The RNA Chaperone Hfq and Small Non-Coding RNAs Modulate the Biofilm Formation of the Fish Pathogen Yersinia ruckeri
Abstract
1. Introduction
2. Results
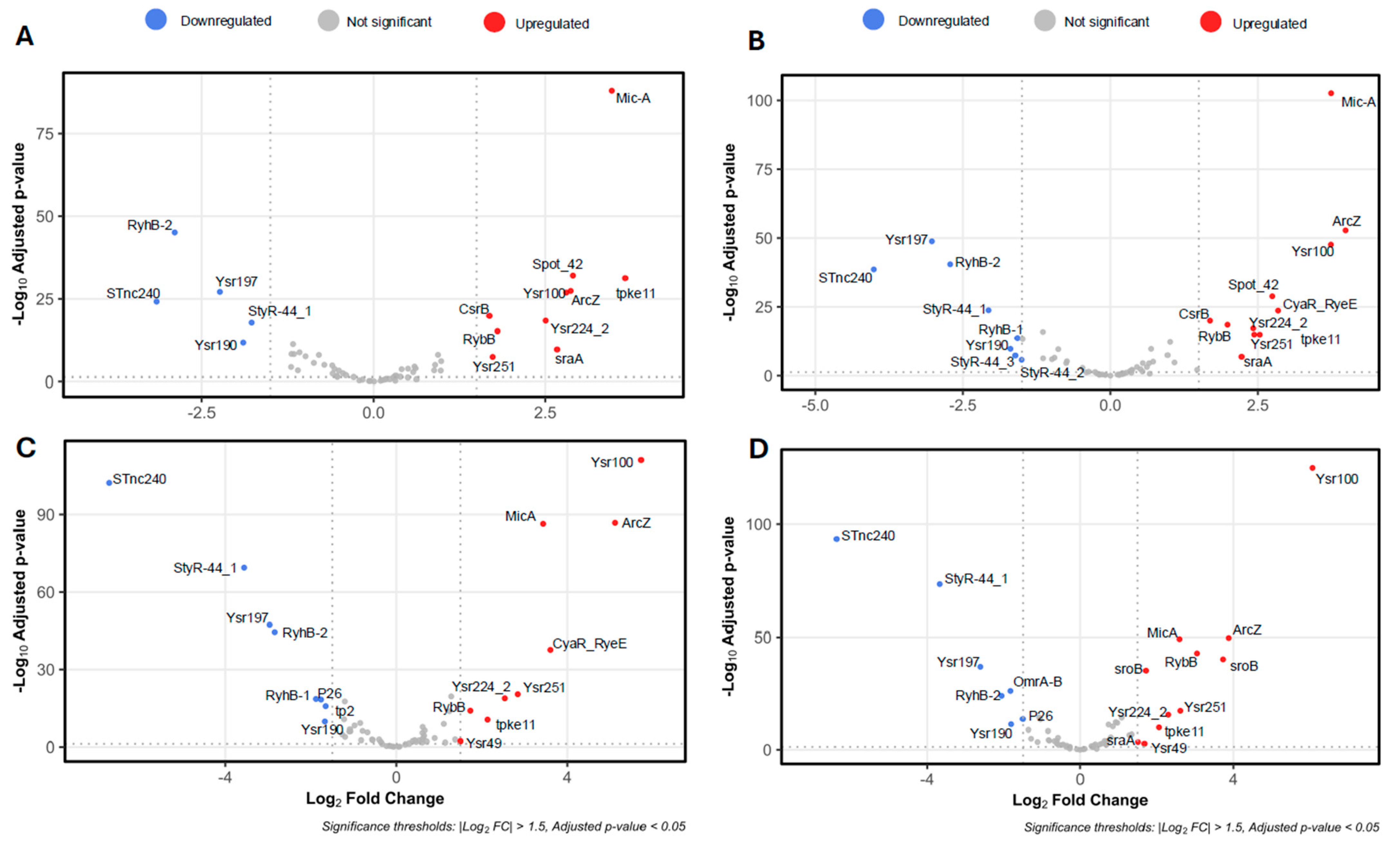
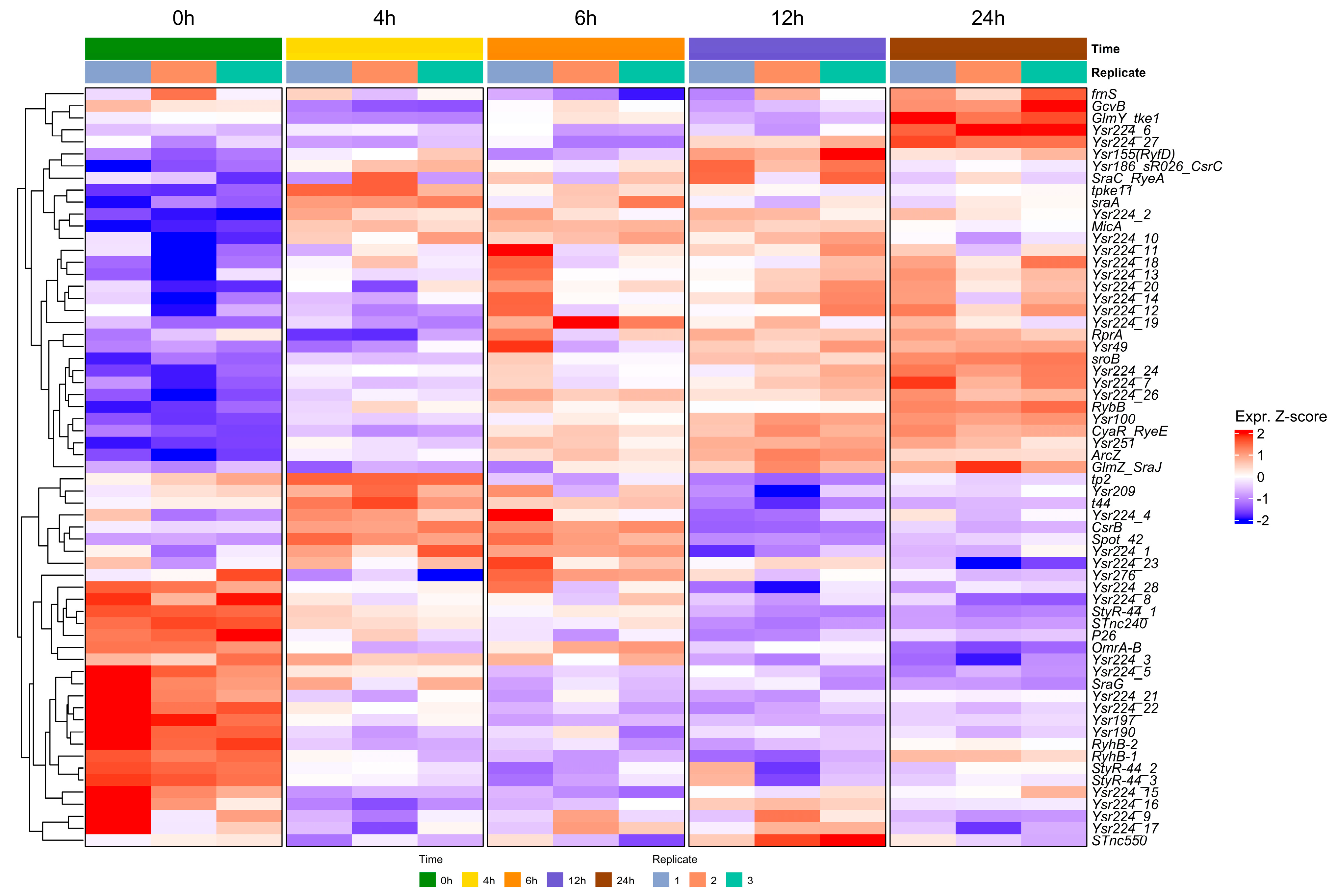
2.1. Determination of the sRNAome of Y. ruckeri That Forms Biofilms on an Abiotic Surface
2.2. Hfq Is Required for the Stability of sRNAs Expressed During the Biofilm Formation of Y. ruckeri
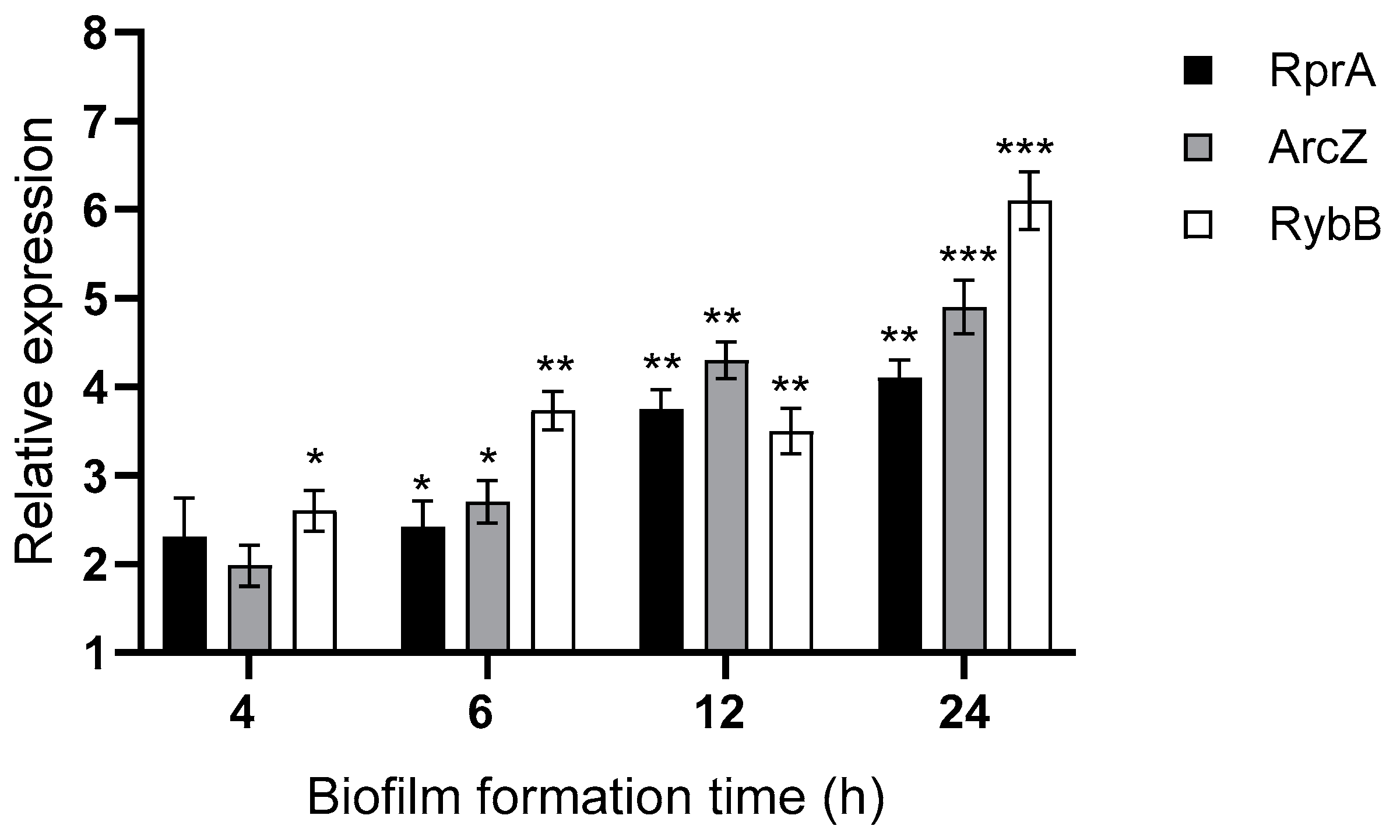
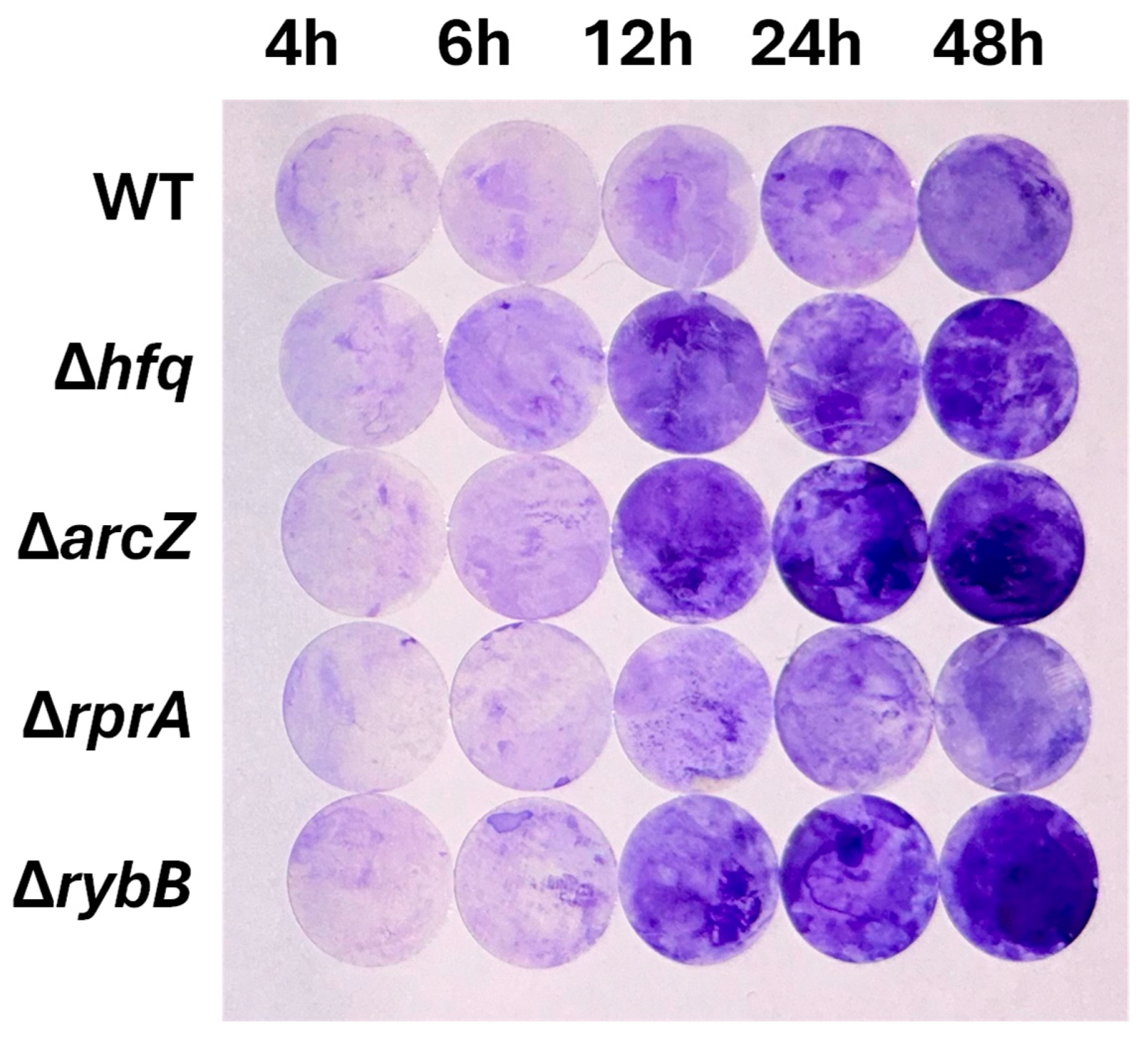
2.3. Hfq and the sRNAs ArcZ, RprA, and RybB Differentially Modulate Biofilm Formation in Y. ruckeri
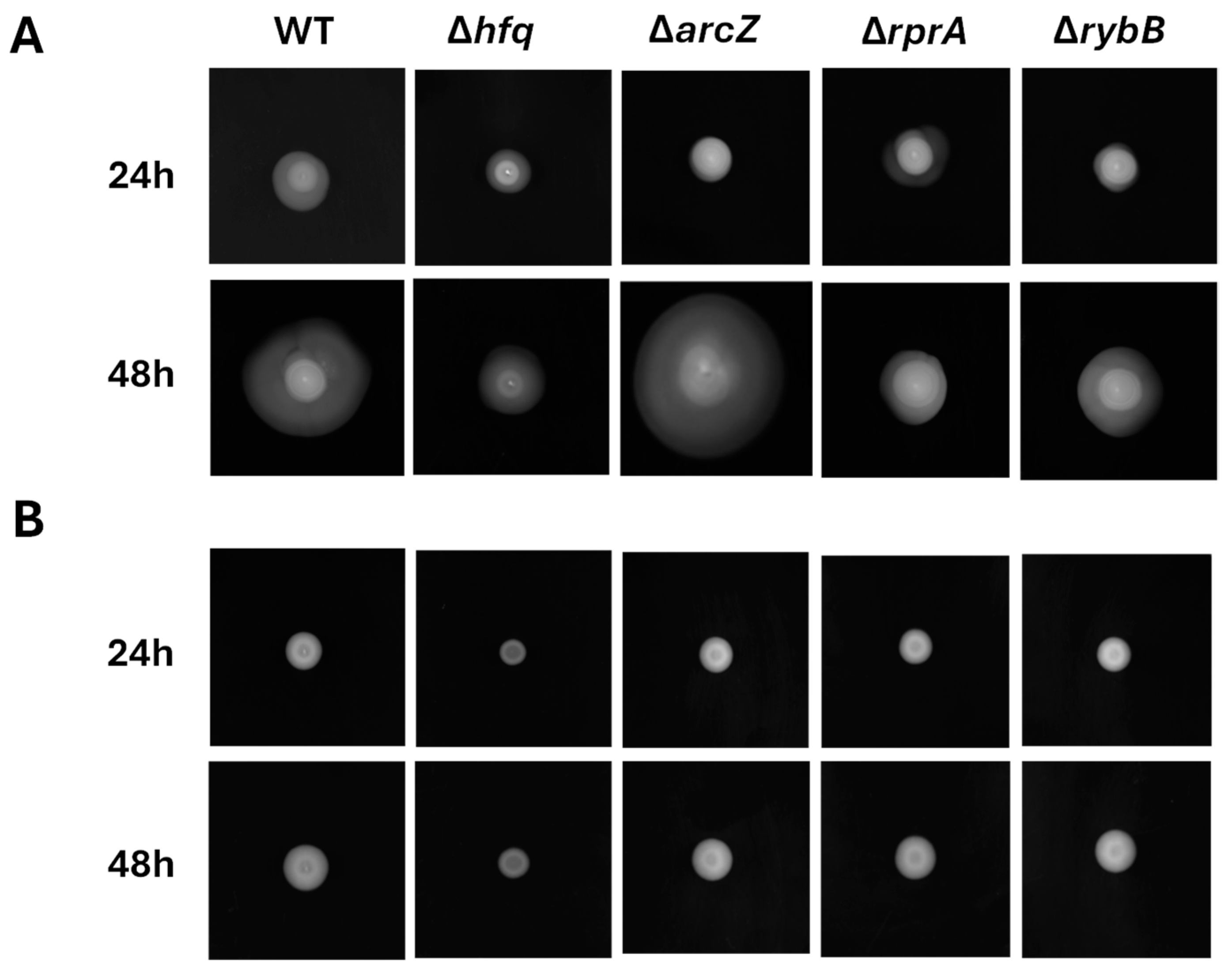
2.3.1. Hfq and the sRNAs ArcZ, RprA, and RybB Are Required for the Motility Regulation of Y. ruckeri, a Pivotal Trait for Specific Stages of Biofilm Formation
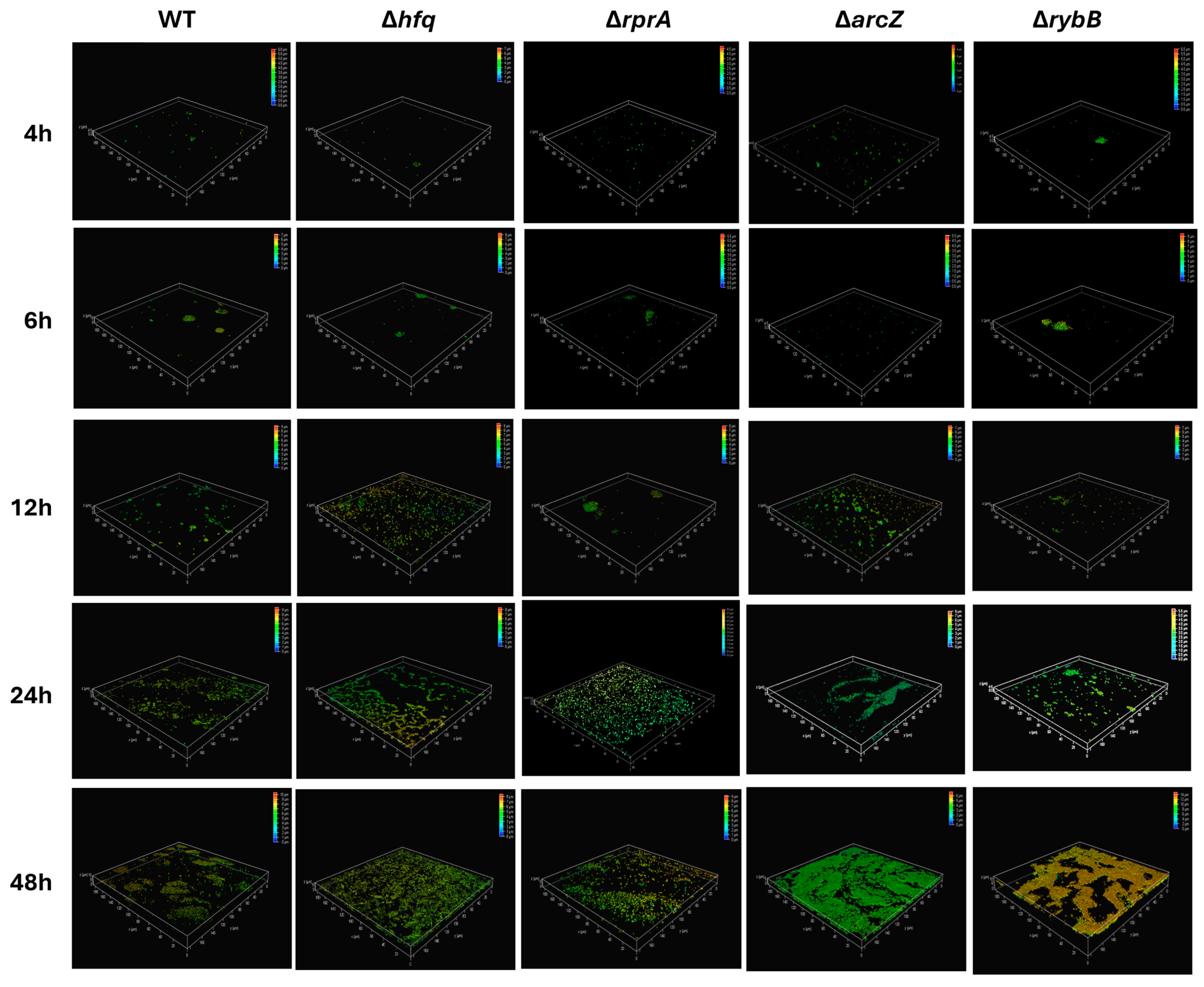
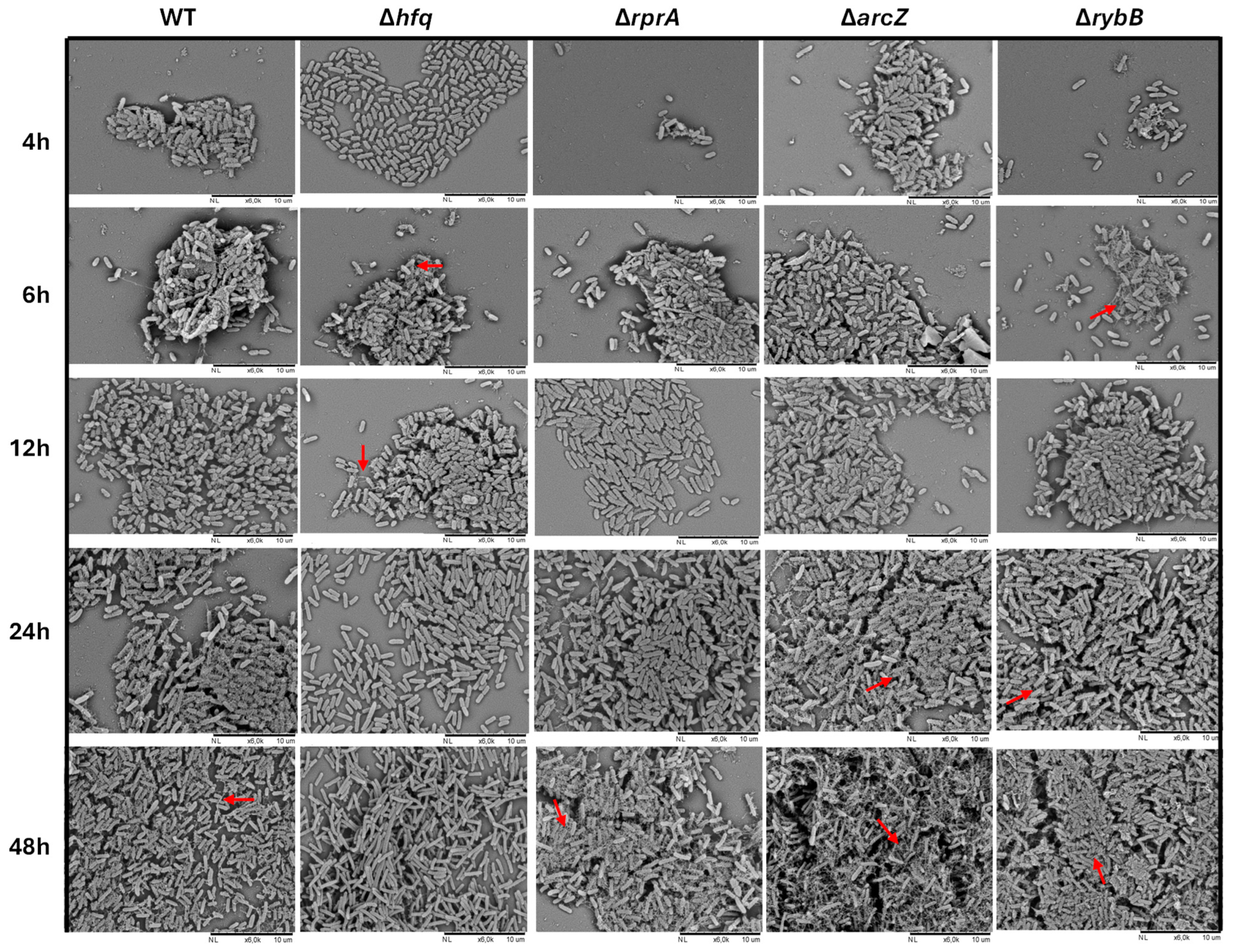
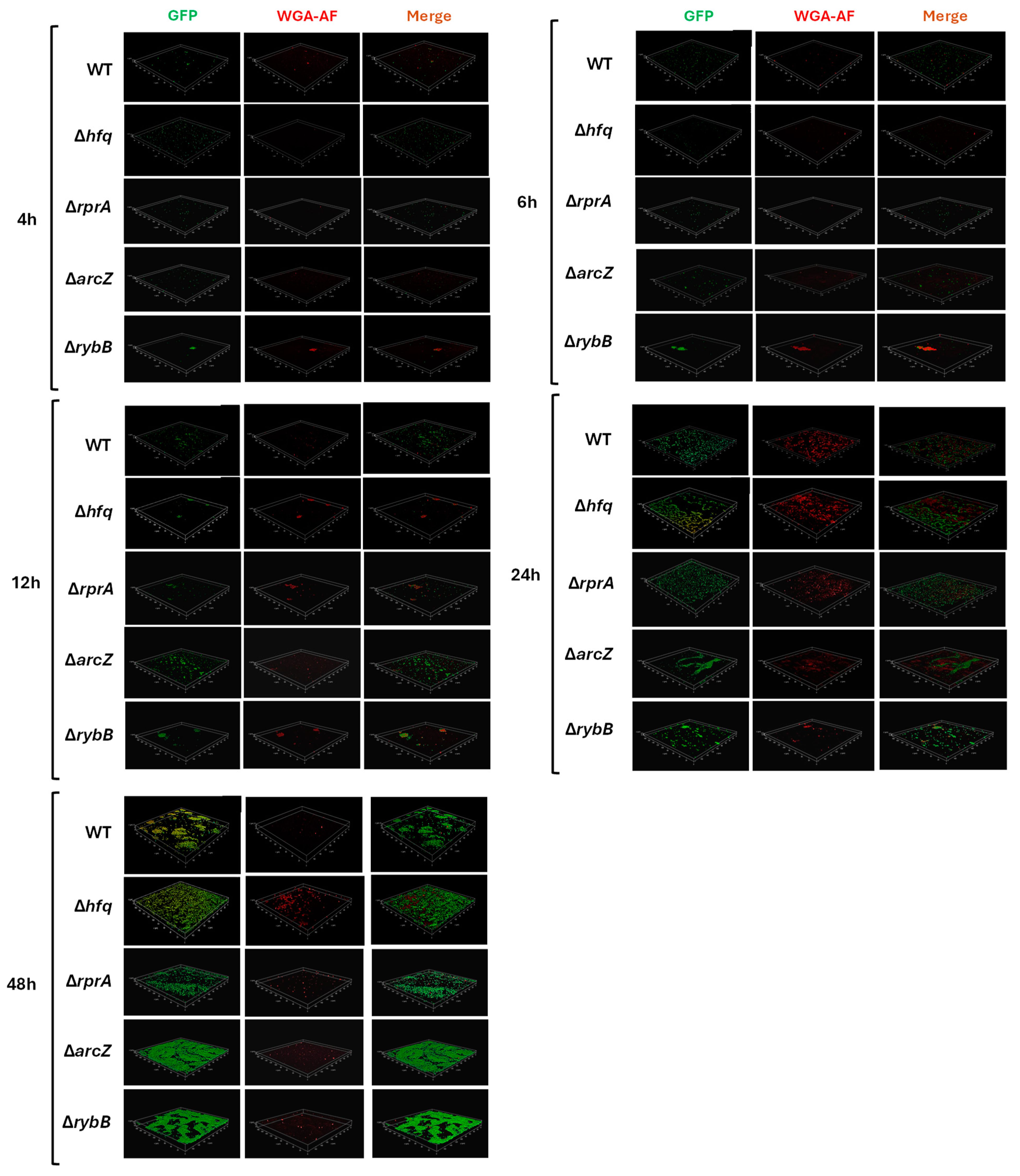
2.3.2. Hfq and the sRNAs ArcZ, RprA, and RybB Participate at Different Stages of Biofilm Formation, Modulating the Attachment and/or the Architecture of Y. ruckeri Conglomerates
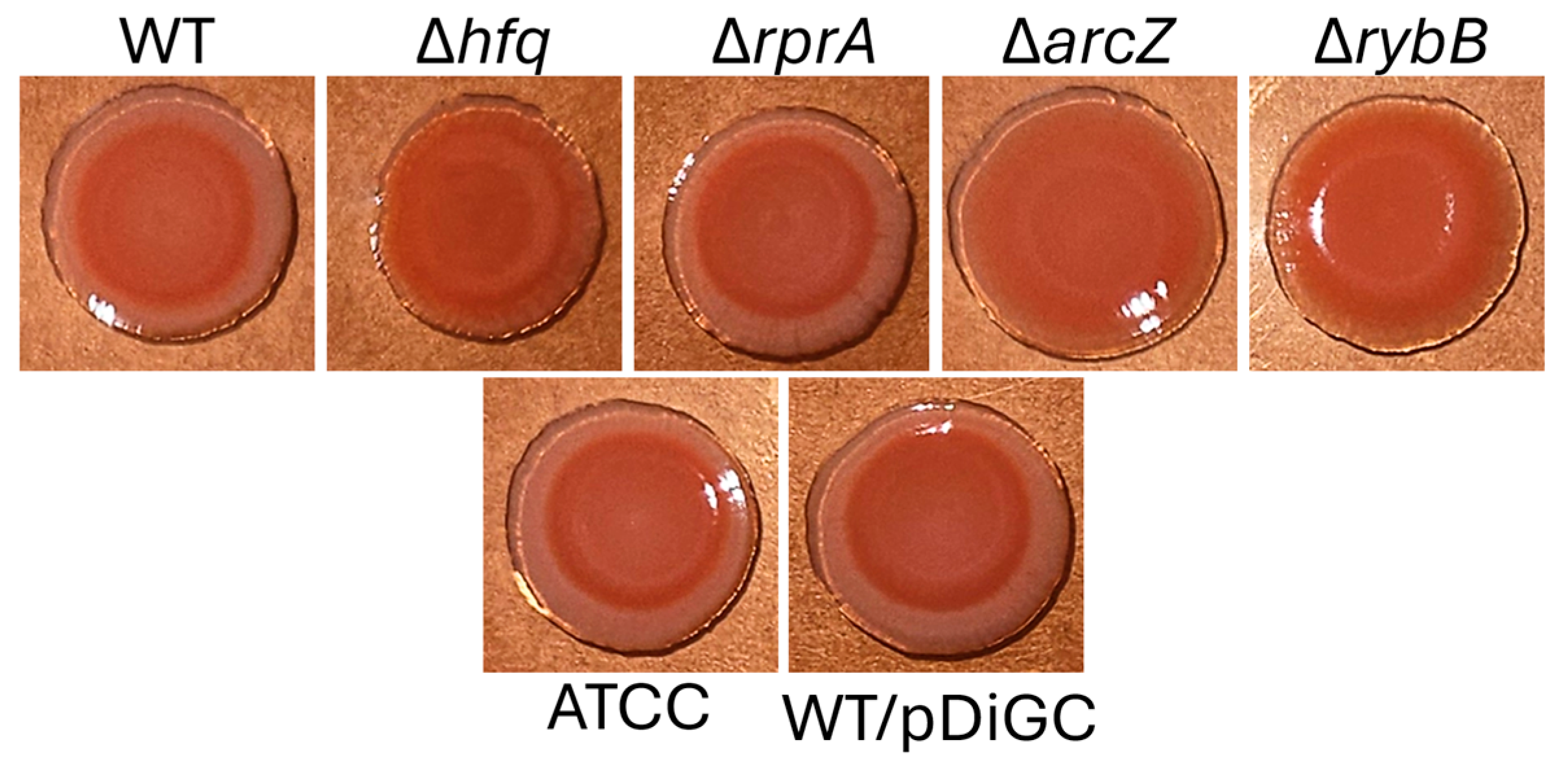
