Optimizing Detection of Circulating Tumor Cells in Breast Cancer: Unveiling New Markers for Clinical Applications
Abstract
1. Background
2. Results
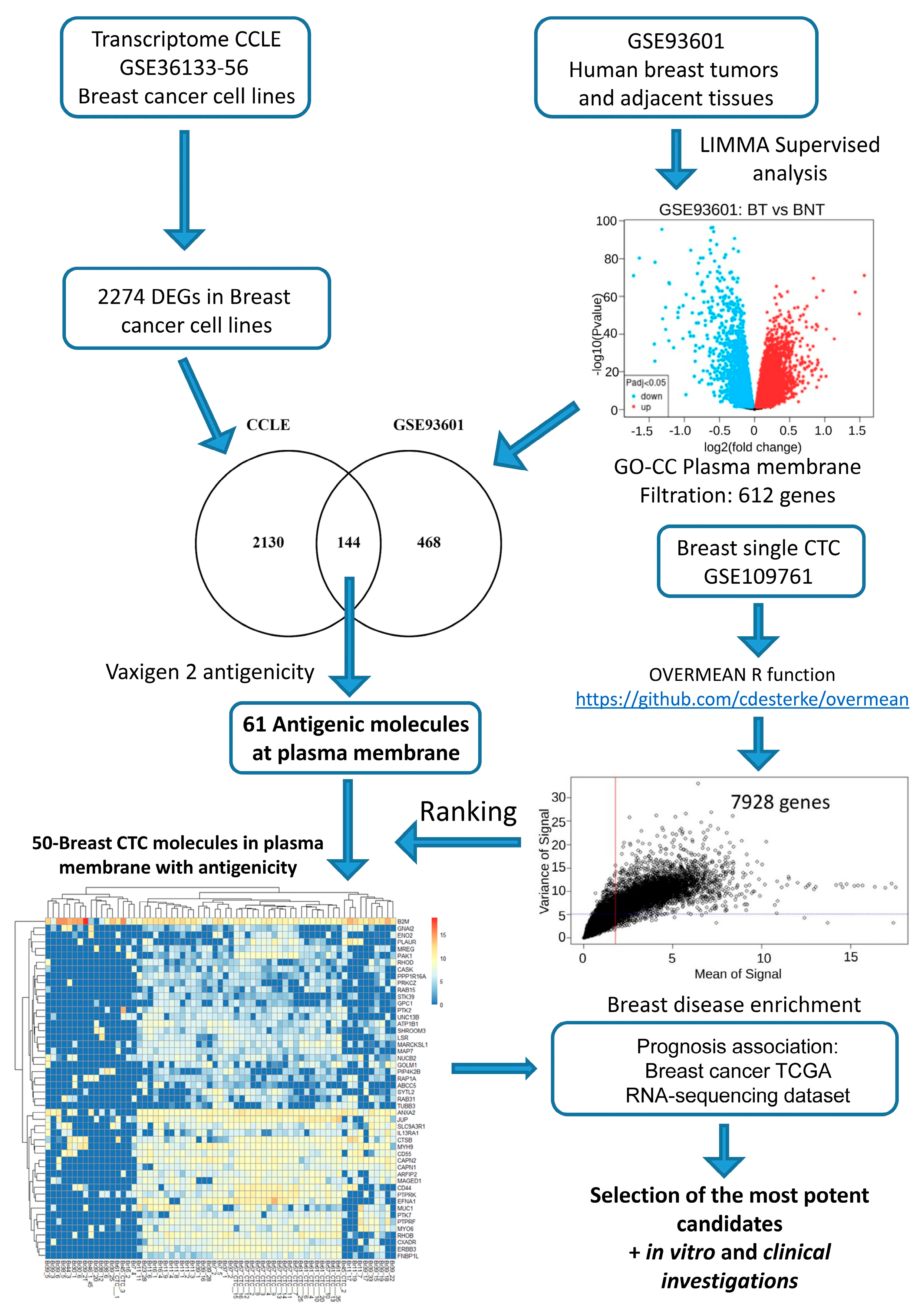
2.1. In Silico Investigations for Finding Novel Markers
2.2. In Silico Filtration to Achieve a Final Blueprint of Potential Markers of Interest
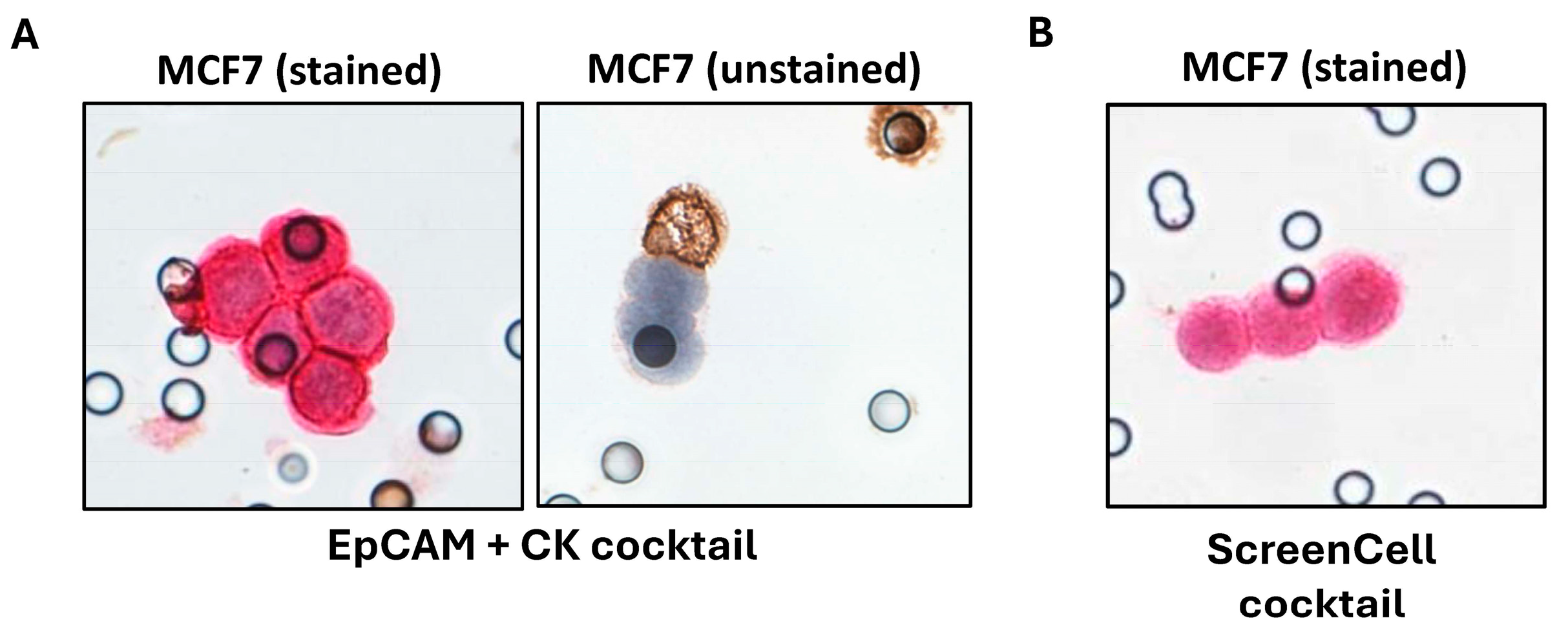
2.3. Investigations on BC Cell Lines Revealed the Targets with the Most Specific and Highest Expression
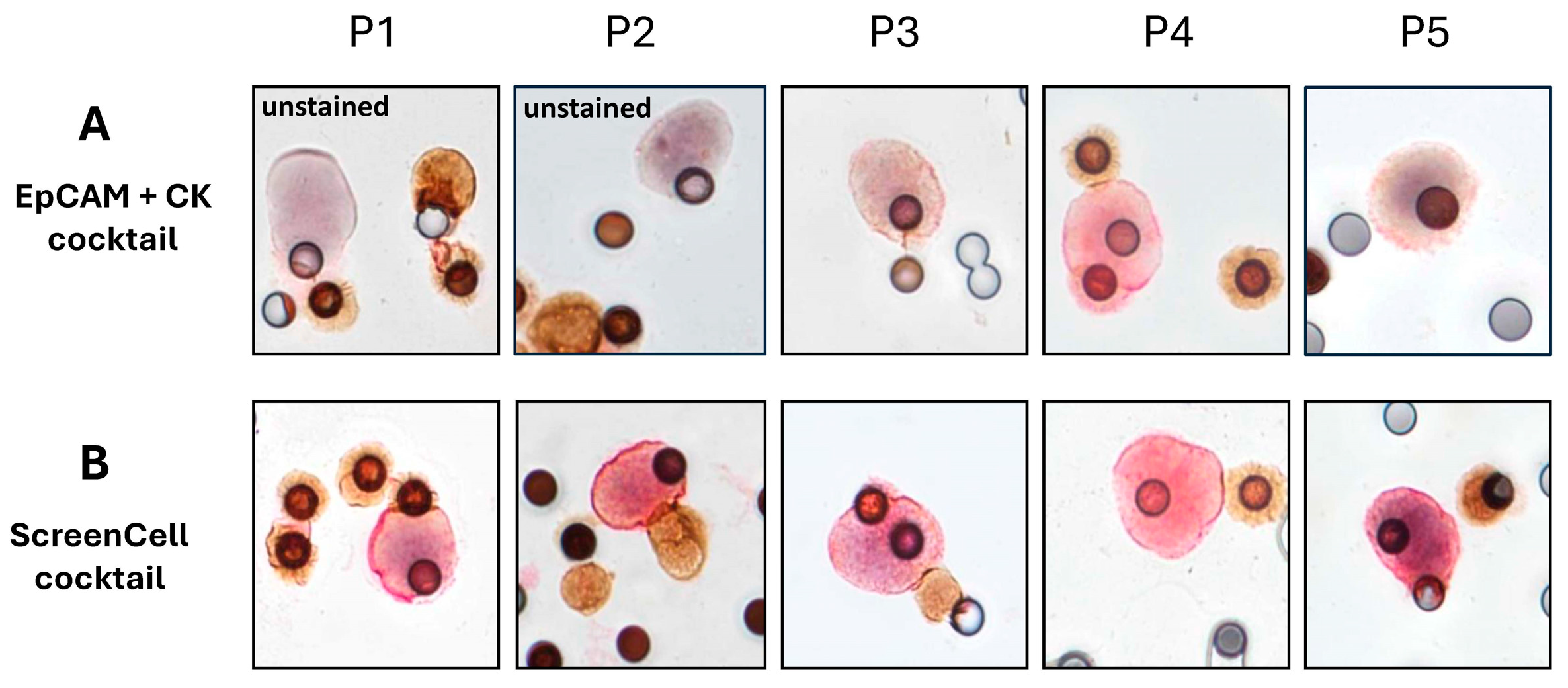
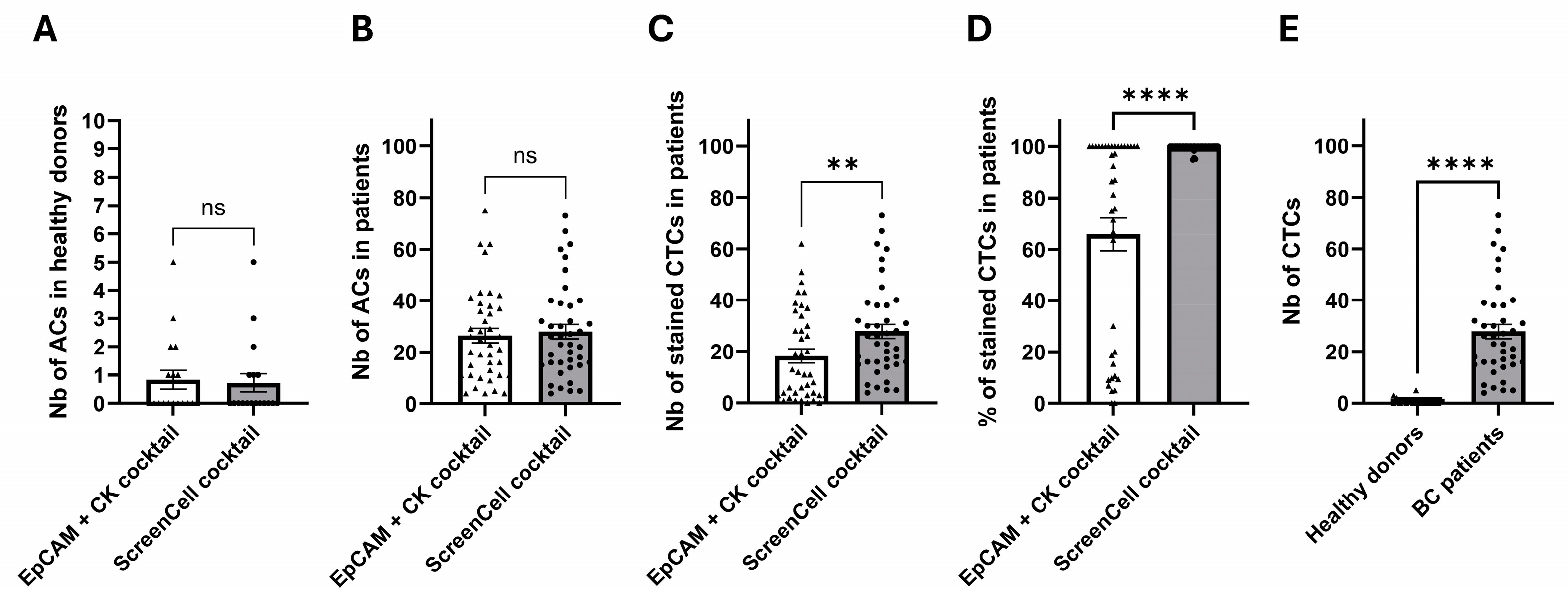
2.4. Clinical Investigations Validated the Efficacy of Novel Markers for CTC Detection
3. Discussion
4. Methods
4.1. Patients, Cohort Description, and Healthy Blood Samples
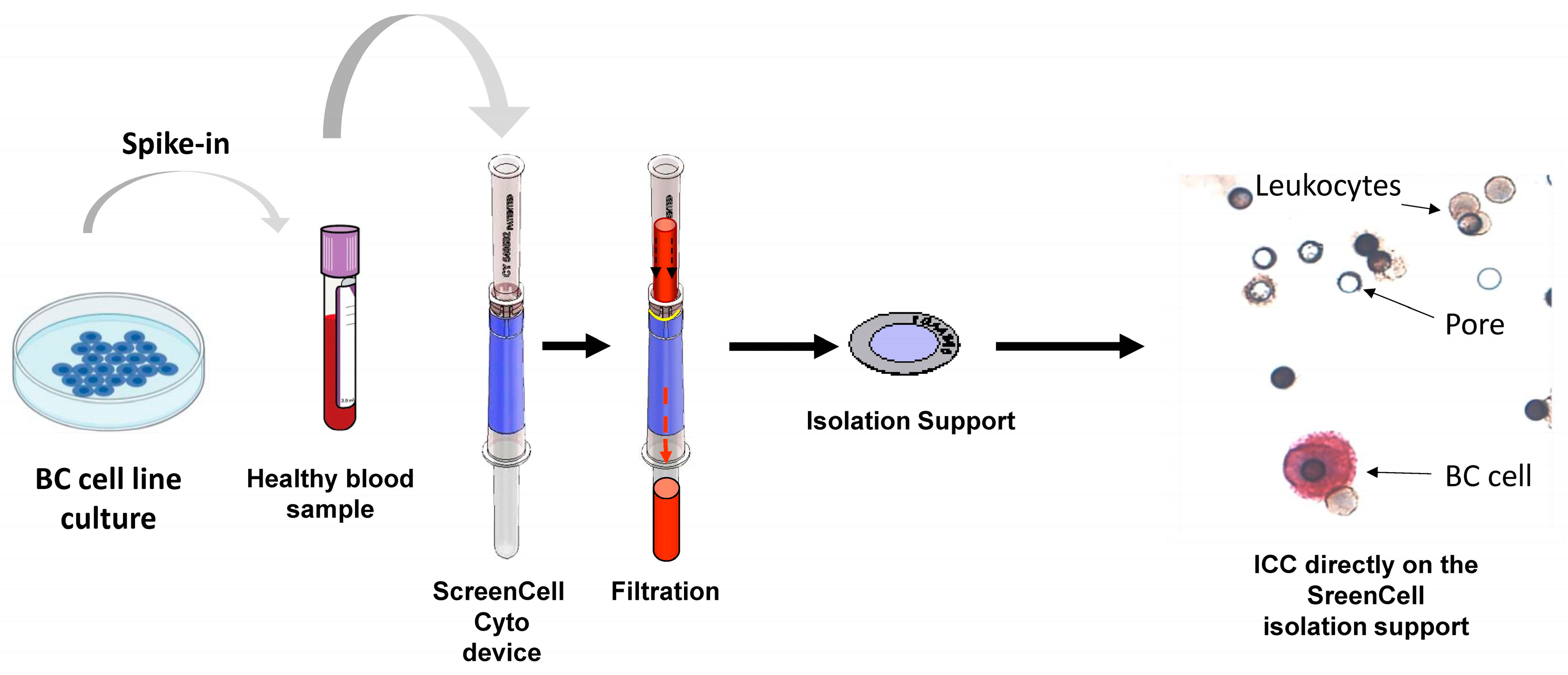
4.2. Preparation of Different Experimental Conditions Using Blood Samples
4.3. Public Omics Datasets
4.3.1. Transcriptome Dataset of Cancer Cell Line Encyclopedia (CCLE) GSE36133
4.3.2. Transcriptome Dataset of Primary Breast Tumors and Breast-Adjacent Tissues GSE93601
4.3.3. Single-Cell Transcriptome Dataset of Circulating Tumor Cells of Breast Cancer GSE109761
4.3.4. Breast Cancer TCGA RNA-Sequencing Dataset Associated with Clinical Data
4.3.5. Bioinformatics Analysis
4.3.6. Selection of Highly Variable Expressed Genes in Transcriptome by Parametric and Unsupervised Approaches
4.3.7. Protein Sequence Antigenicity Estimation
4.3.8. In Silico Sorting
4.3.9. Cell Line Preparation and Culture
4.3.10. ScreenCell Technology
4.3.11. Immunocytochemistry
4.3.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | breast cancer |
| CK | cytokeratin |
| CTC | circulating tumor cell |
| DEG | differentially expressed gene |
| dPCR | digital polymerase chain reaction |
| EMT | epithelial to mesenchymal transition |
| EpCAM | epithelial cell adhesion molecule |
| HER2 | human epidermal growth factor receptor 2 |
| HR | hormone receptor |
| ICC | immunocytochemistry |
| IDC | infiltrating ductal carcinoma |
| IF | immunofluorescence |
| IHC | immunohistochemistry |
| ILC | infiltrating lobular carcinoma |
