Structure of K102 Capsular Polysaccharide from Acinetobacter baumannii KZ-1102 and Its Cleavage by Phage Cato Depolymerase
Abstract
1. Introduction
2. Results
2.1. Characteristics of the A. baumannii KZ-1102 Isolate
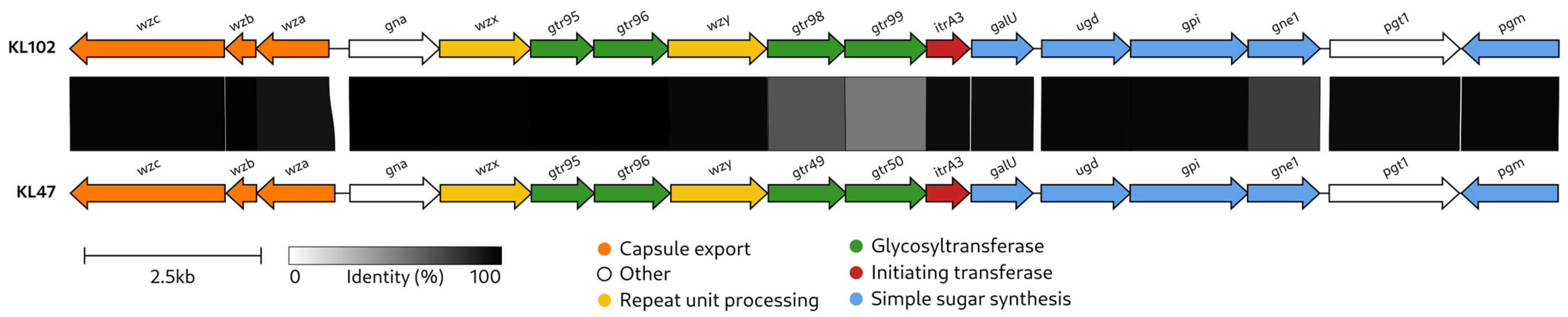
2.2. Characterization of the KL102 CPS Biosynthesis Gene Cluster
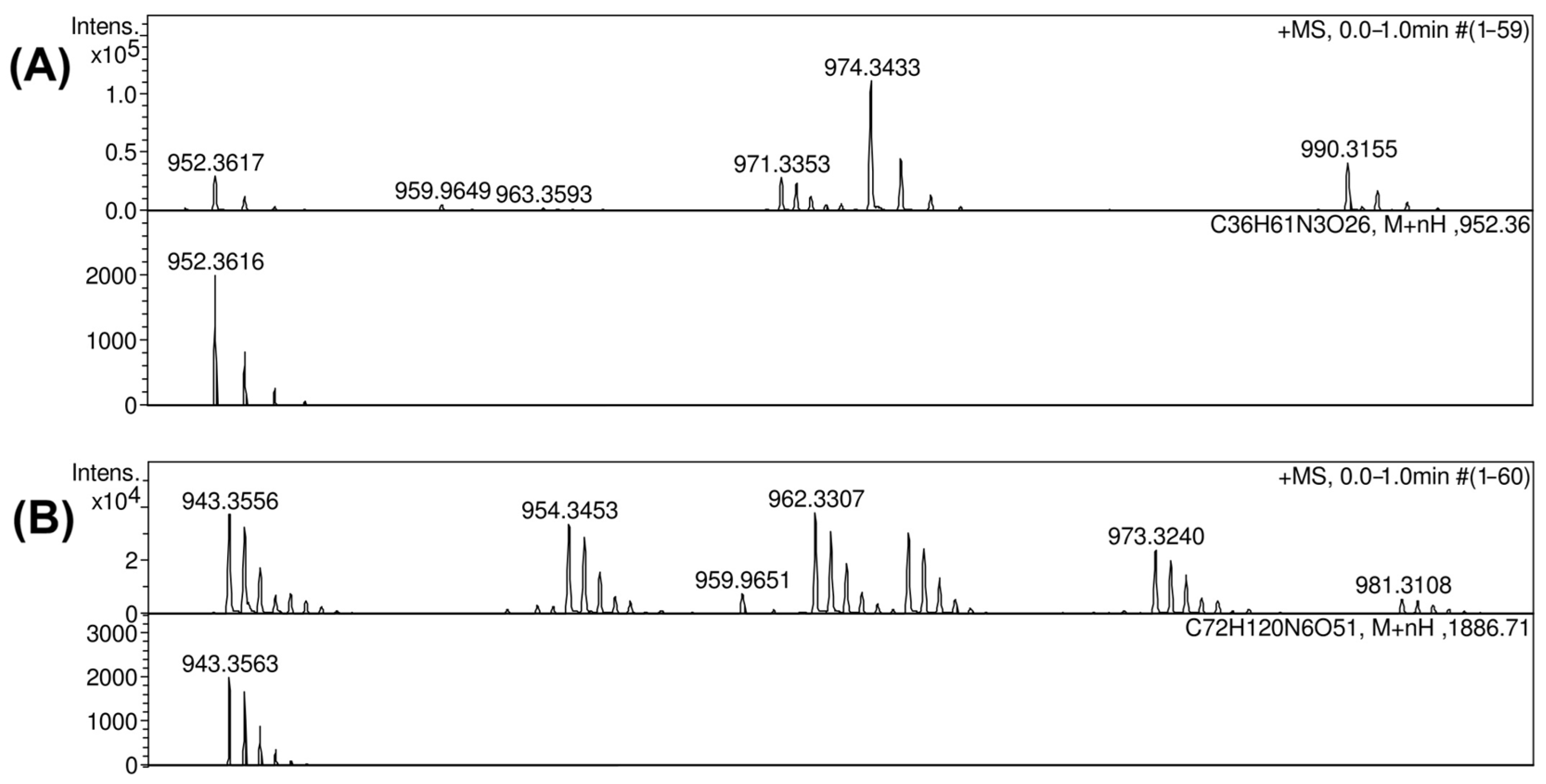
2.3. Resolution of the K102 CPS Structure
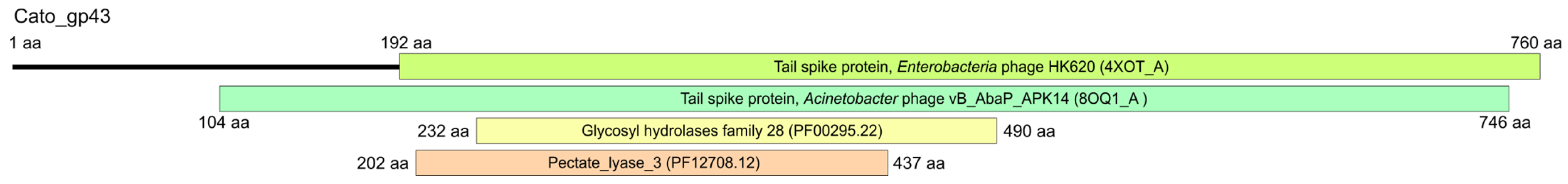
2.4. K102 CPS Cleavage by Specific Depolymerase Cato_gp43
3. Discussion
4. Materials and Methods
4.1. Bacterial Isolate and Antimicrobial Susceptibility
4.2. Sequencing and Bioinformatic Analysis
4.3. Isolation of K102 CPS
4.4. Sugar Analysis
4.5. Smith Degradation
4.6. Preparation of Recombinant Depolymerase Cato_Gp43 for CPS-Depolymerization
4.7. Depolymerization of K102 CPS by Recombinant Depolymerase
4.8. NMR Spectroscopy
4.9. Mass Spectrometry
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Ayoub Moubareck, C.; Hammoudi Halat, D. Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Sarshar, M.; Behzadi, P.; Scribano, D.; Palamara, A.T.; Ambrosi, C. Acinetobacter baumannii: An ancient commensal with weapons of a pathogen. Pathogens 2021, 10, 387. [Google Scholar] [CrossRef]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. BioMed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef]
- Denissen, J.; Reyneke, B.; Waso-Reyneke, M.; Havenga, B.; Barnard, T.; Khan, S.; Khan, W. Prevalence of ESKAPE pathogens in the environment: Antibiotic resistance status, community-acquired infection and risk to human health. Int. J. Hyg. Environ. Health 2022, 244, 114006. [Google Scholar] [CrossRef]