2.3.3. Hfq and the sRNAs ArcZ and RybB Modulate the Production of Extracellular Polysaccharides in Y. ruckeri Biofilms
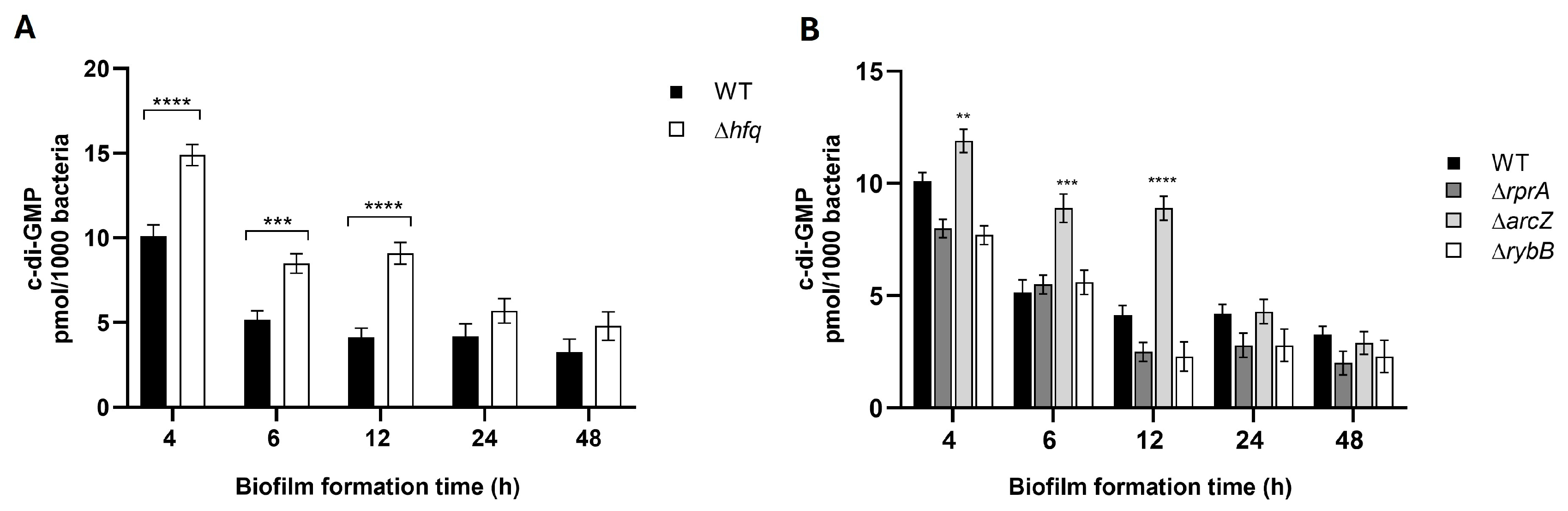
2.4. Hfq and the sRNA ArcZ Modulate the Intracellular Levels of c-di-GMP in Y. ruckeri
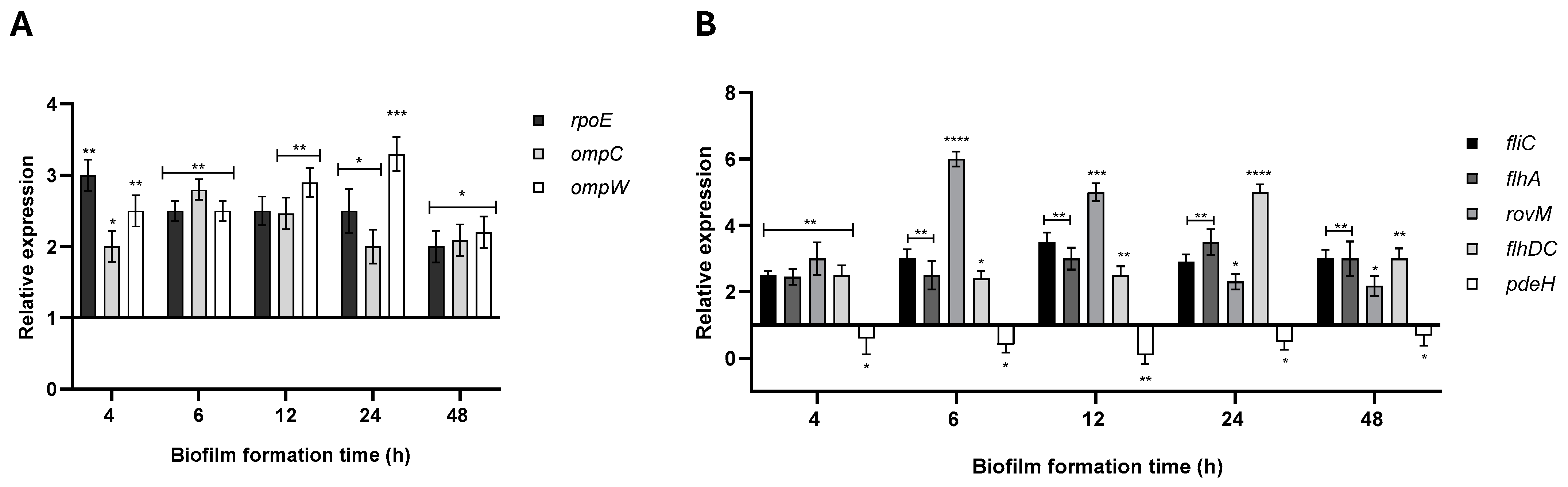
2.5. ArcZ and RybB Regulate the Expression of Biofilm-Related Genes in Y. ruckeri
3. Discussion
3.1. sRNAome of Yersinia ruckeri That Forms Biofilms Revealed Newly and Conserved sRNAs
3.2. Differential Correlation Among the Motility Phenotypes and Biofilm Outputs of Y. ruckeri Strains Indicates That They Operate at Different Nodes of the Regulatory Network
3.3. Hfq and sRNAs Modulate the Biofilm Formation Ability of Yersinia ruckeri by Maintaining the Balance on Gene Expression, Metabolism, and Production of Key Factors Related to Biofilm Development
3.4. Future Perspectives and Challenges
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Motility Assays
4.3. Biofilm Formation Assay
4.4. Crystal Violet Staining Assay
4.5. Macrocolony Formation in Congo Red Plates
4.6. Analysis of Biofilms by Confocal Microscopy
4.7. Biofilm Architecture Analysis by Scanning Electron Microscopy
