Genome-Wide Identification of 109 NAC Genes and Dynamic Expression Profiles Under Cold Stress in Madhuca longifolia
Abstract
1. Introduction
2. Results
2.1. Identification of NAC Transcription Factors and Analysis of Protein Physicochemical Properties in M. longifolia
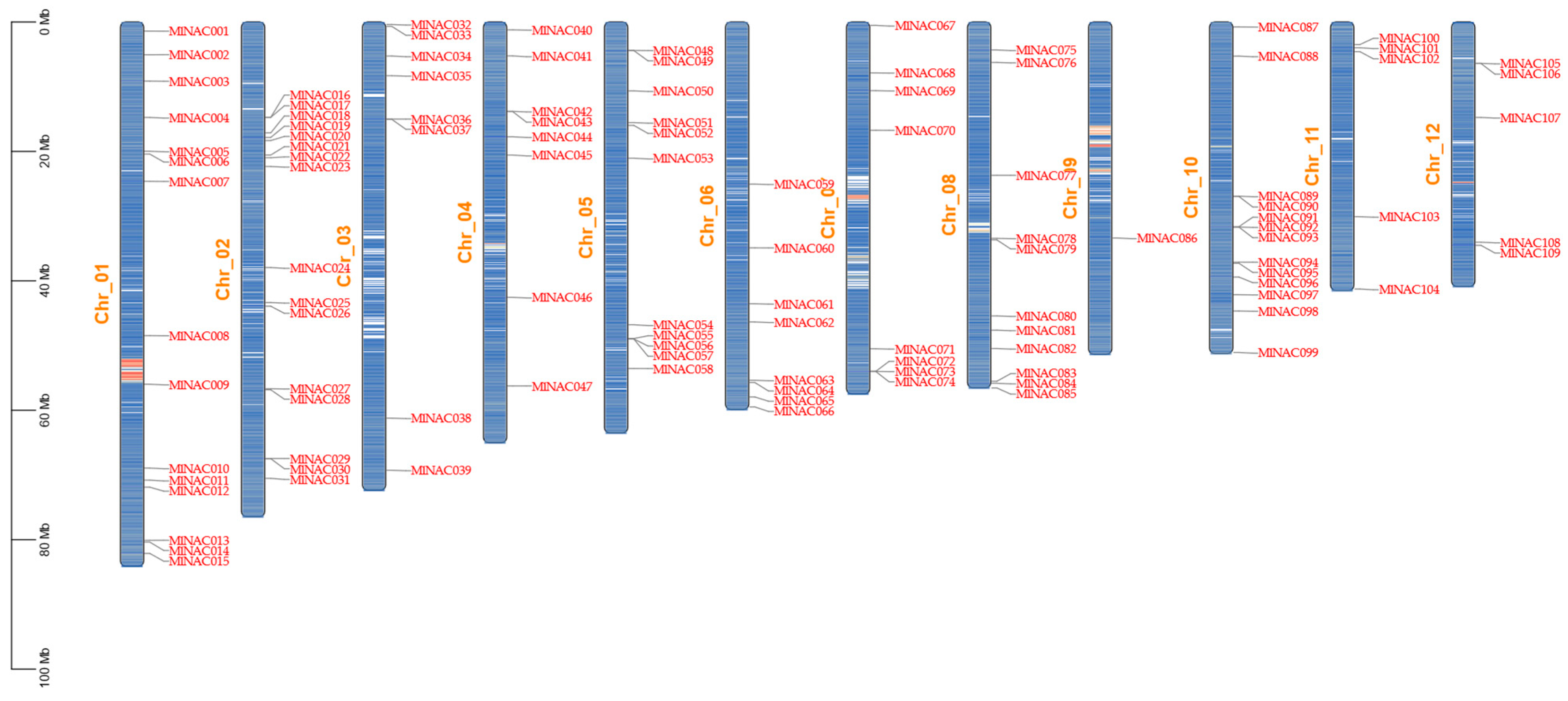
2.2. Chromosomal Localization of MlNAC Gene Family Members
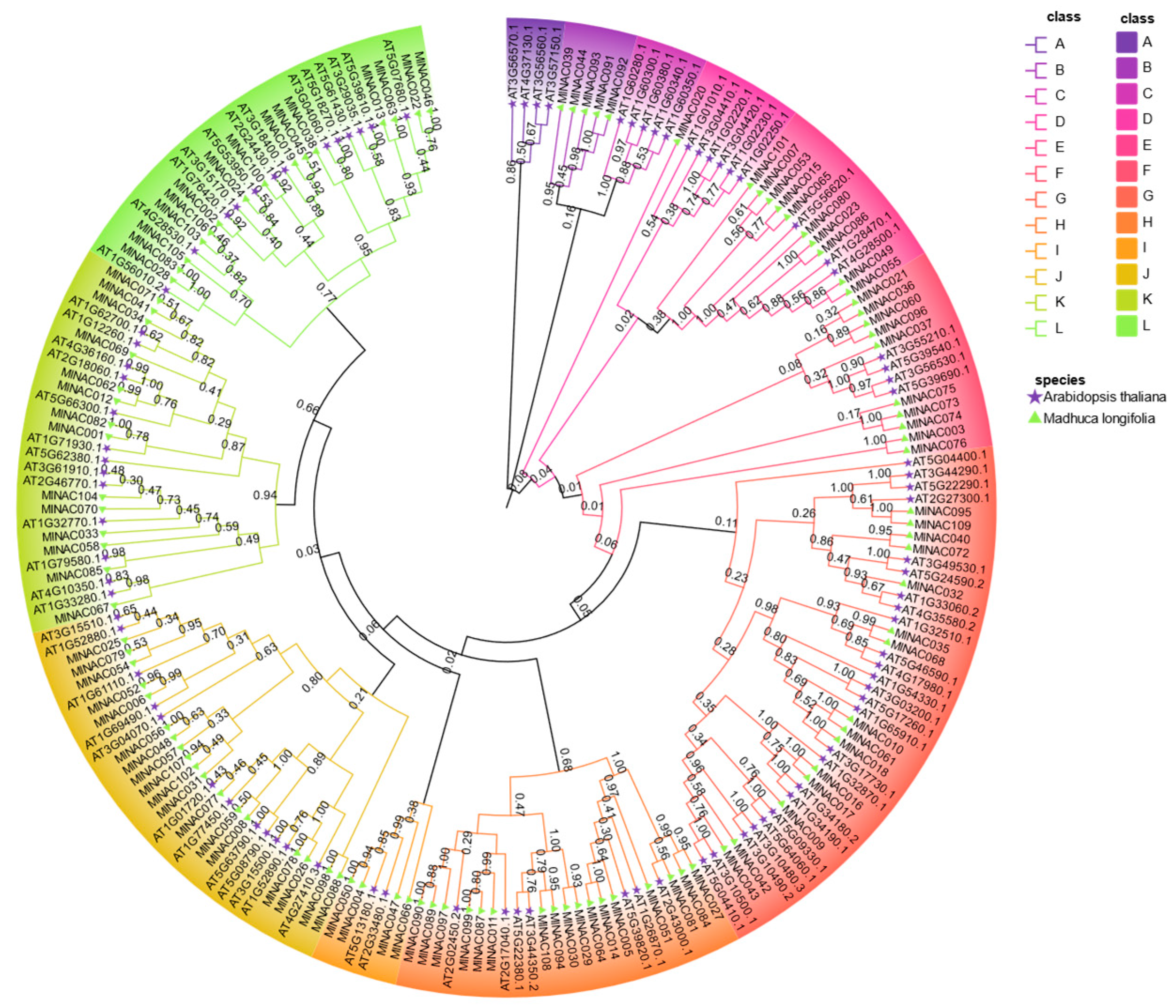
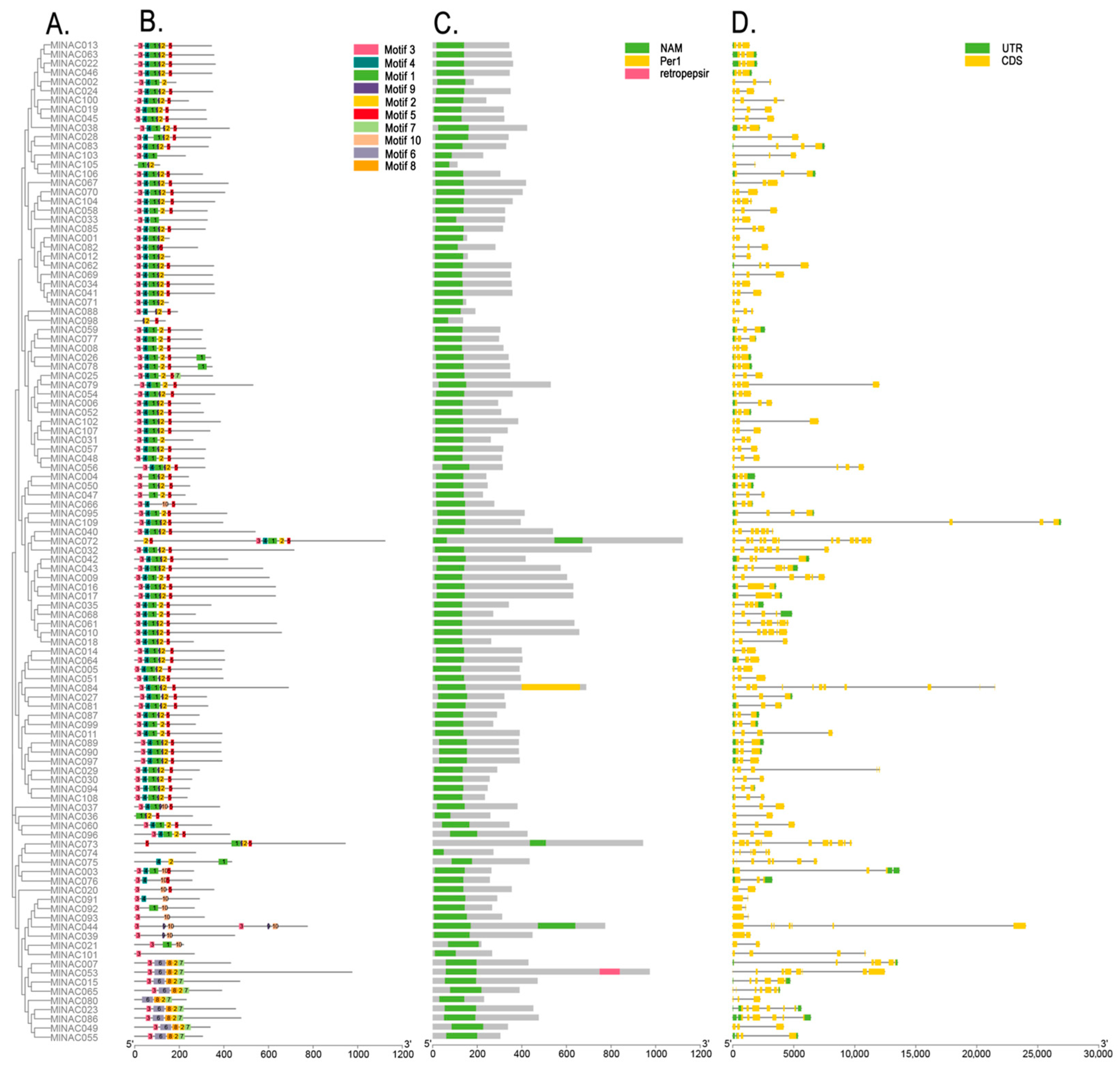
2.3. Phylogenetic Studies of the NAC Transcription Factor Family in M. longifolia
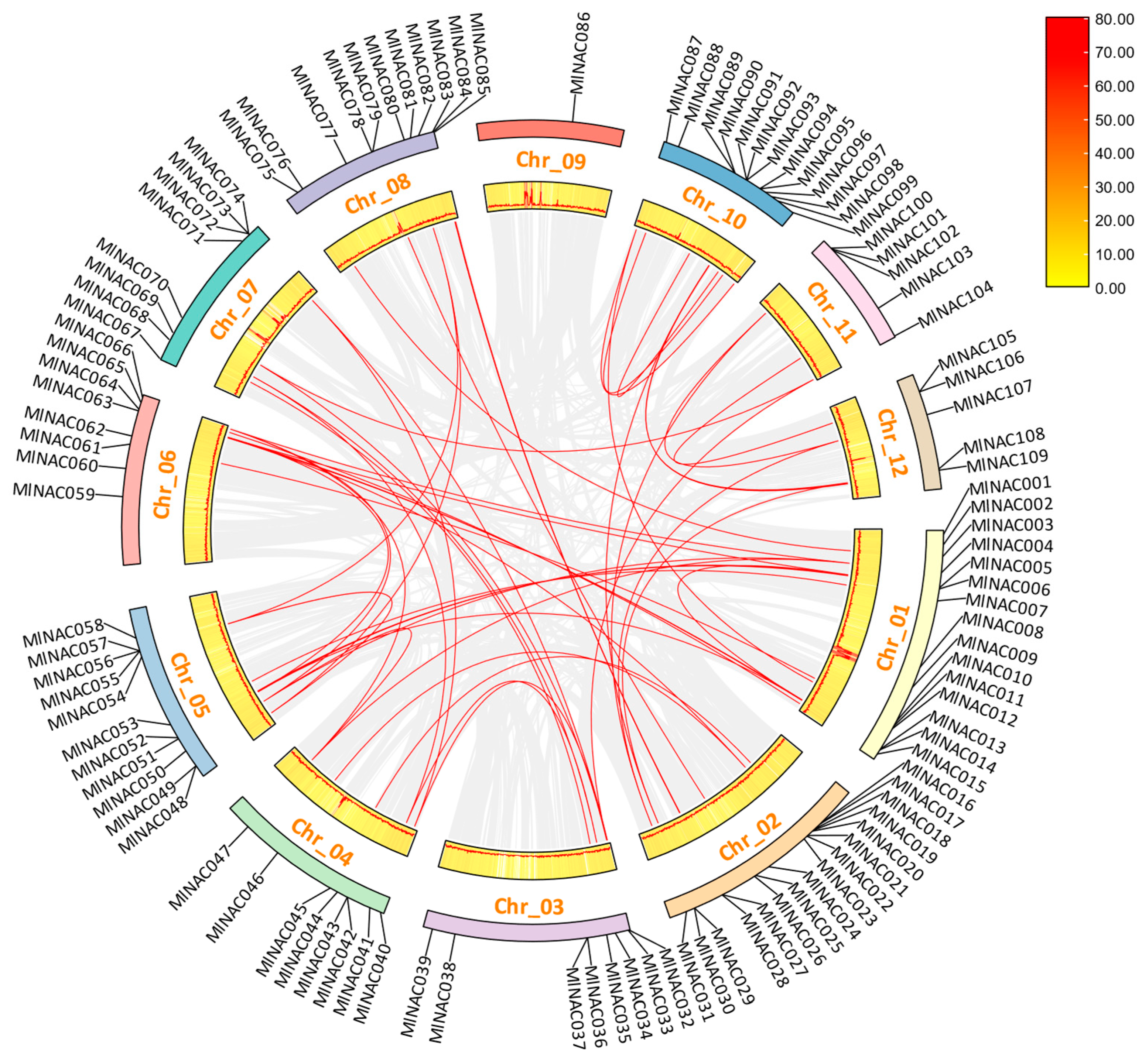
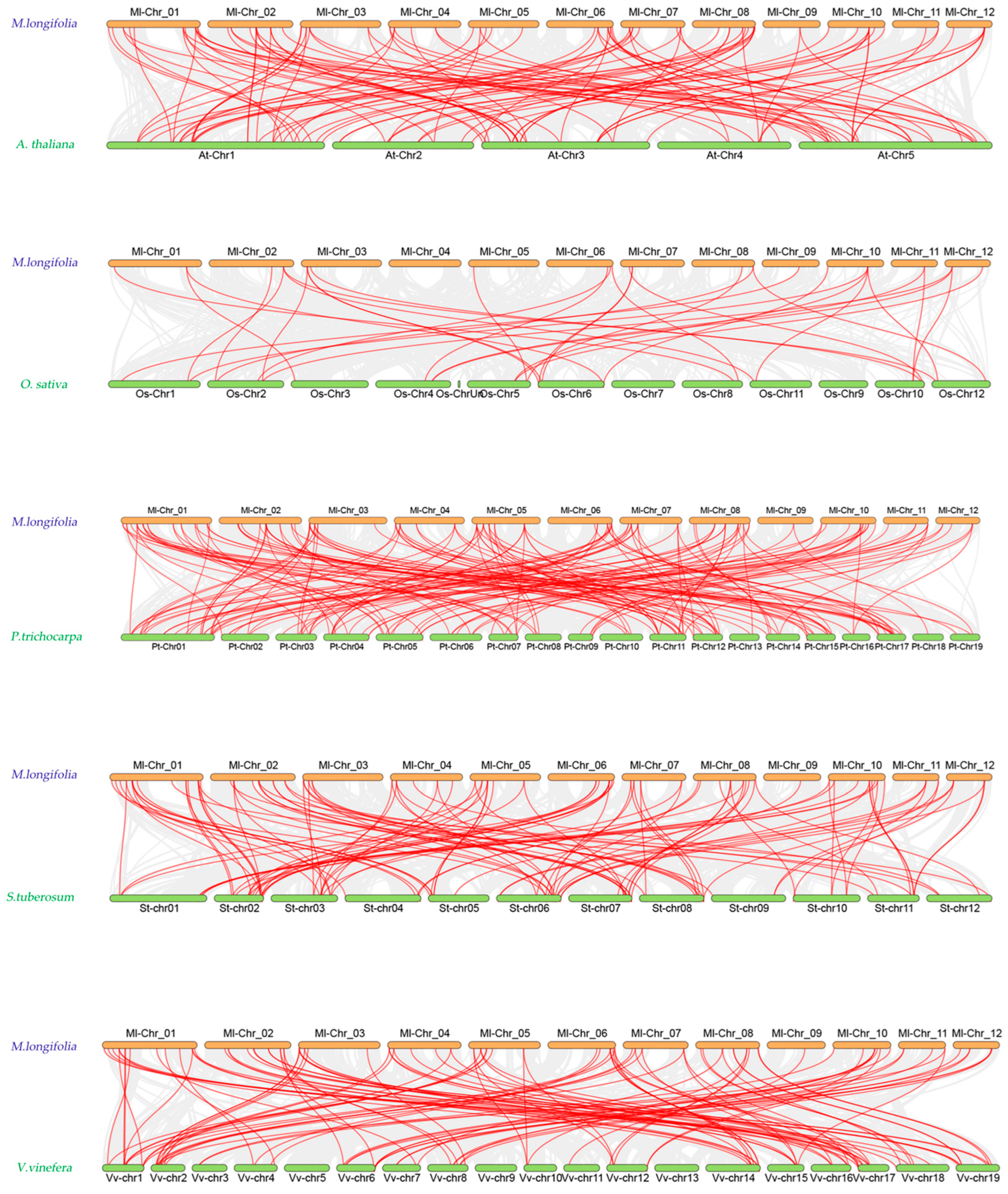
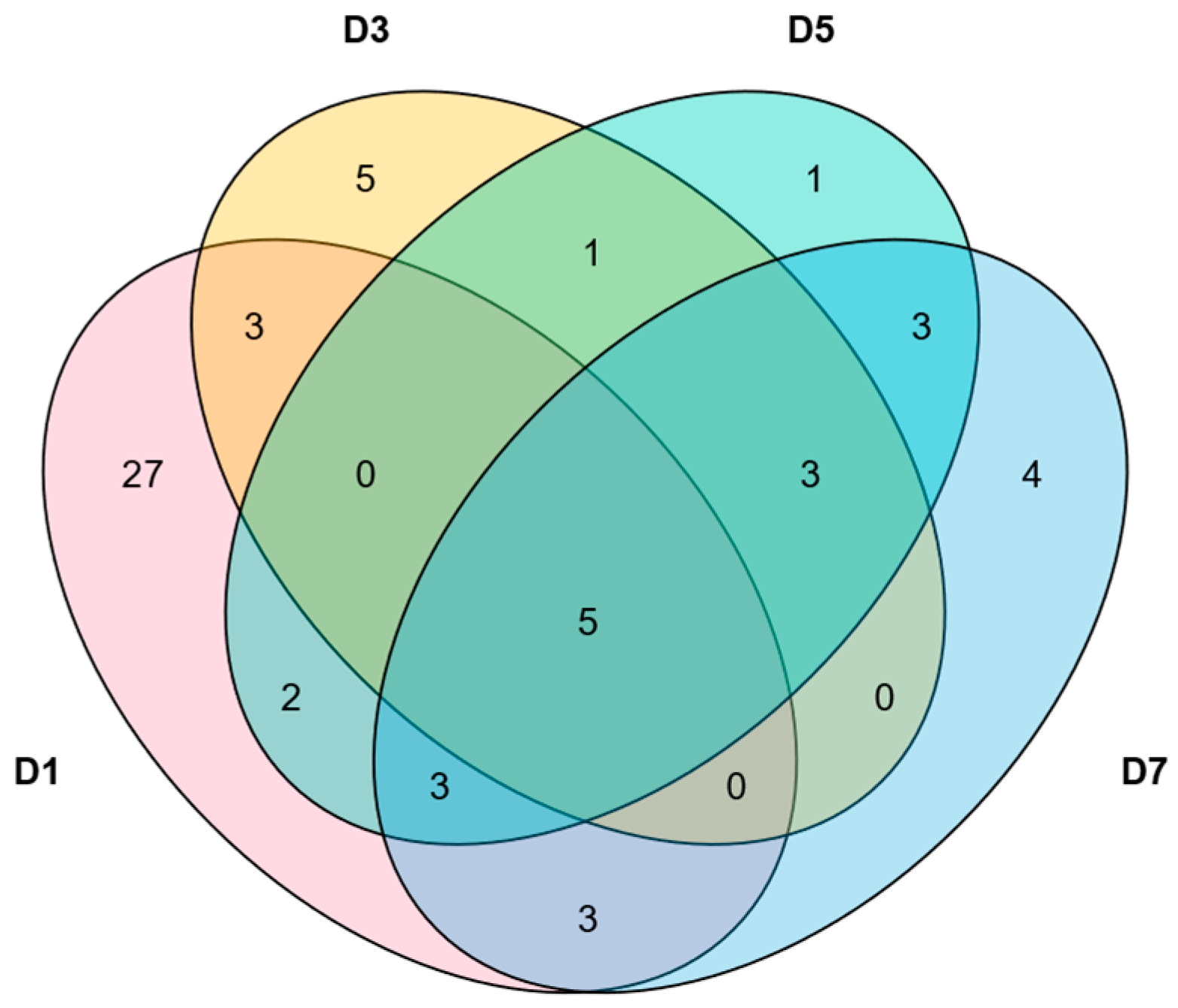
2.4. Analysis of Intraspecific and Interspecific Collinearity of NAC Family Genes in M. longifolia
2.5. Motifs and Gene Structures of the MlNAC Transcription Factor Family
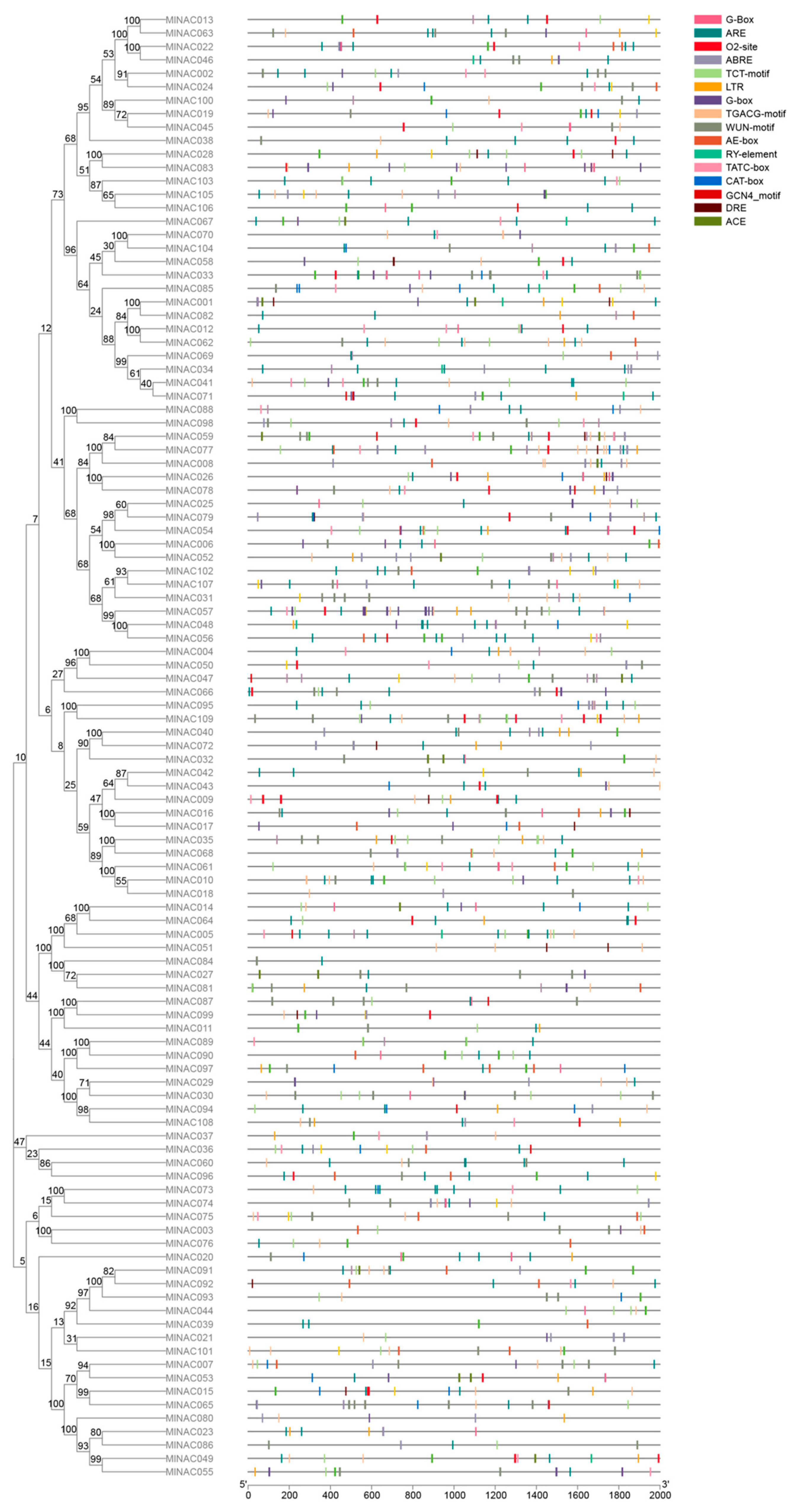
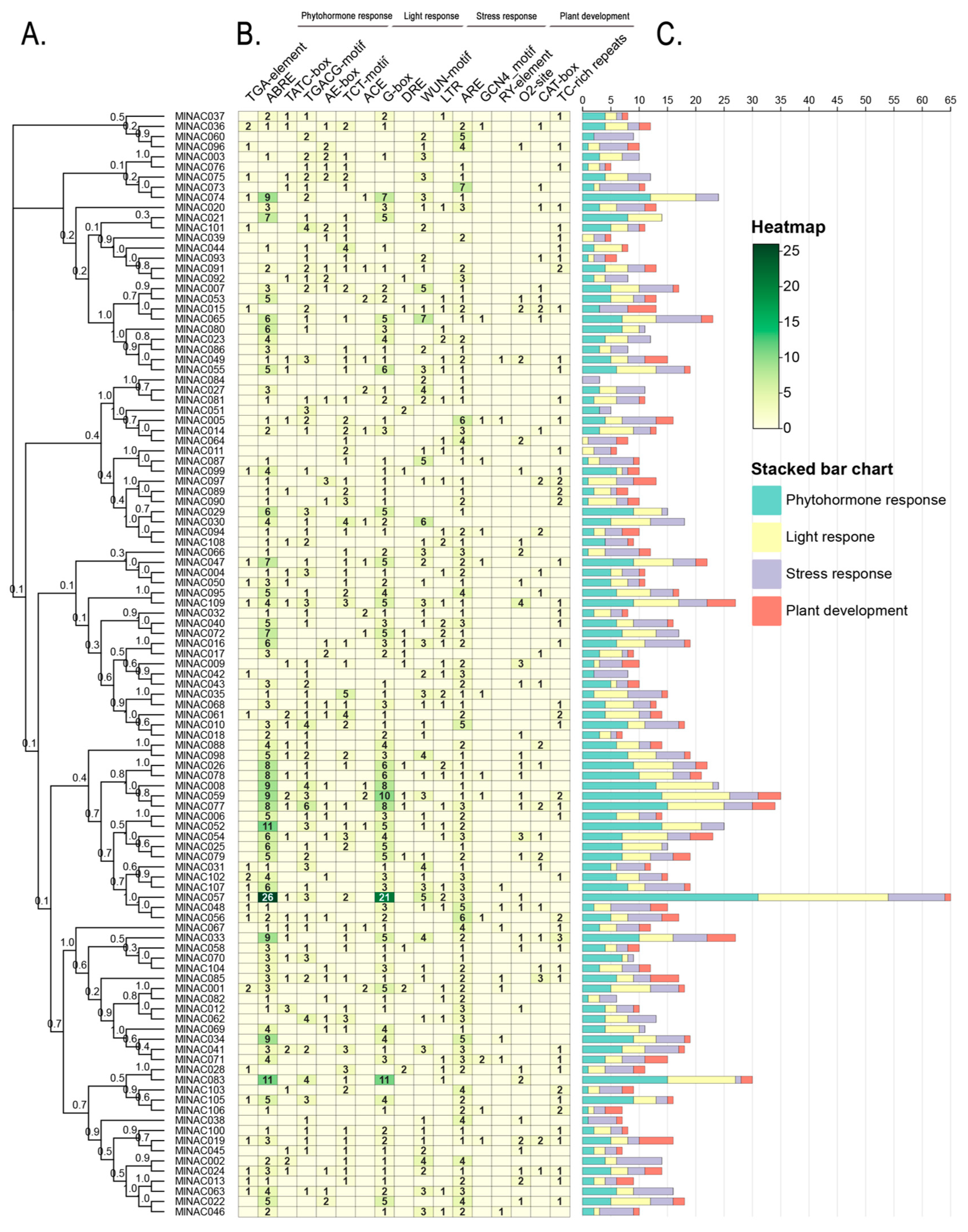
2.6. Analysis of Cis-Acting Elements in the NAC Transcription Factor Family of M. longifolia
2.7. Analysis of the Expression Patterns of MlNAC Genes Under Low Temperature
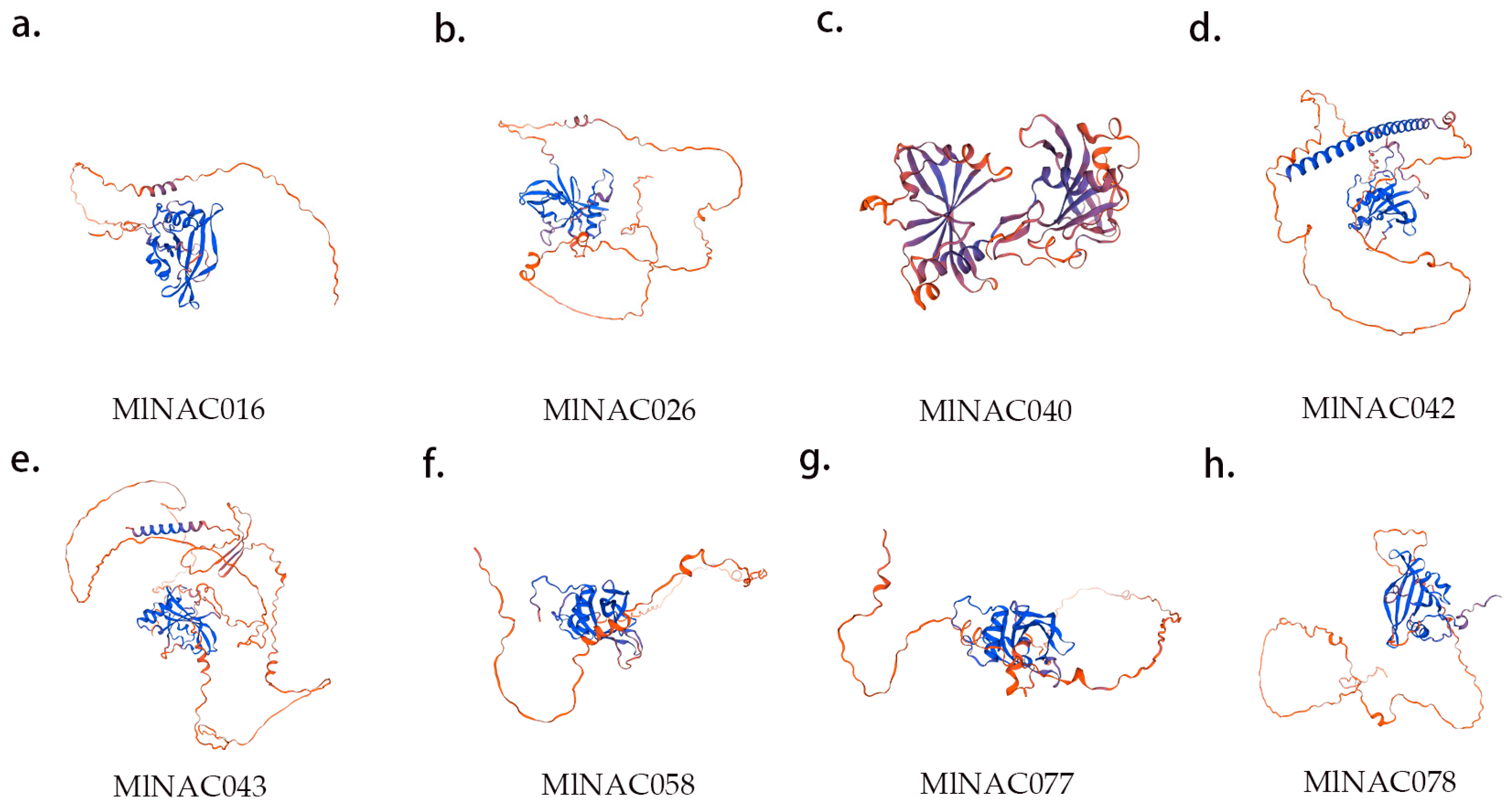
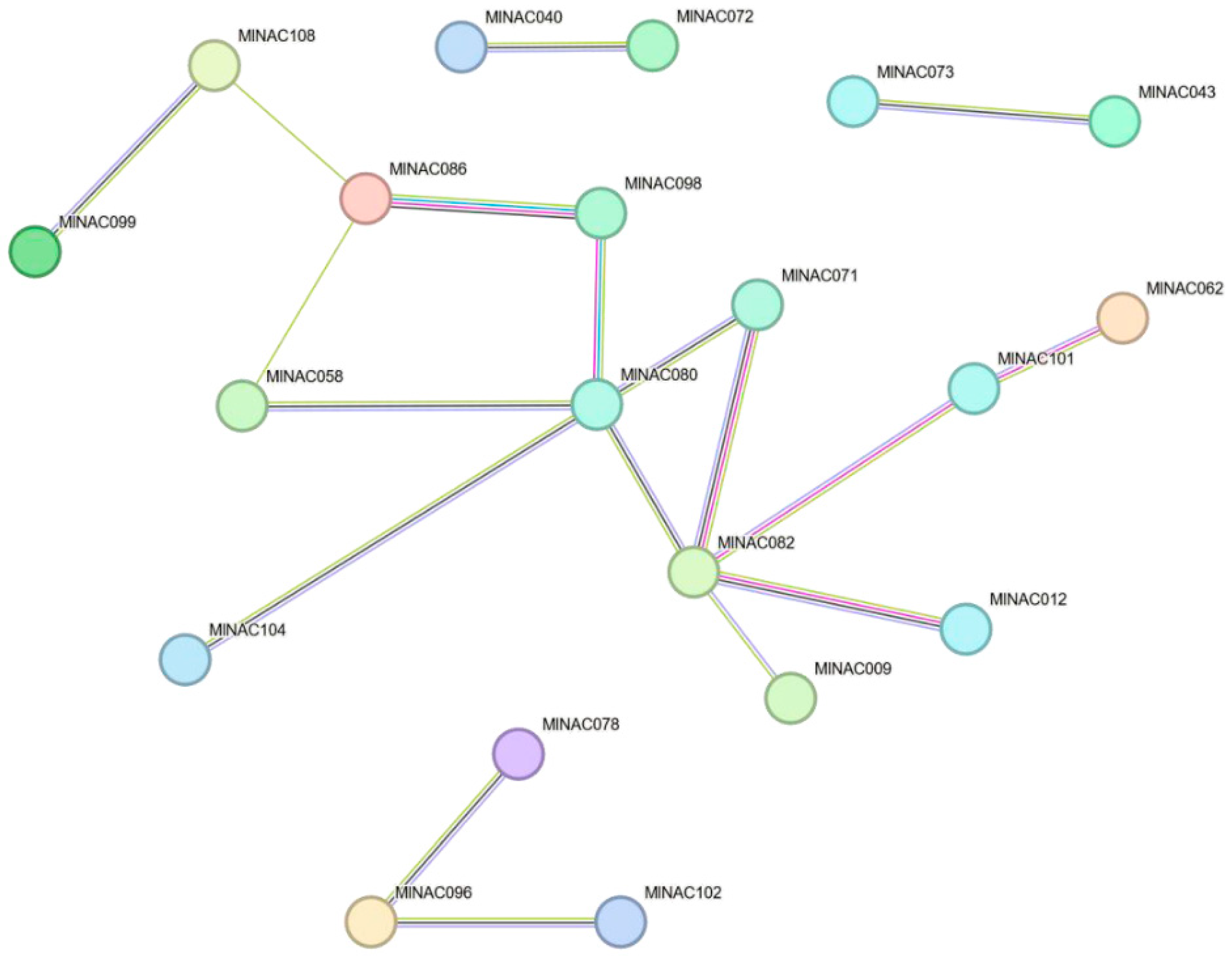
2.8. Key Protein Structure Prediction and NAC Protein Interaction Network Prediction

3. Discussion
3.1. Evolution and Duplication
3.2. Functional Prediction
3.3. Compartive Insights
3.4. Cis-Acting Elements and PPI Analysis
3.5. Future Directions
4. Materials and Methods
4.1. Plant Material
4.2. Genome-Wide Identification of the NAC Transcription Factor Family and Prediction of Physicochemical Properties
4.3. Phylogenetic Analysis of NAC Proteins in M. longifolia
4.4. Chromosomal Distribution
4.5. Collinearity Analysis
4.6. Analysis of NAC Protein Structure and Conserved Motifs
4.7. Analysis of Cis-Acting Elements of NAC Proteins
4.8. Expression Patterns of NAC Proteins Under Low Temperature
4.9. Key Protein Structure and NAC Protein Interaction Network Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, P.; Singh, D.; Mallik, A.; Nayak, S. Madhuca longifolia (Sapotaceae): A review of its traditional uses, phytochemistry and pharmacology. Int. J. Biomed. Res. 2012, 3, 291–305. [Google Scholar] [CrossRef]
- Sinha, J.; Singh, V.; Singh, J.; Rai, A.K. Phytochemistry, ethnomedical uses and future prospects of Mahua (Madhuca longifolia) as a food: A review. J. Nutr. Food Sci. 2017, 7, 1000573. [Google Scholar] [CrossRef]
- Wang, S.; Lin, H.; Ye, S.; Jiao, Z.; Chen, Z.; Ma, Y.; Zhang, L. High-quality chromosome-level genomic insights into molecular adaptation to low-temperature stress in Madhuca longifolia in southern subtropical China. BMC Genom. 2024, 25, 877. [Google Scholar] [CrossRef]