- Cureño-Díaz, M.A.; Plascencia-Nieto, E.S.; Loyola-Cruz, M.Á.; Cruz-Cruz, C.; Nolasco-Rojas, A.E.; Durán-Manuel, E.M.; Ibáñez-Cervantes, G.; Gómez-Zamora, E.; Tamayo-Ordóñez, M.C.; Tamayo-Ordóñez, Y.J.; et al. Gram-negative ESKAPE bacteria surveillance in COVID-19 pandemic exposes high-risk sequence types of Acinetobacter baumannii MDR in a tertiary care hospital. Pathogens 2024, 13, 50. [Google Scholar] [CrossRef]
- Russo, T.A.; Luke, N.R.; Beanan, J.M.; Olson, R.; Sauberan, S.L.; MacDonald, U.; Schultz, L.W.; Umland, T.C.; Campagnari, A.A. The K1 capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect. Immun. 2010, 78, 3993–4000. [Google Scholar] [CrossRef]
- Singh, J.K.; Adams, F.G.; Brown, M.H. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front. Microbiol. 2018, 9, 3301. [Google Scholar] [CrossRef]
- Bjånes, E.; Koh, T.; Qayum, T.; Zurich, R.; McCabe, S.; Hampel, K.; Cartwright, L.; Nizet, V. Exploring roles of the polysaccharide capsule in pathogenesis of hypervirulent Acinetobacter baumannii clinical isolate Lac-4. Antibiotics. 2023, 13, 10. [Google Scholar] [CrossRef]
- Talyansky, Y.; Nielsen, T.B.; Yan, J.; Carlino-Macdonald, U.; Di Venanzio, G.; Chakravorty, S.; Ulhaq, A.; Feldman, M.F.; Russo, T.A.; Vinogradov, E.; et al. Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLoS Pathog. 2021, 17, e1009291. [Google Scholar] [CrossRef]
- Cahill, S.M.; Hall, R.M.; Kenyon, J.J. An update to the database for Acinetobacter baumannii capsular polysaccharide locus typing extends the extensive and diverse repertoire of genes found at and outside the K locus. Microb. Genom. 2022, 8, 000878. [Google Scholar] [CrossRef] [PubMed]