4.8. Quantification of Cyclic Diguanosine Monophosphate (c-di-GMP)
4.9. Genome Assembly and Annotation
4.10. RNA-Seq and sRNAome Analyses
4.11. RT-qPCR Analysis
4.12. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dunne, W.M. Bacterial adhesion: Seen any good biofilms lately? Clin. Microbiol. Rev. 2002, 15, 155–166. [Google Scholar] [CrossRef]
- Zhou, D.; Yang, R. Formation and regulation of Yersinia biofilms. Protein Cell 2011, 2, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Calderón, I.L.; Barros, M.J.; Fernández-Navarro, N.; Acuña, L.G. Detection of Nucleic Acids of the Fish Pathogen Yersinia ruckeri from Planktonic and Biofilm Samples with a CRISPR/Cas13a-Based Assay. Microorganisms 2024, 12, 283. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Bystritskaya, E.; Stenkova, A.; Chistuylin, D.; Chernysheva, N.; Khomenko, V.; Anastyuk, S.; Novikova, O.; Rakin, A.; Isaeva, M. Adaptive responses of outer membrane porin balance of Yersinia ruckeri under different incubation temperature, osmolarity, and oxygen availability. Microbiol. Open 2016, 5, 597–603. [Google Scholar] [CrossRef]
- Wrobel, A.; Saragliadis, A.; Pérez-Ortega, J.; Sittman, C.; Göttig, S.S.; Liskiewicz, K.; Spence, M.H.; Schneider, K.; Leo, J.C.; Arenas, J.; et al. The inverse autotransporters of Yersinia ruckeri, YrInv and YrIlm, contribute to biofilm formation and virulence. Environ. Microbiol. 2020, 22, 2939–2955. [Google Scholar] [CrossRef]
- Wrobel, A.; Leo, J.C.; Linke, D. Overcoming Fish Defences: The Virulence Factors of Yersinia ruckeri. Genes 2019, 10, 700. [Google Scholar] [CrossRef]
- Coquet, L.; Cosette, P.; Quillet, L.; Petit, F.; Junter, G.A.; Jouenne, T. Occurrence and Phenotypic Characterization of Yersinia ruckeri Strains with Biofilm-Forming Capacity in a Rainbow Trout Farm. Appl. Environ. Microbiol. 2002, 68, 470–475. [Google Scholar] [CrossRef]
- Little, J.I.; Singh, P.K.; Zhao, J.; Dunn, S.; Matz, H.; Donnenberg, M.S. Type IV pili of Enterobacteriaceae species. EcoSal Plus 2024, 12, eesp00032023. [Google Scholar] [CrossRef]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- Mika, F.; Hengge, R. Small Regulatory RNAs in the Control of Motility and Biofilm Formation in E. coli and Salmonella. Int. J. Mol. Sci. 2013, 14, 4560–4579. [Google Scholar] [CrossRef] [PubMed]