| IS | isolation support |
| NGS | next-generation sequencing |
| OS | overall survival |
| PDL1 | programmed death-ligand 1 |
| PFS | progression-free survival |
| RBC | red blood cell |
| TNBC | triple-negative breast cancer |
| WHO | World Health Organization |
References
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Kantara, C.; O’Connell, M.R.; Luthra, G.; Gajjar, A.; Sarkar, S.; Ullrich, R.L.; Singh, P. Methods for Detecting Circulating Cancer Stem Cells (CCSCs) as a Novel Approach for Diagnosis of Colon Cancer Relapse/Metastasis. Lab. Investig. J. Tech. Methods Pathol. 2015, 95, 100–112. [Google Scholar] [CrossRef]
- Reduzzi, C.; Di Cosimo, S.; Gerratana, L.; Motta, R.; Martinetti, A.; Vingiani, A.; D’Amico, P.; Zhang, Y.; Vismara, M.; Depretto, C.; et al. Circulating Tumor Cell Clusters Are Frequently Detected in Women with Early-Stage Breast Cancer. Cancers 2021, 13, 2356. [Google Scholar] [CrossRef]
- Braun, S.; Vogl, F.D.; Naume, B.; Janni, W.; Osborne, M.P.; Coombes, R.C.; Schlimok, G.; Diel, I.J.; Gerber, B.; Gebauer, G.; et al. A Pooled Analysis of Bone Marrow Micrometastasis in Breast Cancer. N. Engl. J. Med. 2005, 353, 793–802. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in Diagnosis of Pancreatic Cancer. World J. Gastroenterol. 2018, 24, 2047–2060. [Google Scholar] [CrossRef]
- Lopresti, A.; Acquaviva, C.; Boudin, L.; Finetti, P.; Garnier, S.; Aulas, A.; Liberatoscioli, M.L.; Cabaud, O.; Guille, A.; de Nonneville, A.; et al. Identification of Atypical Circulating Tumor Cells with Prognostic Value in Metastatic Breast Cancer Patients. Cancers 2022, 14, 932. [Google Scholar] [CrossRef]
- Schuster, E.; Taftaf, R.; Reduzzi, C.; Albert, M.K.; Romero-Calvo, I.; Liu, H. Better Together: Circulating Tumor Cell Clustering in Metastatic Cancer. Trends Cancer 2021, 7, 1020–1032. [Google Scholar] [CrossRef]
- Rejniak, K.A. Circulating Tumor Cells: When a Solid Tumor Meets a Fluid Microenvironment. Adv. Exp. Med. Biol. 2016, 936, 93–106. [Google Scholar] [CrossRef]