- Zhou, L.; Ullah, F.; Zou, J.; Zeng, X. Molecular and Physiological Responses of Plants that Enhance Cold Tolerance. Int. J. Mol. Sci. 2025, 26, 1157. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Zheng, L.; Liu, P.; Liu, Q.; Ji, T.; Liu, J.; Gao, Y.; Liu, L.; Dong, J.; Wang, T. The S-acylation cycle of transcription factor MtNAC80 influences cold stress responses in Medicago truncatula. Plant Cell 2024, 36, 2629–2651. [Google Scholar] [CrossRef]
- Figueroa, N.; Gómez, R. Bolstered plant tolerance to low temperatures by overexpressing NAC transcription factors: Identification of critical variables by meta-analysis. Planta 2022, 256, 92. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes. Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef]
- He, X.; Mu, R.; Cao, W.; Zhang, Z.; Zhang, J.; Chen, S. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef]
- Li, F.; Guo, X.; Liu, J.; Zhou, F.; Liu, W.; Wu, J.; Zhang, H.; Cao, H.; Su, H.; Wen, R. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Transcription Factor in Chenopodium quinoa. Genes 2019, 10, 500. [Google Scholar] [CrossRef]
- Su, H.; Zhang, S.; Yuan, X.; Chen, C.; Wang, X.; Hao, Y. Genome-wide analysis and identification of stress-responsive genes of the NAM-ATAF1,2-CUC2 transcription factor family in apple. Plant Physiol. Biochem. 2013, 71, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Jiang, Z.; Fu, H.; Chen, L.; Liao, G.; He, Y.; Huang, C.; Xu, X. Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia). BMC Plant Biol. 2021, 21, 44. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Sharma, V.; Pal, A.K.; Acharya, V.; Ahuja, P.S. Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 2013, 20, 403–423. [Google Scholar] [CrossRef]
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 288. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhao, Y.; Li, X.; Wu, M.; Chai, W.; Sheng, L.; Wang, Y.; Dong, Q.; Jiang, H.; Cheng, B. Genomewide identification, classification and analysis of NAC type gene family in maize. J. Genet. 2015, 94, 377–390. [Google Scholar] [CrossRef]
- Liu, T.; Song, X.; Duan, W.; Huang, Z.; Liu, G.; Li, Y.; Hou, X. Genome-Wide Analysis and Expression Patterns of NAC Transcription Factor Family Under Different Developmental Stages and Abiotic Stresses in Chinese Cabbage. Plant Mol. Biol. Rep. 2014, 32, 1041–1056. [Google Scholar] [CrossRef]
- Abdoulaye, A.H.; Yuhua, C.; Xiaoyan, Z.; Yiwei, Y.; Wang, H.; Yinhua, C. Computational analysis and expression profiling of NAC transcription factor family involved in biotic stress response in Manihot esculenta. Plant Biol. 2024, 26, 1247–1259. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Nisar, M.A.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef]
- Karahalil, B. Overview of Systems Biology and Omics Technologies. Curr. Med. Chem. 2016, 23, 4221–4230. [Google Scholar] [CrossRef]