- Simm, R.; Morr, M.; Kader, A.; Nimtz, M.; Römling, U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004, 53, 1123–1134. [Google Scholar] [CrossRef]
- Yoong, P.; Cywes-Bentley, C.; Pier, G.B. Poly-N-Acetylglucosamine Expression by Wild-Type Yersinia pestis Is Maximal at Mammalian, Not Flea, Temperatures. mBio 2012, 3, 00217-12. [Google Scholar] [CrossRef]
- Wiechmann, A.; Garcia, V.; Elton, L.; Williams, P.; Atkinson, S. Reciprocal regulation of NagC and quorum sensing systems and their roles in hmsHFRS expression and biofilm formation in Yersinia pseudotuberculosis. Microbiology 2023, 169, 001397. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.R.; Sauer, K. Small RNAs and their role in biofilm formation. Trends Microbiolgy 2013, 21, 39–49. [Google Scholar] [CrossRef]
- Condinho, M.; Carvalho, B.; Cruz, A.; Pinto, S.N.; Arraiano, C.M.; Pobre, V. The role of RNA regulators, quorum sensing and c-di-GMP in bacterial biofilm formation. FEBS Open Bio 2022, 13, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Yu, Z.; Zhu, L.; Li, Z.; Yin, W.; Shang, X.; Chou, S.H.; Tan, Q.; He, J. The Multiple Regulatory Relationship Between RNA-Chaperone Hfq and the Second Messenger c-di-GMP. Front. Microbiol. 2021, 12, 689619. [Google Scholar] [CrossRef]
- Torabi, D.; Soltanian, S.; Sharifiyazdi, H.; Bossier, P. Effect of quorum quenching bacteria on growth, virulence factors and biofilm formation of Yersinia ruckeri in vitro and an in vivo evaluation of their probiotic effect in rainbow trout. J. Fish Dis. 2018, 41, 1429–1438. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, M.; Burgess, R.R. Adaptation in bacterial flagellar and motility systems: From regulon members to ’foraging’-like behavior in E. coli. Nucleic Acids Res. 2007, 35, 4441–4452. [Google Scholar] [CrossRef]
- Papenfort, K.; Pfeiffer, V.; Mika, F.; Lucchini, S.; Hinton, J.C.D.; Vogel, J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006, 6, 1674–1688. [Google Scholar] [CrossRef]
- Mika, F.; Busse, S.; Possling, A.; Berkholz, J.; Tschowri, N.; Sommerfeldt, N.N.; Pruteanu, M.; Hengge, R. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol. Microbiol. 2012, 84, 51–65. [Google Scholar] [CrossRef]
- Gu, Z. Complex heatmap visualization. Imeta 2022, 1, e43. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 14 March 2025).