- Wang, W.-C.; Zhang, X.-F.; Peng, J.; Li, X.-F.; Wang, A.-L.; Bie, Y.-Q.; Shi, L.-H.; Lin, M.-B.; Zhang, X.-F. Survival Mechanisms and Influence Factors of Circulating Tumor Cells. BioMed Res. Int. 2018, 2018, 6304701. [Google Scholar] [CrossRef]
- Hayes, B.; Brady, L.; Sheill, G.; Baird, A.-M.; Guinan, E.; Stanfill, B.; Dunne, J.; Holden, D.; Vlajnic, T.; Casey, O.; et al. Circulating Tumour Cell Numbers Correlate with Platelet Count and Circulating Lymphocyte Subsets in Men with Advanced Prostate Cancer: Data from the ExPeCT Clinical Trial (CTRIAL-IE 15-21). Cancers 2021, 13, 4690. [Google Scholar] [CrossRef]
- Mazel, M.; Jacot, W.; Pantel, K.; Bartkowiak, K.; Topart, D.; Cayrefourcq, L.; Rossille, D.; Maudelonde, T.; Fest, T.; Alix-Panabières, C. Frequent Expression of PD-L1 on Circulating Breast Cancer Cells. Mol. Oncol. 2015, 9, 1773–1782. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Q.; Liu, Q.; Wang, C.; Yao, R.; Wang, Y. CTC Immune Escape Mediated by PD-L1. Med. Hypotheses 2016, 93, 138–139. [Google Scholar] [CrossRef]
- König, L.; Kasimir-Bauer, S.; Hoffmann, O.; Bittner, A.-K.; Wagner, B.; Manvailer, L.F.S.; Schramm, S.; Bankfalvi, A.; Giebel, B.; Kimmig, R.; et al. The Prognostic Impact of Soluble and Vesicular HLA-G and Its Relationship to Circulating Tumor Cells in Neoadjuvant Treated Breast Cancer Patients. Hum. Immunol. 2016, 77, 791–799. [Google Scholar] [CrossRef]
- Rossi, E.; Zamarchi, R. Single-Cell Analysis of Circulating Tumor Cells: How Far Have We Come in the -Omics Era? Front. Genet. 2019, 10, 958. [Google Scholar] [CrossRef]
- Thiele, J.-A.; Pitule, P.; Hicks, J.; Kuhn, P. Single-Cell Analysis of Circulating Tumor Cells. Adv. Struct. Saf. Stud. 2019, 1908, 243–264. [Google Scholar] [CrossRef]
- Janni, W.J.; Rack, B.; Terstappen, L.W.M.M.; Pierga, J.-Y.; Taran, F.-A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.-C.; Friedl, T.W.P.; et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef]
- van Dalum, G.; van der Stam, G.J.; Tibbe, A.G.J.; Franken, B.; Mastboom, W.J.B.; Vermes, I.; de Groot, M.R.; Terstappen, L.W.M.M. Circulating Tumor Cells before and during Follow-up after Breast Cancer Surgery. Int. J. Oncol. 2015, 46, 407–413. [Google Scholar] [CrossRef]