- Ling, L.; Song, L.; Wang, Y.; Guo, C. Genome-wide analysis and expression patterns of the NAC transcription factor family in Medicago truncatula. Physiol. Mol. Biol. Plants 2017, 23, 343–356. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Di, G.; Zhao, Q.; Gao, C.; Zhang, D.; Wang, J.; Shen, Z.; Han, W. Identification of Cold Tolerance Transcriptional Regulatory Genes in Seedlings of Medicago sativa L. and Medicago falcata L. Int. J. Mol. Sci. 2024, 25, 10345. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Wu, S.; Han, B.; Jiao, Y. Genetic Contribution of Paleopolyploidy to Adaptive Evolution in Angiosperms. Mol. Plant 2020, 13, 59–71. [Google Scholar] [CrossRef]
- Landis, J.B.; Soltis, D.E.; Li, Z.; Marx, H.E.; Barker, M.S.; Tank, D.C.; Soltis, P.S. Impact of whole-genome duplication events on diversification rates in angiosperms. Am. J. Bot. 2018, 105, 348–363. [Google Scholar] [CrossRef]
- Clark, J.W.; Donoghue, P.C.J. Whole-Genome Duplication and Plant Macroevolution. Trends Plant Sci. 2018, 23, 933–945. [Google Scholar] [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol. Plant 2018, 11, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Zhao, Y.; Sun, Y.; Li, Y. NACs, generalist in plant life. Plant Biotechnol. J. 2023, 21, 2433–2457. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, D.; Zhang, H.; Song, F. Functions of NAC transcription factors in biotic and abiotic stress responses in plants. Yi Chuan 2012, 34, 993–1002. [Google Scholar] [CrossRef]
- Diao, W.; Snyder, J.C.; Wang, S.; Liu, J.; Pan, B.; Guo, G.; Ge, W.; Dawood, M.H.S.A. Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network. Int. J. Mol. Sci. 2018, 19, 1028. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, H.; Yang, Y.; Tang, K.; Yang, Y.; Ouyang, W.; Du, G. Genome-Wide Identification of NAC Family Genes and Their Expression Analyses in Response to Osmotic Stress in Cannabis sativa L. Int. J. Mol. Sci. 2024, 25, 9466. [Google Scholar] [CrossRef]
- Wang, X.; Li, D.; Jiang, J.; Dong, Z.; Ma, Y. Soybean NAC gene family: Sequence analysis and expression under low nitrogen supply. Biol. Plant. 2017, 61, 473–482. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Abd Allah, E.F.; Al-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4-and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Huang, J.; Piater, L.A.; Dubery, I.A. The NAC transcription factor gene ANAC072 is differentially expressed in Arabidopsis thaliana in response to microbe-associated molecular pattern (MAMP) molecules. Physiol. Mol. Plant Pathol. 2012, 80, 19–27. [Google Scholar] [CrossRef]