- Ontiveros-Palacios, N.; Cooke, E.; Nawrocki, E.P.; Triebel, S.; Marz, M.; Rivas, E.; Griffiths-Jones, S.; Petrov, A.I.; Bateman, A.; Sweeney, B. Rfam 15: RNA families database in 2025. Nucleic Acids Res. 2025, 53, D258–D267. [Google Scholar] [CrossRef] [PubMed]
- Helaine, S.; Thompson, J.A.; Watson, K.G.; Liu, M.; Boyle, C.; Holden, D.W. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. USA 2010, 107, 3746–3751. [Google Scholar] [CrossRef]
- Lu, X.; Li, G.; Pang, J.; Yang, X.; Cywes-Bentley, C.; You, X.B. Gerald. Antibodies Targeting a Conserved Surface Polysaccharide Are Protective Against a Wide Range of Microbial Pathogens Producing β-1–6-Linked Poly-N-Acetylglucosamine (PNAG). Engineering 2024, 38, 69–76. [Google Scholar] [CrossRef]
- Flannery, A.; Le Berre, M.; Pier, G.B.; O’Gara, J.P.; Kilcoyne, M. Glycomics Microarrays Reveal Differential In Situ Presentation of the Biofilm Polysaccharide Poly-N-acetylglucosamine on Acinetobacter baumannii and Staphylococcus aureus Cell Surfaces. Int. J. Mol. Sci. 2020, 21, 2465. [Google Scholar] [CrossRef]
- Burton, E.; Yakandawala, N.; LoVetri, K.; Madhyastha, M.S. A microplate spectrofluorometric assay for bacterial biofilms. J. Ind. Microbiol. Biotechnol. 2007, 34, 1–4. [Google Scholar] [CrossRef]
- Wright, P.R.; Georg, J.; Mann, M.; Sorescu, D.A.; Richter, A.S.; Lott, S.; Kleinkauf, R.; Hess, W.R.; Backofen, R. CopraRNA and IntaRNA: Predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014, 42, W119–W123. [Google Scholar] [CrossRef]
- Wright, P.R.; Richter, A.S.; Papenfort, K.; Mann, M.; Vogel, J.; Hess, W.R.; Backofen, R.; Georg, J. Comparative genomics boosts target prediction for bacterial small RNAs. Proc. Natl. Acad. Sci. USA 2013, 110, E3487–E3496. [Google Scholar] [CrossRef]
- Raden, M.; Ali, S.M.; Alkhnbashi, O.S.; Busch, A.; Costa, F.; Davis, J.A.; Eggenhofer, F.; Elhausen, R.; Georg, J.; Heyne, S.; et al. Freiburg RNA tools: A central online resource for RNA-focused research and teaching. Nucleic Acids Res. 2018, 46, W25–W29. [Google Scholar] [CrossRef]
- Zhao, R.; Song, Y.; Dai, Q.; Kang, Y.; Pan, J.; Zhu, L.L.; Zhang, L.; Wang, Y.; Shen, X. A starvation-induced regulator, RovM, acts as a switch for planktonic/biofilm state transition in Yersinia pseudotuberculosis. Sci. Rep. 2017, 7, 639. [Google Scholar] [CrossRef]
- Pesavento, C.; Becker, G.; Sommerfeldt, N.; Possling, A.; Tschowri, N.; Mehlis, A.; Hengge, R. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 2008, 22, 2434–2446. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Hennelly, S.P.; Stubben, C.J.; Micheva-Viteva, S.; Hu, B.; Shou, Y.; Vuyisich, M.; Tung, C.; Chain, P.S.; Sanbonmatsu, K.Y.; et al. Functional and Structural Analysis of a Highly-Expressed Yersinia pestis Small RNA following Infection of Cultured Macrophages. PLoS ONE 2016, 11, e0168915. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Su, S.; Meng, X.; Ji, X.; Qu, Y.; Liu, Z.; Zhang, J.; Wang, Q.; Zhang, Z.; Wang, J.; et al. Determination of sRNA expressions by RNA-seq in Yersinia pestis grown in vitro and during infection. PLoS ONE 2013, 8, e74495. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.T.; Lathem, W.W. Global discovery of small noncoding RNAs in pathogenic Yersinia species. In Advances in Yersinia Research; de Almeida, A.M.P., Leal, N.C., Eds.; Springer: New York, NY, USA, 2012; Volume 2, pp. 305–314. [Google Scholar]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial biofilm: A review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Zhao, A.; Sun, J.; Liu, Y. Understanding bacterial biofilms: From definition to treatment strategies. Front. Cell. Infect. Microbiol. 2023, 13, 1137947. [Google Scholar] [CrossRef]
- Chen, P.E.; Cook, C.; Stewart, A.C.; Nagarajan, N.; Sommer, D.D.; Pop, M.; Thomason, S.G.; Thomason, K.M.; Hogg, J.F.; Radune, G.; et al. Genomic characterization of the Yersinia genus. Genome Biol. 2010, 11, R1. [Google Scholar] [CrossRef]
- Verstraeten, N.; Braeken, K.; Debkumari, B.; Fauvart, M.; Fransaer, J.; Vermant, J.; Michiels, J. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008, 16, 496–506. [Google Scholar] [CrossRef]
- Deng, Y.; Zang, S.; Lin, Z.; Xu, L.; Cheng, C.; Feng, J. The pleiotropic phenotypes caused by an hfq null mutation in Vibrio harveyi. Microorganisms 2023, 11, 2741. [Google Scholar] [CrossRef]
- Schiano, C.A.; Bellows, L.E.; Lathem, W.W. The small RNA chaperone Hfq is required for the virulence of Yersinia pseudotuberculosis. Infect. Immun. 2010, 78, 2034–2044. [Google Scholar] [CrossRef]
- Bak, G.; Lee, J.; Suk, S.; Kim, D.; Lee, J.Y.; Kim, K.S. Identification of novel sRNAs involved in biofilm formation, motility and fimbriae formation in Escherichia coli. Sci. Rep. 2015, 5, 15287. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schachterle, J.K.; Sundin, G.W. Orchestration of virulence factor expression and modulation of biofilm dispersal in Erwinia amylovora through activation of the Hfq-dependent small RNA RprA. Mol. Plant Pathol. 2020, 22, 255–270. [Google Scholar] [CrossRef]
- Schachterle, J.K.; Zeng, Q.; Sundin, G.W. Three Hfq-dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora. Mol. Microbiol. 2019, 111, 1476–1492. [Google Scholar] [CrossRef] [PubMed]
- Felden, B.; Augagneur, Y. Diversity and versatility in small RNA-mediated regulation in bacterial pathogens. Front. Microbiol. 2021, 12, 719977. [Google Scholar] [CrossRef]
- De Lay, N.; Gottesman, S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012, 86, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Svensson, S.L.; Sharma, C.M. Small RNAs in bacterial virulence and communication. Microbiol. Spectr. 2016, 4, 3. [Google Scholar] [CrossRef]
- Bellows, L.E.; Koestler, B.J.; Karaba, S.M.; Waters, C.M.; Lathem, W.W. Hfq-dependent, coordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 2012, 86, 661–674. [Google Scholar] [CrossRef]
- Frasch, C.E. Role of lipopolysaccharide in wheat germ agglutinin-mediated agglutination of Neisseria meningitidis and Neisseria gonorrhoeae. J. Clin. Microbiol. 1980, 12, 498–501. [Google Scholar] [CrossRef]
- Papenfort, K.; Vogel, J. Regulatory RNA in bacterial pathogens. Cell Host Microbe 2010, 8, 116–127. [Google Scholar] [CrossRef]
- Chen, H.; Ahsan, S.S.; Santiago-Berrios, M.B.; Abruña, H.D.; Webb, W.W. Mechanisms of quenching of Alexa fluorophores by natural amino acids. J. Am. Chem. Soc. 2010, 132, 7244–7245. [Google Scholar] [CrossRef]
- Zambrano, N.; Guichard, P.; Bi, Y.; Cayrol, B.; Marco, S.; Arluison, V. Involvement of HFq protein in the post-transcriptional regulation of E. coli bacterial cytoskeleton and cell division proteins. Cell Cycle 2009, 8, 2470–2472. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle–expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Calderón, I.L.; Barros, M.J.; Montt, F.; Gil, F.; Fuentes, J.A.; Acuña, L.G. The RNA chaperone Hfq participates in persistence to multiple antibiotics in the fish pathogen Yersinia ruckeri. Microorganisms 2021, 9, 1404. [Google Scholar] [CrossRef] [PubMed]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The green tea polyphenol EGCG inhibits E. coli biofilm formation by impairing amyloid curli fibre assembly and downregulating the biofilm regulator CsgD via the σ(E)-dependent sRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar]
- Römling, U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 2005, 62, 1234–1246. [Google Scholar] [CrossRef]
