidoreductase activity | Catalysis of an oxidation–reduction reaction in which an iron-sulfur protein acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor. | ↓ | |
| GO.0015667 | DNA methyltransferase (cytosine-N4-specific) activity | DNA-modification, catalysis of the reaction: S-adenosyl-L-methionine + DNA cytosine = S-adenosyl-L-homocysteine + DNA N4-methylcytosine. | ↓ ** | |
| Models | Included Features | AUROC | ||
|---|---|---|---|---|
| Baseline (CI, %) | 1yFU (CI, %) | 4yFU (CI, %) | ||
| Taxonomic (Genera) | 38 | 72% (60–84) | 70% (59–82) | 58% (45–71) |
| Functional (Gene Ontology [GO]) | 14 | 84% (75–93) | 78% (68–88) | 75% (63–87) |
| Clinical characteristics (Age, BMI, gender, and ApoE4) | 4 | 57% (44–71) | 54% (40–68) | 55% (41–68) |
| Ensemble learning model | 84% (75–93) | 78% (68–88) | 75% (63–87) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauch, A.; Baur, J.; Honold, I.; Willmann, M.; Weber, G.L.; Müller, S.; Sodenkamp, S.; Peter, S.; Schoppmeier, U.; Laske, C. Prognostic Value of a Multivariate Gut Microbiome Model for Progression from Normal Cognition to Mild Cognitive Impairment Within 4 Years. Int. J. Mol. Sci. 2025, 26, 4735. https://doi.org/10.3390/ijms26104735
Bauch A, Baur J, Honold I, Willmann M, Weber GL, Müller S, Sodenkamp S, Peter S, Schoppmeier U, Laske C. Prognostic Value of a Multivariate Gut Microbiome Model for Progression from Normal Cognition to Mild Cognitive Impairment Within 4 Years. International Journal of Molecular Sciences. 2025; 26(10):4735. https://doi.org/10.3390/ijms26104735
Chicago/Turabian StyleBauch, Anne, Julia Baur, Iris Honold, Matthias Willmann, Greta Louise Weber, Stephan Müller, Sebastian Sodenkamp, Silke Peter, Ulrich Schoppmeier, and Christoph Laske. 2025. "Prognostic Value of a Multivariate Gut Microbiome Model for Progression from Normal Cognition to Mild Cognitive Impairment Within 4 Years" International Journal of Molecular Sciences 26, no. 10: 4735. https://doi.org/10.3390/ijms26104735
APA StyleBauch, A., Baur, J., Honold, I., Willmann, M., Weber, G. L., Müller, S., Sodenkamp, S., Peter, S., Schoppmeier, U., & Laske, C. (2025). Prognostic Value of a Multivariate Gut Microbiome Model for Progression from Normal Cognition to Mild Cognitive Impairment Within 4 Years. International Journal of Molecular Sciences, 26(10), 4735. https://doi.org/10.3390/ijms26104735