- Roshini, J.; Patro, L.P.P.; Sundaresan, S.; Rathinavelan, T. Structural diversity among Acinetobacter baumannii K-antigens and its implication in the in silico serotyping. Front. Microbiol. 2023, 14, 1191542. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.J.; Speciale, I.; Hall, R.M.; De Castro, C. Structure of repeating unit of the capsular polysaccharide from Acinetobacter baumannii D78 and assignment of the K4 gene cluster. Carbohydr. Res. 2016, 434, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Evseev, P.; Gornostal, E.; Shneider, M.; Mikhaylova, Y.; Shelenkov, A.; Popova, A.; Miroshnikov, K. A novel Acinetobacter phage Cato: Lytic myovirus containing tailspike depolymerase. In Proceedings of the 2022 Ural-Siberian Conference on Computational Technologies in Cognitive Science, Genomics and Biomedicine (CSGB), Novosibirsk, Russia, 7–8 July 2022; pp. 110–114. [Google Scholar] [CrossRef]
- Evseev, P.V.; Shneider, M.M.; Kolupaeva, L.V.; Kasimova, A.A.; Timoshina, O.Y.; Perepelov, A.V.; Shpirt, A.M.; Shelenkov, A.A.; Mikhailova, Y.V.; Suzina, N.E.; et al. New Obolenskvirus phages Brutus and Scipio: Biology, evolution, and phage-host interaction. Int. J. Mol. Sci. 2024, 25, 2074. [Google Scholar] [CrossRef]
- ISO 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices—Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. ISO: Geneva, Switzerland, 2019.
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. V.15.0. 2025. Available online: http://www.eucast.org (accessed on 12 February 2025).