- Goodman, C.R.; Seagle, B.-L.L.; Friedl, T.W.P.; Rack, B.; Lato, K.; Fink, V.; Cristofanilli, M.; Donnelly, E.D.; Janni, W.; Shahabi, S.; et al. Association of Circulating Tumor Cell Status With Benefit of Radiotherapy and Survival in Early-Stage Breast Cancer. JAMA Oncol. 2018, 4, e180163. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical Validity of Circulating Tumour Cells in Patients with Metastatic Breast Cancer: A Pooled Analysis of Individual Patient Data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Hendrix, L.H.; Hyslop, T.; Barry, W.T.; Winer, E.P.; Hudis, C.; Toppmeyer, D.; Carey, L.A.; Partridge, A.H.; Pierga, J.-Y.; et al. Serial Analysis of Circulating Tumor Cells in Metastatic Breast Cancer Receiving First-Line Chemotherapy. J. Natl. Cancer Inst. 2021, 113, 443–452. [Google Scholar] [CrossRef]
- Carneiro, A.; Piairo, P.; Matos, B.; Santos, D.A.R.; Palmeira, C.; Santos, L.L.; Lima, L.; Diéguez, L. Minimizing False Positives for CTC Identification. Anal. Chim. Acta 2024, 1288, 342165. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in Cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Brown, T.C.; Sankpal, N.V.; Gillanders, W.E. Functional Implications of the Dynamic Regulation of EpCAM during Epithelial-to-Mesenchymal Transition. Biomolecules 2021, 11, 956. [Google Scholar] [CrossRef]
- Kuburich, N.A.; Den Hollander, P.; Pietz, J.T.; Mani, S.A. Vimentin and Cytokeratin: Good Alone, Bad Together. Semin. Cancer Biol. 2022, 86, 816–826. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Koo, G.-B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.-I.; Jung, H.-I.; Kim, Y.-S. Epithelial-to-Mesenchymal Transition Leads to Loss of EpCAM and Different Physical Properties in Circulating Tumor Cells from Metastatic Breast Cancer. Oncotarget 2016, 7, 24677–24687. [Google Scholar] [CrossRef]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Alix-Panabières, C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells 2020, 9, 1836. [Google Scholar] [CrossRef]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A New Device for Rapid Isolation by Size and Characterization of Rare Circulating Tumor Cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar]