- Li, S.; Gao, J.; Yao, L.; Ren, G.; Zhu, X.; Gao, S.; Qiu, K.; Zhou, X.; Kuai, B. The role of ANAC072 in the regulation of chlorophyll degradation during age- and dark-induced leaf senescence. Plant Cell Rep. 2016, 35, 1729–1741. [Google Scholar] [CrossRef]
- Sukiran, N.L.; Ma, J.C.; Ma, H.; Su, Z. ANAC019 is required for recovery of reproductive development under drought stress in Arabidopsis. Plant Mol. Biol. 2019, 99, 161–174. [Google Scholar] [CrossRef]
- Bu, Q.; Jiang, H.; Li, C.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef]
- Kang, M.; Kim, S.; Kim, H.J.; Shrestha, P.; Yun, J.; Phee, B.; Lee, W.; Nam, H.G.; Chang, I. The C-Domain of the NAC Transcription Factor ANAC019 Is Necessary for pH-Tuned DNA Binding through a Histidine Switch in the N-Domain. Cell Rep. 2018, 22, 1141–1150. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, Y.; Zhao, L.; Li, C.; Yu, J.; Li, T.; Yang, W.; Zhang, S.; Su, H.; Wang, L. A novel NAC transcription factor, MdNAC42, regulates anthocyanin accumulation in red-fleshed apple by interacting with MdMYB10. Tree Physiol. 2020, 40, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; He, H.; Chang, Y.; Miao, B.; Liu, Z.; Wang, Q.; Dong, F.; Xiong, L. Multiple roles of NAC transcription factors in plant development and stress responses. J. Integr. Plant Biol. 2025, 67, 510–538. [Google Scholar] [CrossRef]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys Acta 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Mathew, I.E.; Das, S.; Mahto, A.; Agarwal, P. Three Rice NAC Transcription Factors Heteromerize and Are Associated with Seed Size. Front. Plant Sci. 2016, 7, 1638. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Andreeva, A.; Blum, M.; Chuguransky, S.R.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Llinares-Lopez, F.; Meng-Papaxanthos, L.; et al. The Pfam protein families database: Embracing AI/ML. Nucleic Acids Res. 2025, 53, D523–D534. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]



| Sequence ID | Number of Amino Acid | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| MlNAC001 | 154 | 18,067.56 | 9.02 | 30.78 | 62.01 | −0.736 | cyto |
| MlNAC002 | 184 | 21,329.25 | 8.64 | 38.55 | 62.55 | −0.723 | cyto |
| MlNAC003 | 263 | 29,405.18 | 8.18 | 43.99 | 70.08 | −0.488 | cyto |
| MlNAC004 | 240 | 27,362.78 | 8.65 | 43.88 | 60.88 | −0.789 | nucl |
| MlNAC005 | 389 | 43,528.92 | 8.66 | 59.18 | 59.46 | −0.54 | nucl |
| MlNAC006 | 293 | 33,207.45 | 9.17 | 32.8 | 62.25 | −0.68 | nucl |
| MlNAC007 | 429 | 48,232.66 | 5.04 | 50.32 | 67.67 | −0.796 | chlo |
| MlNAC008 | 317 | 36,579.04 | 5.8 | 55.1 | 61.32 | −0.792 | cyto |
| MlNAC009 | 602 | 66,549.72 | 5.11 | 41.47 | 73.22 | −0.361 | nucl |
| MlNAC010 | 657 | 74,134.14 | 5.87 | 51.85 | 67.84 | −0.615 | plas |
| MlNAC011 | 390 | 44,480.42 | 6.19 | 49.78 | 72.97 | −0.482 | nucl |