- Diancourt, L.; Passet, V.; Nemec, A.; Dijkshoorn, L.; Brisse, S. The population structure of Acinetobacter baumannii: Expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 2010, 5, e10034. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390. [Google Scholar] [CrossRef]
- Meumann, E.M.; Anstey, N.M.; Currie, B.J.; Piera, K.A.; Kenyon, J.J.; Hall, R.M.; Davis, J.S.; Sarovich, D.S. Genomic epidemiology of severe community-onset Acinetobacter baumannii infection. Microb. Genom. 2019, 5, e000258. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Kenyon, J.J.; Arbatsky, N.P.; Shneider, M.M.; Popova, A.V.; Miroshnikov, K.A.; Hall, R.M.; Knirel, Y.A. Related structures of neutral capsular polysaccharides of Acinetobacter baumannii isolates that carry related capsule gene clusters KL43, KL47, and KL88. Carbohydr. Res. 2016, 435, 173–179. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Chooi, Y.-H. Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, 37, 2473–2475. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides extraction with phenol-water and further applications of the proce-dure. Methods Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Lipkind, G.M.; Shashkov, A.S.; Knirel, Y.A.; Vinogradov, E.V.; Kochetkov, N.K. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr. Res. 1988, 175, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, L.; Stephens, A.; Nam, S.