- Ye, F.; Wechsler, J.; Bouzidi, A.; Uzan, G.; Naserian, S. Fast and Efficient Isolation of Murine Circulating Tumor Cells Using Screencell Technology for Pre-Clinical Analyzes. Sci. Rep. 2024, 14, 15019. [Google Scholar] [CrossRef]
- Drucker, A.; Teh, E.M.; Kostyleva, R.; Rayson, D.; Douglas, S.; Pinto, D.M. Comparative Performance of Different Methods for Circulating Tumor Cell Enrichment in Metastatic Breast Cancer Patients. PLoS ONE 2020, 15, e0237308. [Google Scholar] [CrossRef]
- Mu, Z.; Benali-Furet, N.; Uzan, G.; Znaty, A.; Ye, Z.; Paolillo, C.; Wang, C.; Austin, L.; Rossi, G.; Fortina, P.; et al. Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer. Int. J. Mol. Sci. 2016, 17, 1665. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Kruspe, S.; Dickey, D.D.; Urak, K.T.; Blanco, G.N.; Miller, M.J.; Clark, K.C.; Burghardt, E.; Gutierrez, W.R.; Phadke, S.D.; Kamboj, S.; et al. Rapid and Sensitive Detection of Breast Cancer Cells in Patient Blood with Nuclease-Activated Probe Technology. Mol. Ther. Nucleic Acids 2017, 8, 542–557. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; To, K.K.W.; Cui, C.; Luo, M.; Wu, S.; Huang, L.; Fu, K.; Pan, C.; Liu, Z.; et al. Circulating Tumor Cells Shielded with Extracellular Vesicle-Derived CD45 Evade T Cell Attack to Enable Metastasis. Signal Transduct. Target. Ther. 2024, 9, 84. [Google Scholar] [CrossRef]
- Doghri, R.; Manai, M.; Finetti, P.; Driss, M.; Agavnian, E.; Lopez, M.; Elghardallou, M.; Charafe-Jauffret, E.; Manai, M.; Chaffanet, M.; et al. Stromal Expression of MARCKS Protein in Ovarian Carcinomas Has Unfavorable Prognostic Value. Int. J. Mol. Sci. 2017, 19, 41. [Google Scholar] [CrossRef]