| MlNAC012 | 157 | 18,572.19 | 8.66 | 38.97 | 62.74 | −0.743 | mito |
| MlNAC013 | 343 | 38,898.95 | 5.44 | 46.91 | 60.52 | −0.678 | nucl |
| MlNAC014 | 399 | 44,718.46 | 6.03 | 45.34 | 59.67 | −0.603 | nucl |
| MlNAC015 | 470 | 52,209.88 | 4.85 | 49.96 | 64.96 | −0.683 | nucl |
| MlNAC016 | 630 | 70,689.88 | 4.75 | 50.07 | 64.83 | −0.647 | nucl |
| MlNAC017 | 630 | 70,690.55 | 4.9 | 49.07 | 67.33 | −0.588 | nucl |
| MlNAC018 | 262 | 30,223.44 | 5.43 | 47.5 | 59.16 | −0.858 | nucl |
| MlNAC019 | 318 | 35,802.99 | 6.14 | 41.55 | 63.24 | −0.615 | nucl |
| MlNAC020 | 354 | 38,681.94 | 4.46 | 33.94 | 74.72 | −0.421 | cyto |
| MlNAC021 | 218 | 24,918.81 | 4.84 | 47.88 | 60.78 | −0.753 | nucl |
| MlNAC022 | 360 | 40,498.04 | 8.13 | 33.3 | 70.42 | −0.523 | nucl |
| MlNAC023 | 451 | 50,548.99 | 6.33 | 46.38 | 59.02 | −0.908 | nucl |
| MlNAC024 | 349 | 38,608.43 | 7.11 | 48.73 | 64.04 | −0.494 | nucl |
| MlNAC025 | 348 | 38,494.09 | 7.81 | 42.29 | 69.83 | −0.63 | nucl |
| MlNAC026 | 340 | 38,058.8 | 8.91 | 41.23 | 62.5 | −0.586 | nucl |
| MlNAC027 | 321 | 36,823.03 | 6.61 | 31.9 | 64.39 | −0.856 | cysk |
| MlNAC028 | 340 | 37,832.75 | 6.98 | 48.35 | 61.59 | −0.518 | nucl |
| MlNAC029 | 289 | 33,036.78 | 5.38 | 53.9 | 63.77 | −0.684 | nucl |
| MlNAC030 | 255 | 29,119.4 | 6.46 | 58.44 | 49.33 | −0.908 | nucl |
| MlNAC031 | 260 | 29,481.4 | 8.9 | 42.33 | 63.04 | −0.669 | nucl |
| MlNAC032 | 713 | 79,667.29 | 5.56 | 57.45 | 69.16 | −0.651 | nucl |
| MlNAC033 | 324 | 36,182.55 | 5.02 | 43.57 | 59.26 | −0.8 | nucl |
| MlNAC034 | 354 | 41,066.81 | 6.19 | 55.27 | 57.32 | −0.891 | pero |
| MlNAC035 | 341 | 38,750.11 | 5.12 | 48.41 | 48.53 | −0.734 | nucl |
| MlNAC036 | 258 | 29,094.2 | 6.55 | 45.8 | 53.64 | −0.99 | nucl |
| MlNAC037 | 380 | 43,909.44 | 5.19 | 56.13 | 58.71 | −1.016 | nucl |
| MlNAC038 | 423 | 48,492.12 | 5.85 | 45.2 | 52.03 | −0.861 | nucl |
| MlNAC039 | 447 | 50,537.86 | 5.13 | 48.39 | 76.51 | −0.599 | cyto |
| MlNAC040 | 539 | 60,286.63 | 5.44 | 40.63 | 67.46 | −0.619 | nucl |
| MlNAC041 | 357 | 41,689.57 | 6.01 | 53.29 | 63.05 | −0.889 | pero |
| MlNAC042 | 416 | 46,467.24 | 5.57 | 44.99 | 75.31 | −0.6 | chlo |
| MlNAC043 | 573 | 63,561.84 | 4.56 | 38.94 | 72.67 | −0.472 | chlo |
| MlNAC044 | 773 | 85,627.26 | 5.91 | 52.35 | 73.95 | −0.446 | nucl |
| MlNAC045 | 321 | 36,140.6 | 8.32 | 47.4 | 67.13 | −0.621 | nucl_plas |
| MlNAC046 | 345 | 38,756.19 | 8.66 | 34.91 | 73.71 | −0.492 | nucl |
| MlNAC047 | 225 | 25,293.48 | 8.96 | 46.15 | 63.24 | −0.644 | cyto_nucl |
| MlNAC048 | 310 | 35,162.77 | 6.33 | 41.45 | 71.71 | −0.555 | nucl |
| MlNAC049 | 337 | 37,586.41 | 8.54 | 41.57 | 71.1 | −0.69 | nucl |
| MlNAC050 | 246 | 27,882.26 | 8.97 | 43.17 | 54.63 | −0.784 | nucl |
| MlNAC051 | 395 | 45,035.59 | 7.64 | 45.53 | 60.99 | −0.701 | nucl |