-Z.; Rau, D.; Kübler, J.; Lozajic, M.; Gabler, F.; Söding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Shchurova, A.S.; Shneider, M.M.; Arbatsky, N.P.; Shashkov, A.S.; Chizhov, A.O.; Skryabin, Y.P.; Mikhaylova, Y.V.; Sokolova, O.S.; Shelenkov, A.A.; Miroshnikov, K.A.; et al. Novel Acinetobacter baumannii myovirus TaPaz encoding two tailspike depolymerases: Characterization and host-recognition strategy. Viruses 2021, 13, 978. [Google Scholar] [CrossRef]
- Knirel, Y.A.; Shneider, M.M.; Popova, A.V.; Kasimova, A.A.; Senchenkova, S.N.; Shashkov, A.S.; Chizhov, A.O. Mechanisms of Acinetobacter baumannii capsular polysaccharide cleavage by phage depolymerases. Biochemistry 2020, 85, 567–574. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020, 6, e000339. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Taylor, N.M.I.; Prokhorov, N.S.; Guerrero-Ferreira, R.C.; Shneider, M.M.; Browning, C.; Goldie, K.N.; Stahlberg, H.; Leiman, P.G. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 2016, 533, 346–352. [Google Scholar] [CrossRef]
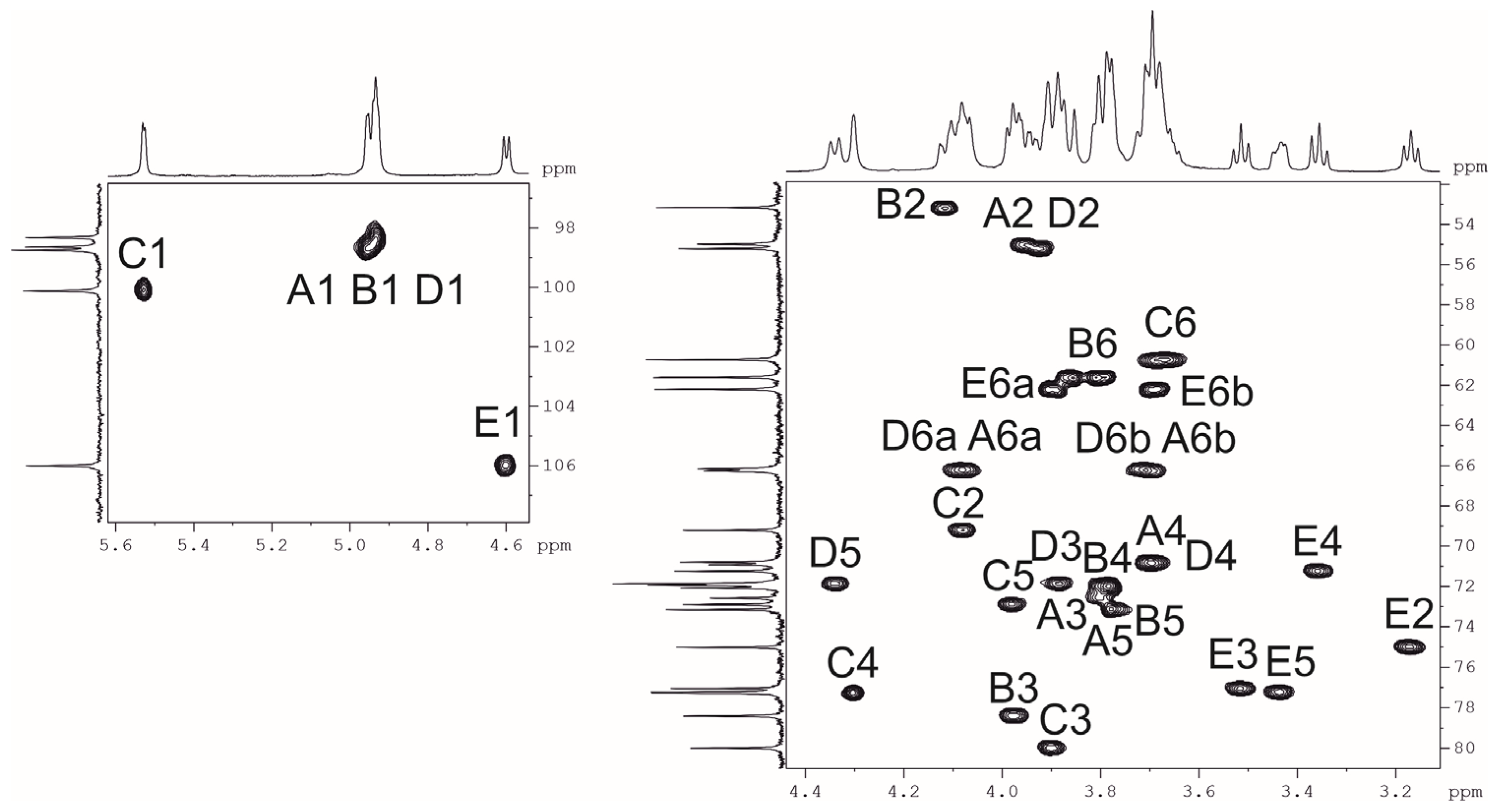


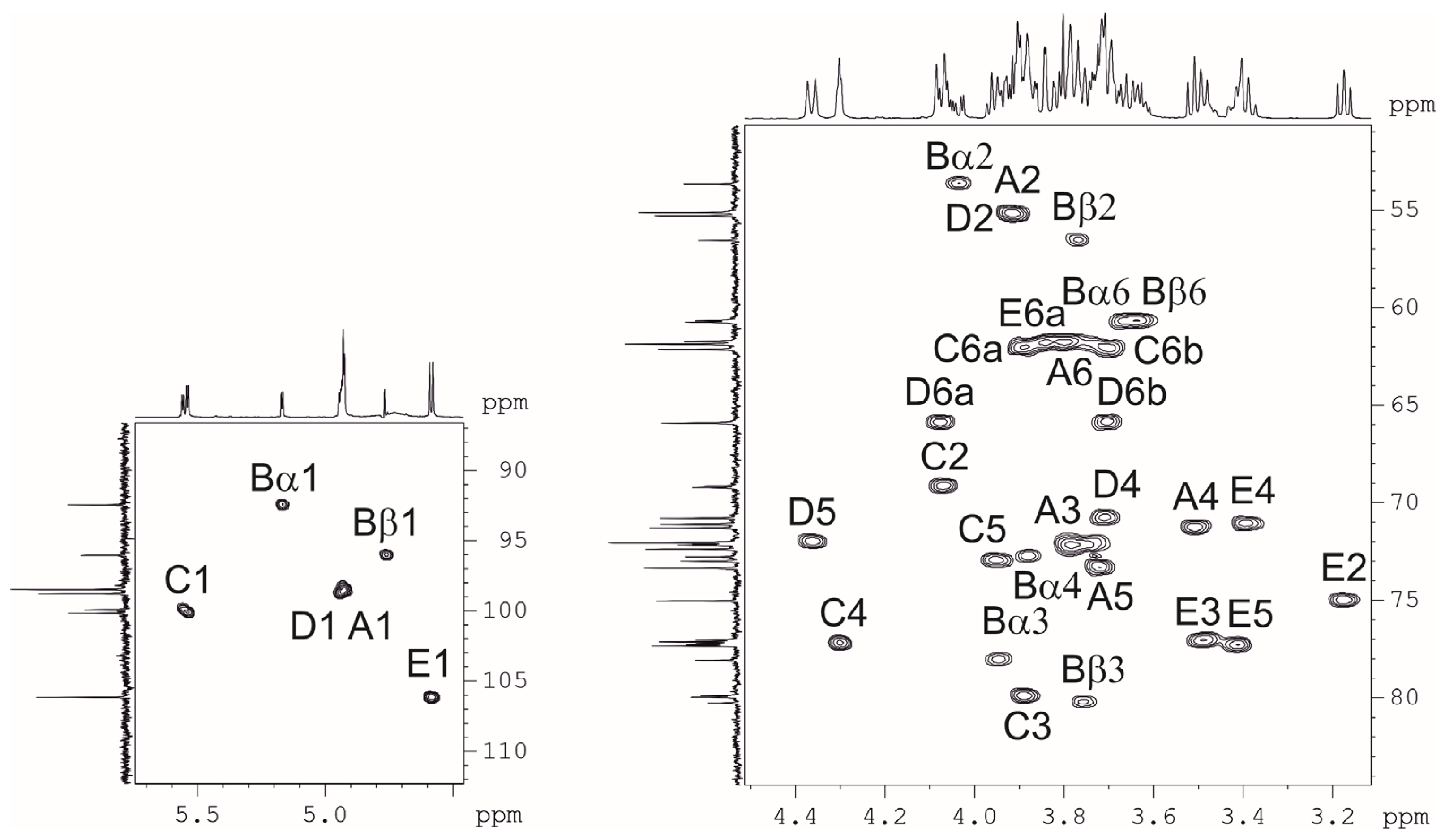

| Sugar Residue | C-1 H-1 | C-2 H-2 | C-3 H-3 | C-4 H-4 | C-5 H-5 | C-6 H-6 (6a,6b) |
|---|---|---|---|---|---|---|
| CPS A. baumannii K102 | ||||||
| →6)-α-d-GlcpNAc-(1→ | 98.3 | 55.0 | 72.6 | 70.9 | 71.9 | 66.2 |
| A | 4.94 | 3.95 | 3.81 | 3.69 | 3.80 | 3.71, 4.08 |
| →3)-α-d-GlcpNAc-(1→ | 98.3 | 53.2 | 78.4 | 72.1 | 73.2 | 61.6 |
| B | 4.93 | 4.12 | 3.97 | 3.78 | 3.77 | 3.81, 3.86 |
| →3,4)-α-d-Galp-(1→ | 100.1 | 69.2 | 80.0 | 77.3 | 72.9 | 60.7 |
| C | 5.53 | 4.08 | 3.90 | 4.31 | 3.98 | 3.67, 3.69 |
| →6)-α-d-GlcpNAc-(1→ | 98.7 | 55.2 | 71.9 | 70.9 | 71.9 | 66.4 |
| D | 4.96 | 3.92 | 3.88 | 3.69 | 4.34 | 3.71, 4.08 |