- Manai, M.; Thomassin-Piana, J.; Gamoudi, A.; Finetti, P.; Lopez, M.; Eghozzi, R.; Ayadi, S.; Lamine, O.B.; Manai, M.; Rahal, K.; et al. MARCKS Protein Overexpression in Inflammatory Breast Cancer. Oncotarget 2017, 8, 6246–6257. [Google Scholar] [CrossRef]
- Manai, M.; ELBini-Dhouib, I.; Finetti, P.; Bichiou, H.; Reduzzi, C.; Aissaoui, D.; Ben-Hamida, N.; Agavnian, E.; Srairi-Abid, N.; Lopez, M.; et al. MARCKS as a Potential Therapeutic Target in Inflammatory Breast Cancer. Cells 2022, 11, 2926. [Google Scholar] [CrossRef]
- Liang, W.; Gao, R.; Yang, M.; Wang, X.; Cheng, K.; Shi, X.; He, C.; Li, Y.; Wu, Y.; Shi, L.; et al. MARCKSL1 Promotes the Proliferation, Migration and Invasion of Lung Adenocarcinoma Cells. Oncol. Lett. 2020, 19, 2272–2280. [Google Scholar] [CrossRef]
- Dorris, E.; O’Neill, A.; Hanrahan, K.; Treacy, A.; Watson, R.W. MARCKS Promotes Invasion and Is Associated with Biochemical Recurrence in Prostate Cancer. Oncotarget 2017, 8, 72021–72030. [Google Scholar] [CrossRef]
- Bretscher, A.; Chambers, D.; Nguyen, R.; Reczek, D. ERM-Merlin and EBP50 Protein Families in Plasma Membrane Organization and Function. Annu. Rev. Cell Dev. Biol. 2000, 16, 113–143. [Google Scholar] [CrossRef]
- Weinman, E.J.; Steplock, D.; Wang, Y.; Shenolikar, S. Characterization of a Protein Cofactor That Mediates Protein Kinase A Regulation of the Renal Brush Border Membrane Na(+)-H+ Exchanger. J. Clin. Investig. 1995, 95, 2143–2149. [Google Scholar] [CrossRef]
- Ma, Q.; Jiao, Y.; Hao, Y.; Yan, S.; Lyu, N.; Gao, H.; Li, D.; Liu, Q.; Zheng, J.; Song, N. Targeting of NHERF1 through RNA Interference Inhibits the Proliferation and Migration of Metastatic Prostate Cancer Cells. Oncol. Lett. 2016, 11, 1149–1154. [Google Scholar] [CrossRef]
- Fraenzer, J.-T.; Pan, H.; Minimo, L.; Smith, G.M.; Knauer, D.; Hung, G. Overexpression of the NF2 Gene Inhibits Schwannoma Cell Proliferation through Promoting PDGFR Degradation. Int. J. Oncol. 2003, 23, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Feng, D.; Bian, W.; Yang, L.; Li, Y.; Yang, Z.; Xiong, Y.; Zheng, J.; Zhai, R.; He, J. EBP50 Inhibits EGF-Induced Breast Cancer Cell Proliferation by Blocking EGFR Phosphorylation. Amino Acids 2012, 43, 2027–2035. [Google Scholar] [CrossRef]