| MlNAC052 | 307 | 34,989.1 | 7.65 | 34.33 | 61.01 | −0.777 | cyto_nucl |
| MlNAC053 | 973 | 108,909.9 | 5.71 | 45.2 | 77.81 | −0.561 | nucl |
| MlNAC054 | 358 | 39,441.27 | 7.79 | 35.24 | 63.24 | −0.649 | nucl |
| MlNAC055 | 303 | 33,915.89 | 6.67 | 48.77 | 68.84 | −0.76 | nucl |
| MlNAC056 | 314 | 35,957.94 | 6.84 | 48.63 | 75.13 | −0.624 | nucl |
| MlNAC057 | 316 | 35,537.07 | 6.96 | 43.92 | 66.36 | −0.552 | nucl |
| MlNAC058 | 324 | 36,754.93 | 5.08 | 42.15 | 63.77 | −0.741 | chlo |
| MlNAC059 | 303 | 35,187.92 | 7.12 | 47.26 | 59.27 | −0.753 | nucl |
| MlNAC060 | 344 | 39,365.82 | 5.05 | 52.97 | 61.22 | −0.802 | nucl |
| MlNAC061 | 635 | 72,052.48 | 5.36 | 44.37 | 69.24 | −0.613 | chlo |
| MlNAC062 | 353 | 40,551.19 | 5.25 | 53.57 | 63.77 | −0.775 | nucl |
| MlNAC063 | 354 | 40,320.37 | 5.34 | 37.02 | 64.44 | −0.626 | nucl |
| MlNAC064 | 402 | 45,228.78 | 6.46 | 44.77 | 60.65 | −0.607 | cyto |
| MlNAC065 | 389 | 43,220.1 | 5.57 | 41.79 | 65.86 | −0.74 | nucl |
| MlNAC066 | 276 | 31,309.61 | 9.27 | 52.02 | 64.2 | −0.704 | nucl |
| MlNAC067 | 418 | 47,169.16 | 7.12 | 42.9 | 57.18 | −0.877 | nucl |
| MlNAC068 | 271 | 31,244.46 | 8.94 | 33.21 | 60.74 | −0.65 | nucl |
| MlNAC069 | 348 | 40,141.64 | 5.92 | 56.3 | 66.41 | −0.791 | nucl |
| MlNAC070 | 403 | 45,594.45 | 6.01 | 52.55 | 63.37 | −0.831 | nucl |
| MlNAC071 | 150 | 17,312.42 | 8.49 | 38.25 | 59.8 | −0.808 | pero |
| MlNAC072 | 1121 | 123,393.1 | 4.93 | 47.12 | 68.22 | −0.622 | nucl |
| MlNAC073 | 943 | 104,588.3 | 4.8 | 44.69 | 66.31 | −0.674 | chlo |
| MlNAC074 | 272 | 30,613.57 | 5.27 | 36.7 | 70.26 | −0.526 | nucl |
| MlNAC075 | 434 | 47,312.69 | 5.84 | 49.08 | 71.27 | −0.54 | nucl |
| MlNAC076 | 256 | 28,836.65 | 8.67 | 54.68 | 79.61 | −0.308 | cyto |
| MlNAC077 | 297 | 33,866.32 | 6.67 | 49.76 | 64.07 | −0.669 | nucl |
| MlNAC078 | 346 | 38,621.3 | 8.17 | 33.99 | 67.08 | −0.575 | nucl |
| MlNAC079 | 529 | 58,847.39 | 9.97 | 52.83 | 64.18 | −0.792 | nucl |
| MlNAC080 | 230 | 26,459.87 | 9.76 | 42.92 | 60.04 | −0.957 | nucl |
| MlNAC081 | 327 | 36,894.07 | 8.2 | 46.76 | 67.09 | −0.654 | cyto_nucl |
| MlNAC082 | 281 | 32,135.79 | 5.55 | 24.8 | 63.49 | −0.721 | cyto |
| MlNAC083 | 329 | 37,165.14 | 7.93 | 46.79 | 59.85 | −0.561 | nucl |
| MlNAC084 | 688 | 79,660.6 | 8.62 | 40.58 | 85.17 | −0.189 | plas |
| MlNAC085 | 315 | 36,187.75 | 6.84 | 39.11 | 63.78 | −0.714 | pero |
| MlNAC086 | 475 | 53,104.02 | 6.47 | 47.46 | 62.25 | −0.829 | nucl |
| MlNAC087 | 288 | 33,471.35 | 6.42 | 39.15 | 73.82 | −0.59 | cyto_nucl |
| MlNAC088 | 191 | 21,771.36 | 4.93 | 45.15 | 66.81 | −0.57 | nucl |
| MlNAC089 | 386 | 43,746.99 | 6.77 | 57 | 65.52 | −0.697 | nucl |
| MlNAC090 | 386 | 43,742.12 | 6.81 | 57 | 65 | −0.688 | nucl |