- Mika, F.; Hengge, R. Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol. 2014, 11, 494–507. [Google Scholar] [CrossRef]
- Monteiro, C.; Papenfort, K.; Hentrich, K.; Ahmad, I.; Le Guyon, S.; Reimann, R.; Grantcharova, N.; Römling, U. Hfq and Hfq-dependent small RNAs are major contributors to multicellular development in Salmonella enterica serovar Typhimurium. RNA Biol. 2012, 9, 489–502. [Google Scholar] [CrossRef]
- Schachterle, J.K.; Stewart, R.M.; Schachterle, M.B.; Calder, J.T.; Kang, H.; Prince, J.T.; Erickson, D.L. Yersinia pseudotuberculosis BarA-UvrY two-component regulatory system represses biofilms via CsrB. Front. Cell. Infect. Microbiol. 2018, 8, 323. [Google Scholar] [CrossRef] [PubMed]
- Goller, C.; Wang, X.; Itoh, Y.; Romeo, T. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesin poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 2006, 188, 8022–8032. [Google Scholar] [CrossRef]
- Cerca, N.; Jefferson, K.K. Effect of growth conditions on poly-N-acetylglucosamine expression and biofilm formation in Escherichia coli. FEMS Microbiol. Lett. 2008, 283, 36–41. [Google Scholar] [CrossRef]
- Riborg, A.; Gulla, S.; Fiskebeck, E.Z.; Ryder, D.; Verner-Jeffreys, D.W.; Colquhoun, D.J.; Welch, T.J. Pan-genome survey of the fish pathogen Yersinia ruckeri links accessory- and amplified genes to virulence. PLoS ONE 2023, 18, e0285257. [Google Scholar] [CrossRef] [PubMed]
- Petchiappan, A.; Naik, S.Y.; Chatterji, D. Tracking the homeostasis of second messenger cyclic-di-GMP in bacteria. Biophys. Rev. 2020, 12, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Heroven, A.K.; Dersch, P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol. Microbiol. 2006, 62, 1469–1483. [Google Scholar] [CrossRef]
- Liu, L.; Fang, H.; Yang, H.; Zhang, Y.; Han, Y.; Zhou, D.; Yang, R. Reciprocal regulation of Yersinia pestis biofilm formation and virulence by RovM and RovA. Open Biol. 2016, 6, 150198. [Google Scholar] [CrossRef]
- Sarenko, O.; Klauck, G.; Wilke, F.M.; Pfiffer, V.; Richter, A.M.; Herbst, S.; Kaever, V.; Hengge, R. More than enzymes that make or break cyclic di-GMP—Local signaling in the interactome of GGDEF/EAL domain proteins of Escherichia coli. mBio 2017, 8, e01639-17. [Google Scholar] [CrossRef]
- Fang, N.; Qu, S.; Yang, H.; Fang, H.; Liu, L.; Zhang, Y.; Zhou, D.; Yang, R. HmsB enhances biofilm formation in Yersinia pestis. Front. Microbiol. 2014, 5, 685. [Google Scholar] [CrossRef][Green Version]
- Liu, L.; Fang, H.; Yang, H.; Zhang, Y.; Han, Y.; Zhou, D.; Yang, R. CRP is an activator of Yersinia pestis biofilm formation that operates via a mechanism involving gmhA and waaAE-coaD. Front. Microbiol. 2016, 7, 295. [Google Scholar] [CrossRef]
- Schachterle, J.K.; Sundin, G.W. The leucine-responsive regulatory protein Lrp participates in virulence regulation downstream of small RNA ArcZ in Erwinia amylovora. mBio 2019, 10, e00757-19. [Google Scholar] [CrossRef]
- Thompson, K.M.; Rhodius, V.A.; Gottesman, S. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007, 189, 4243–4256. [Google Scholar] [CrossRef] [PubMed]
- Gogol, E.B.; Rhodius, V.A.; Papenfort, K.; Vogel, J.; Gross, C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA 2011, 108, 12875–12880. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.; Rasmussen, A.A.; Overgaard, M.; Valentin-Hansen, P. Conserved small non-coding RNAs that belong to the sigmaE regulon: Role in down-regulation of outer membrane proteins. J. Mol. Biol. 2006, 364, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Bossi, N.; Lemire, S.; Maloriol, D.; Balbontín, R.; Casadesús, J.; Bossi, L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006, 62, 838–852. [Google Scholar] [CrossRef]
- de Dios, R.; Santero, E.; Reyes-Ramírez, F. Extracytoplasmic Function σ Factors as Tools for Coordinating Stress Responses. Int. J. Mol. Sci. 2021, 22, 3900. [Google Scholar] [CrossRef]
- Costanzo, A.; Sarah, E.A. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 2006, 188, 4627–4634. [Google Scholar] [CrossRef]
- Vidovic, S.; Medihala, P.; Dynes, J.J.; Daida, P.; Vujanovic, V.; Hitchcock, A.P.; Shetty, D.; Zhang, H.; Brown, D.R.; Lawrence, J.R.; et al. Importance of the RpoE Regulon in Maintaining the Lipid Bilayer during Antimicrobial Treatment with the Polycationic Agent, Chlorhexidine. Proteomics 2018, 18, 1700285. [Google Scholar] [CrossRef]
- Potapova, A.; Garvey, W.; Dahl, P.; Guo, S.; Chang, Y.; Schwechheimer, C.; Trebino, M.A.; Floyd, K.A.; Phinney, B.S.; Liu, J.; et al. Outer membrane vesicles and the outer membrane protein OmpU govern Vibrio cholerae biofilm matrix assembly. mBio 2024, 15, e0330423. [Google Scholar] [CrossRef]
- Saxena, D.; Maitra, R.; Bormon, R.; Czekanska, M.; Meiers, J.; Titz, A.; Verma, S.; Chopra, S. Tackling the outer membrane: Facilitating compound entry into Gram-negative bacterial pathogens. NPJ Antimicrob. Resist. 2023, 1, 17. [Google Scholar] [CrossRef]
- Figueroa-Bossi, N.; Bossi, L. Sponges and Predators in the Small RNA World. Microbiol. Spectr. 2018, 6, RWR-0021-2018. [Google Scholar] [CrossRef]
- Park, S.; Prévost, K.; Heideman, E.M.; Carrier, M.C.; Azam, M.S.; Reyer, M.A.; Liu, W.; Massé, E.; Fei, J. Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. eLife 2021, 10, e64207. [Google Scholar] [CrossRef] [PubMed]
- Acuña, L.G.; Barros, M.J.; Montt, F.; Peñaloza, D.; Núñez, P.; Valdés, I.; Gil, F.F.; Fuentes, J.D.G.; Calderón, I.L. Participation of two sRNA RyhB homologs from the fish pathogen Yersinia ruckeri in bacterial physiology. Microbiol. Res. 2021, 242, 126629. [Google Scholar] [CrossRef]
- Cianfanelli, F.R.; Cunrath, O.; Bumann, D. Efficient dual-negative selection for bacterial genome editing. BMC Microbiol. 2020, 20, 129. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barros, M.J.; Acuña, L.G.; Hernández-Vera, F.; Vásquez-Arriagada, P.; Peñaloza, D.; Moya-Beltrán, A.; Cabezas-Mera, F.; Parra, F.; Gil, F.; Fuentes, J.A.; et al. The RNA Chaperone Hfq and Small Non-Coding RNAs Modulate the Biofilm Formation of the Fish Pathogen Yersinia ruckeri. Int. J. Mol. Sci. 2025, 26, 4733. https://doi.org/10.3390/ijms26104733
Barros MJ, Acuña LG, Hernández-Vera F, Vásquez-Arriagada P, Peñaloza D, Moya-Beltrán A, Cabezas-Mera F, Parra F, Gil F, Fuentes JA, et al. The RNA Chaperone Hfq and Small Non-Coding RNAs Modulate the Biofilm Formation of the Fish Pathogen Yersinia ruckeri. International Journal of Molecular Sciences. 2025; 26(10):4733. https://doi.org/10.3390/ijms26104733
Chicago/Turabian StyleBarros, María J., Lillian G. Acuña, Felipe Hernández-Vera, Pía Vásquez-Arriagada, Diego Peñaloza, Ana Moya-Beltrán, Fausto Cabezas-Mera, Francisco Parra, Fernando Gil, Juan A. Fuentes, and et al. 2025. "The RNA Chaperone Hfq and Small Non-Coding RNAs Modulate the Biofilm Formation of the Fish Pathogen Yersinia ruckeri" International Journal of Molecular Sciences 26, no. 10: 4733. https://doi.org/10.3390/ijms26104733
APA StyleBarros, M. J., Acuña, L. G., Hernández-Vera, F., Vásquez-Arriagada, P., Peñaloza, D., Moya-Beltrán, A., Cabezas-Mera, F., Parra, F., Gil, F., Fuentes, J. A., & Calderón, I. L. (2025). The RNA Chaperone Hfq and Small Non-Coding RNAs Modulate the Biofilm Formation of the Fish Pathogen Yersinia ruckeri. International Journal of Molecular Sciences, 26(10), 4733. https://doi.org/10.3390/ijms26104733