- Cardone, R.A.; Greco, M.R.; Capulli, M.; Weinman, E.J.; Busco, G.; Bellizzi, A.; Casavola, V.; Antelmi, E.; Ambruosi, B.; Dell’Aquila, M.E.; et al. NHERF1 Acts as a Molecular Switch to Program Metastatic Behavior and Organotropism via Its PDZ Domains. Mol. Biol. Cell 2012, 23, 2028–2040. [Google Scholar] [CrossRef]
- Jang, Y.; Cheong, W.; Park, G.; Kim, Y.; Ha, J.; Ahn, S. Tumor Microenvironment and Genes Affecting the Prognosis of Temozolomide-Treated Glioblastoma. J. Pers. Med. 2023, 13, 188. [Google Scholar] [CrossRef]
- Kazmi, N.; Robinson, T.; Zheng, J.; Kar, S.; Martin, R.M.; Ridley, A.J. Rho GTPase Gene Expression and Breast Cancer Risk: A Mendelian Randomization Analysis. Sci. Rep. 2022, 12, 1463. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Beck, A.H.; Collins, L.C.; Chen, W.Y.; Tamimi, R.M.; Hazra, A.; Brown, M.; Rosner, B.; Hankinson, S.E. Alcohol Consumption and Risk of Breast Cancer by Tumor Receptor Expression. Horm. Cancer 2015, 6, 237–246. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils Escort Circulating Tumour Cells to Enable Cell Cycle Progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, X.; Li, T.; Luo, J.; Dong, D. ctcRbase: The Gene Expression Database of Circulating Tumor Cells and Microemboli. Database J. Biol. Databases Curation 2020, 2020, baaa020. [Google Scholar] [CrossRef]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for Gene List Enrichment Analysis and Candidate Gene Prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2019, 48, D845–D855. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A Server for Prediction of Protective Antigens, Tumour Antigens and Subunit Vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]

| Protein | Breast Cancer Cell Line | Expression Profile | Expression by Leukocytes |
|---|---|---|---|
| CD55 | SKBR3 | Low | No |
| LSR | MCF7 | Medium | No |
| MARCKSL1 | MCF7 | Medium/High | No |
| GPC1 | SKBR3 | Low | No |
| SLC9A3R1 | MCF7 | High | No |
| CXADR | SKBR3 | Low | No |