| MlNAC091 | 289 | 33,014.05 | 5.64 | 49.48 | 73.49 | −0.762 | nucl |
| MlNAC092 | 266 | 29,885.38 | 5.64 | 38.25 | 67.78 | −0.759 | nucl |
| MlNAC093 | 311 | 35,639.33 | 5.49 | 49.19 | 74.6 | −0.662 | nucl |
| MlNAC094 | 246 | 27,940.25 | 7.01 | 58.59 | 53.94 | −0.78 | nucl |
| MlNAC095 | 412 | 45,789.47 | 6.63 | 62.95 | 74.51 | −0.592 | E.R._plas |
| MlNAC096 | 425 | 48,121.78 | 6.25 | 43.38 | 63.72 | −0.82 | nucl |
| MlNAC097 | 390 | 44,041.14 | 6.6 | 55.27 | 70.51 | −0.613 | nucl |
| MlNAC098 | 136 | 15,415.93 | 5.56 | 55.69 | 50.07 | −0.828 | nucl |
| MlNAC099 | 271 | 31,334.04 | 6.97 | 42.29 | 69.11 | −0.616 | nucl |
| MlNAC100 | 240 | 27,139.7 | 9.33 | 29.86 | 58.58 | −0.668 | mito |
| MlNAC101 | 266 | 31,598.32 | 8.68 | 45.92 | 73.61 | −0.719 | cyto |
| MlNAC102 | 383 | 43,051.05 | 5.8 | 44.58 | 64.73 | −0.652 | nucl |
| MlNAC103 | 226 | 25,452.48 | 5.12 | 44.08 | 71.15 | −0.496 | chlo |
| MlNAC104 | 358 | 40,926.24 | 5.81 | 48.39 | 60.75 | −0.846 | nucl_cyto |
| MlNAC105 | 111 | 12,421.07 | 10.7 | 24.26 | 46.58 | −0.636 | chlo |
| MlNAC106 | 303 | 34,512.5 | 6.06 | 44.6 | 65.58 | −0.684 | chlo |
| MlNAC107 | 336 | 38,148.58 | 5.42 | 43.98 | 67.65 | −0.739 | nucl |
| MlNAC108 | 234 | 26,713.28 | 8.89 | 47.81 | 64.96 | −0.63 | nucl_plas |
| MlNAC109 | 394 | 44,477.22 | 7.53 | 53.14 | 69.52 | −0.608 | golg |
| Gene Name | Gene Name | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| MlNAC016 | MlNAC017 | 0.19366 | 0.54 | 0.357643 |
| MlNAC056 | MlNAC057 | 0.37887 | 1.94 | 0.194812 |
| MlNAC072 | MlNAC073 | 0.26521 | 0.43 | 0.613301 |
| MlNAC073 | MlNAC074 | 0.17947 | 0.22 | 0.812852 |
| MlNAC092 | MlNAC093 | 0.18383 | 0.44 | 0.415911 |
| MlNAC105 | MlNAC106 | 0.36074 | 0.46 | 0.779525 |
| MlNAC005 | MlNAC014 | 0.2542 | 1.85 | 0.137437 |
| MlNAC004 | MlNAC047 | 0.30535 | 2.1 | 0.145468 |
| MlNAC004 | MlNAC050 | 0.10598 | 0.83 | 0.128115 |
| MlNAC005 | MlNAC051 | 0.26097 | 1.13 | 0.231595 |
| MlNAC006 | MlNAC052 | 0.18814 | 0.9 | 0.209433 |
| MlNAC007 | MlNAC053 | 0.14392 | 0.58 | 0.247552 |
| MlNAC014 | MlNAC051 | 0.313 | 1.99 | 0.15738 |
| MlNAC010 | MlNAC061 | 0.19589 | 0.89 | 0.2205 |
| MlNAC013 | MlNAC063 | 0.12992 | 0.7 | 0.18503 |
| MlNAC014 | MlNAC064 | 0.1225 | 0.45 | 0.27049 |
| MlNAC015 | MlNAC065 | 0.18074 | 0.63 | 0.287519 |
| MlNAC004 | MlNAC066 | 0.40389 | 2.42 | 0.166574 |
| MlNAC005 | MlNAC064 | 0.25499 | 1.58 | 0.161317 |
| MlNAC003 | MlNAC076 | 0.18673 | 0.44 | 0.427734 |
| MlNAC011 | MlNAC087 | 0.24609 | 3.03 | 0.081343 |
| MlNAC022 | MlNAC046 | 0.11964 | 0.6 | 0.199585 |
| MlNAC019 | MlNAC045 | 0.14077 | 0.76 | 0.184086 |
| MlNAC022 | MlNAC063 | 0.25031 | 1.1 | 0.228258 |
| MlNAC027 | MlNAC084 | 0.14583 | 0.77 | 0.189329 |
| MlNAC028 | MlNAC083 | 0.18056 | 0.64 | 0.28242 |