| β-d-Glcp-(1→ | 106.0 | 75.0 | 77.0 | 71.3 | 77.3 | 62.2 |
| E | 4.60 | 3.17 | 3.51 | 3.36 | 3.44 | 3.69, 3.89 |
| Sugar Residue | C-1 H-1 | C-2 H-2 | C-3 H-3 | C-4 H-4 | C-5 H-5 | C-6 H-6 (6a,6b) |
|---|---|---|---|---|---|---|
| OS1 After Smith Degradation | ||||||
| α-d-Galp-(1→ | 100.4 | 69.8 | 70.6 | 70.3 | 71.7 | 61.9 |
| C | 5.43 | 3.83 | 3.78 | 3.99 | 3.95 | 3.75, 3.75 |
| →3)-α- d-GlcpNAc-(1→ | 98.5 | 53.2 | 78.7 | 72.1 | 73.2 | 61.6 |
| B | 4.87 | 4.09 | 3.94 | 3.75 | 3.72 | 3.87, 3.79 |
| →1)-Gro | 69.8 | 72.1 | 63.8 | |||
| A’ | 3.58, 3.76 | 3.88 | 3.69, 3.65 | |||
| Sugar Residue | C-1 H-1 | C-2 H-2 | C-3 H-3 | C-4 H-4 | C-5 H-5 | C-6 H-6 (6a,6b) |
|---|---|---|---|---|---|---|
| OS2 | ||||||
| →6)-α-d-GlcpNAc-(1→ | 98.6 | 55.2 | 72.2 | 71.3 | 73.3 | 61.9 |
| A | 4.92 | 3.90 | 3.79 | 3.51 | 3.72 | 3.81, 3.81 |
| →3)-α-d-GlcpNAc | 92.5 | 53.7 | 78.1 | 72.8 | 73.3 | 60.7 |
| Bα | 5.17 | 4.03 | 3.95 | 3.88 | 3.72 | 3.74, 3.87 |
| →3)-β-d-GlcpNAc | 96.0 | 56.5 | 80.2 | 72.3 | 73.3 | 60.7 |
| Bβ | 4.76 | 3.77 | 3.76 | 3.78 | 3.72 | 3.74, 3.87 |
| →3,4)-α-d-Galp-(1→ | 100.1 | 69.2 | 79.9 | 77.2 | 72.9 | 61.8 |
| C | 5.55 | 4.07 | 3.88 | 4.31 | 3.95 | 3.82, 3.82 |
| →6)-α-d-GlcpNAc-(1→ | 98.8 | 55.2 | 72.3 | 70.7 | 71.9 | 65.9 |
| D | 4.94 | 3.91 | 3.77 | 3.71 | 4.36 | 3.70, 4.08 |
| β-d-Glcp-(1→ | 106.2 | 75.0 | 77.1 | 71.1 | 77.4 | 62.2 |