| SHROOM3 | SKBR3 | Low | No |
| MUC1 | MCF7 | Low/Medium | No |
| PPP1R16A | MCF7 | Low | No |
| ATP1B1 | SKBR3 | Medium | No |
| RHOD | MCF7 | High | No |
| SYTL2 | MCF7 | Medium/High | Yes |
| CK | EPCAM | CK + EPCAM COCKTAIL | SLC9A3R1 | MARCKSL1 | RHOD | SCREENCELL COCKTAIL | |
|---|---|---|---|---|---|---|---|
| NEGATIVE | 34.40% | 69.70% | 30.80% | 11.50% | 40% | 5.12% | 0% |
| LOW | 16.40% | 22.70% | 25% | 37.70% | 13.30% | 41% | 1.90% |
| HIGH | 49.20% | 7.60% | 44.20% | 50.10% | 46.60% | 53.80% | 98.10% |
| POSITIVE | 65.60% | 30.30% | 69.20% | 87.80% | 59.90% | 94.80% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mehtar, A.; Wechsler, J.; Desterke, C.; Giron-Michel, J.; Bouzidi, A.; Burlion, A.; Louache, F.; Kahia-Tani, S.; Uzan, G.; Naserian, S. Optimizing Detection of Circulating Tumor Cells in Breast Cancer: Unveiling New Markers for Clinical Applications. Int. J. Mol. Sci. 2025, 26, 4714. https://doi.org/10.3390/ijms26104714
Mehtar A, Wechsler J, Desterke C, Giron-Michel J, Bouzidi A, Burlion A, Louache F, Kahia-Tani S, Uzan G, Naserian S. Optimizing Detection of Circulating Tumor Cells in Breast Cancer: Unveiling New Markers for Clinical Applications. International Journal of Molecular Sciences. 2025; 26(10):4714. https://doi.org/10.3390/ijms26104714
Chicago/Turabian StyleMehtar, Amira, Janine Wechsler, Christophe Desterke, Julien Giron-Michel, Amira Bouzidi, Aude Burlion, Fawzia Louache, Samira Kahia-Tani, Georges Uzan, and Sina Naserian. 2025. "Optimizing Detection of Circulating Tumor Cells in Breast Cancer: Unveiling New Markers for Clinical Applications" International Journal of Molecular Sciences 26, no. 10: 4714. https://doi.org/10.3390/ijms26104714
APA StyleMehtar, A., Wechsler, J., Desterke, C., Giron-Michel, J., Bouzidi, A., Burlion, A., Louache, F., Kahia-Tani, S., Uzan, G., & Naserian, S. (2025). Optimizing Detection of Circulating Tumor Cells in Breast Cancer: Unveiling New Markers for Clinical Applications. International Journal of Molecular Sciences, 26(10), 4714. https://doi.org/10.3390/ijms26104714