| MlNAC026 | MlNAC078 | 0.09175 | 0.7 | 0.131836 |
| MlNAC029 | MlNAC094 | 0.32689 | 2.29 | 0.142457 |
| MlNAC031 | MlNAC102 | 0.39016 | 1.71 | 0.228424 |
| MlNAC031 | MlNAC107 | 0.40584 | 3.57 | 0.113673 |
| MlNAC029 | MlNAC108 | 0.34506 | 1.5 | 0.230004 |
| MlNAC032 | MlNAC040 | 0.49147 | 2.36 | 0.208597 |
| MlNAC034 | MlNAC041 | 0.18919 | 1.49 | 0.126941 |
| MlNAC033 | MlNAC070 | 0.19103 | 1.2 | 0.158538 |
| MlNAC034 | MlNAC069 | 0.11823 | 0.87 | 0.135643 |
| MlNAC035 | MlNAC068 | 0.1636 | 0.89 | 0.183472 |
| MlNAC032 | MlNAC072 | 0.6226 | 1.98 | 0.314718 |
| MlNAC033 | MlNAC104 | 0.28246 | 1.95 | 0.144742 |
| MlNAC047 | MlNAC050 | 0.36597 | NaN | NaN |
| MlNAC047 | MlNAC066 | 0.52199 | 2.02 | 0.258471 |
| MlNAC040 | MlNAC072 | 0.34701 | 0.76 | 0.45545 |
| MlNAC041 | MlNAC069 | 0.2156 | 1.8 | 0.119882 |
| MlNAC048 | MlNAC056 | 0.16141 | 1.04 | 0.155259 |
| MlNAC049 | MlNAC055 | 0.12889 | 0.68 | 0.189038 |
| MlNAC050 | MlNAC066 | 0.40728 | 2.88 | 0.141278 |
| MlNAC051 | MlNAC064 | 0.34092 | 2.04 | 0.167518 |
| MlNAC055 | MlNAC080 | 0.26642 | 1.74 | 0.152886 |
| MlNAC049 | MlNAC080 | 0.25589 | 1.97 | 0.129971 |
| MlNAC070 | MlNAC104 | 0.29509 | 1.88 | 0.157362 |
| MlNAC088 | MlNAC098 | 0.13961 | 1.11 | 0.125641 |
| MlNAC087 | MlNAC099 | 0.1541 | 1.49 | 0.103642 |
| MlNAC089 | MlNAC097 | 0.1766 | 0.92 | 0.192336 |
| MlNAC094 | MlNAC108 | 0.17362 | 1.09 | 0.159737 |
| MlNAC095 | MlNAC109 | 0.20989 | 0.67 | 0.315408 |
| MlNAC103 | MlNAC106 | 0.23434 | 0.65 | 0.361058 |
| MlNAC102 | MlNAC107 | 0.1665 | 0.73 | 0.226942 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Qin, J.; Wang, Z.; Lin, H.; Ye, S.; Wei, J.; Wang, S.; Zhang, L. Genome-Wide Identification of 109 NAC Genes and Dynamic Expression Profiles Under Cold Stress in Madhuca longifolia. Int. J. Mol. Sci. 2025, 26, 4713. https://doi.org/10.3390/ijms26104713
Chen Y, Qin J, Wang Z, Lin H, Ye S, Wei J, Wang S, Zhang L. Genome-Wide Identification of 109 NAC Genes and Dynamic Expression Profiles Under Cold Stress in Madhuca longifolia. International Journal of Molecular Sciences. 2025; 26(10):4713. https://doi.org/10.3390/ijms26104713
Chicago/Turabian StyleChen, Yule, Jiayu Qin, Ziyao Wang, Haoyou Lin, Shuiyun Ye, Jichen Wei, Shuyu Wang, and Lu Zhang. 2025. "Genome-Wide Identification of 109 NAC Genes and Dynamic Expression Profiles Under Cold Stress in Madhuca longifolia" International Journal of Molecular Sciences 26, no. 10: 4713. https://doi.org/10.3390/ijms26104713
APA StyleChen, Y., Qin, J., Wang, Z., Lin, H., Ye, S., Wei, J., Wang, S., & Zhang, L. (2025). Genome-Wide Identification of 109 NAC Genes and Dynamic Expression Profiles Under Cold Stress in Madhuca longifolia. International Journal of Molecular Sciences, 26(10), 4713. https://doi.org/10.3390/ijms26104713