| E | 4.58 | 3.17 | 3.49 | 3.39 | 3.41 | 3.71, 3.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasimova, A.A.; Arbatsky, N.P.; Gornostal, E.A.; Shneider, M.M.; Sheck, E.A.; Shashkov, A.S.; Shelenkov, A.A.; Mikhailova, Y.V.; Azizov, I.S.; Edelstein, M.V.; et al. Structure of K102 Capsular Polysaccharide from Acinetobacter baumannii KZ-1102 and Its Cleavage by Phage Cato Depolymerase. Int. J. Mol. Sci. 2025, 26, 4727. https://doi.org/10.3390/ijms26104727
Kasimova AA, Arbatsky NP, Gornostal EA, Shneider MM, Sheck EA, Shashkov AS, Shelenkov AA, Mikhailova YV, Azizov IS, Edelstein MV, et al. Structure of K102 Capsular Polysaccharide from Acinetobacter baumannii KZ-1102 and Its Cleavage by Phage Cato Depolymerase. International Journal of Molecular Sciences. 2025; 26(10):4727. https://doi.org/10.3390/ijms26104727
Chicago/Turabian StyleKasimova, Anastasia A., Nikolay P. Arbatsky, Ekaterina A. Gornostal, Mikhail M. Shneider, Eugene A. Sheck, Alexander S. Shashkov, Andrey A. Shelenkov, Yulia V. Mikhailova, Ilya S. Azizov, Mikhail V. Edelstein, and et al. 2025. "Structure of K102 Capsular Polysaccharide from Acinetobacter baumannii KZ-1102 and Its Cleavage by Phage Cato Depolymerase" International Journal of Molecular Sciences 26, no. 10: 4727. https://doi.org/10.3390/ijms26104727
APA StyleKasimova, A. A., Arbatsky, N. P., Gornostal, E. A., Shneider, M. M., Sheck, E. A., Shashkov, A. S., Shelenkov, A. A., Mikhailova, Y. V., Azizov, I. S., Edelstein, M. V., Perepelov, A. V., Shpirt, A. M., Miroshnikov, K. A., Popova, A. V., & Knirel, Y. A. (2025). Structure of K102 Capsular Polysaccharide from Acinetobacter baumannii KZ-1102 and Its Cleavage by Phage Cato Depolymerase. International Journal of Molecular Sciences, 26(10), 4727. https://doi.org/10.3390/ijms26104727







