Genome-Wide Identification and Expression Analysis of the FAR1-RELATED SEQUENCE (FRS) Gene Family in Grape (Vitis vinifera L.)
Abstract
1. Introduction
2. Results
2.1. Identification of VvFRS Family Members in Grape
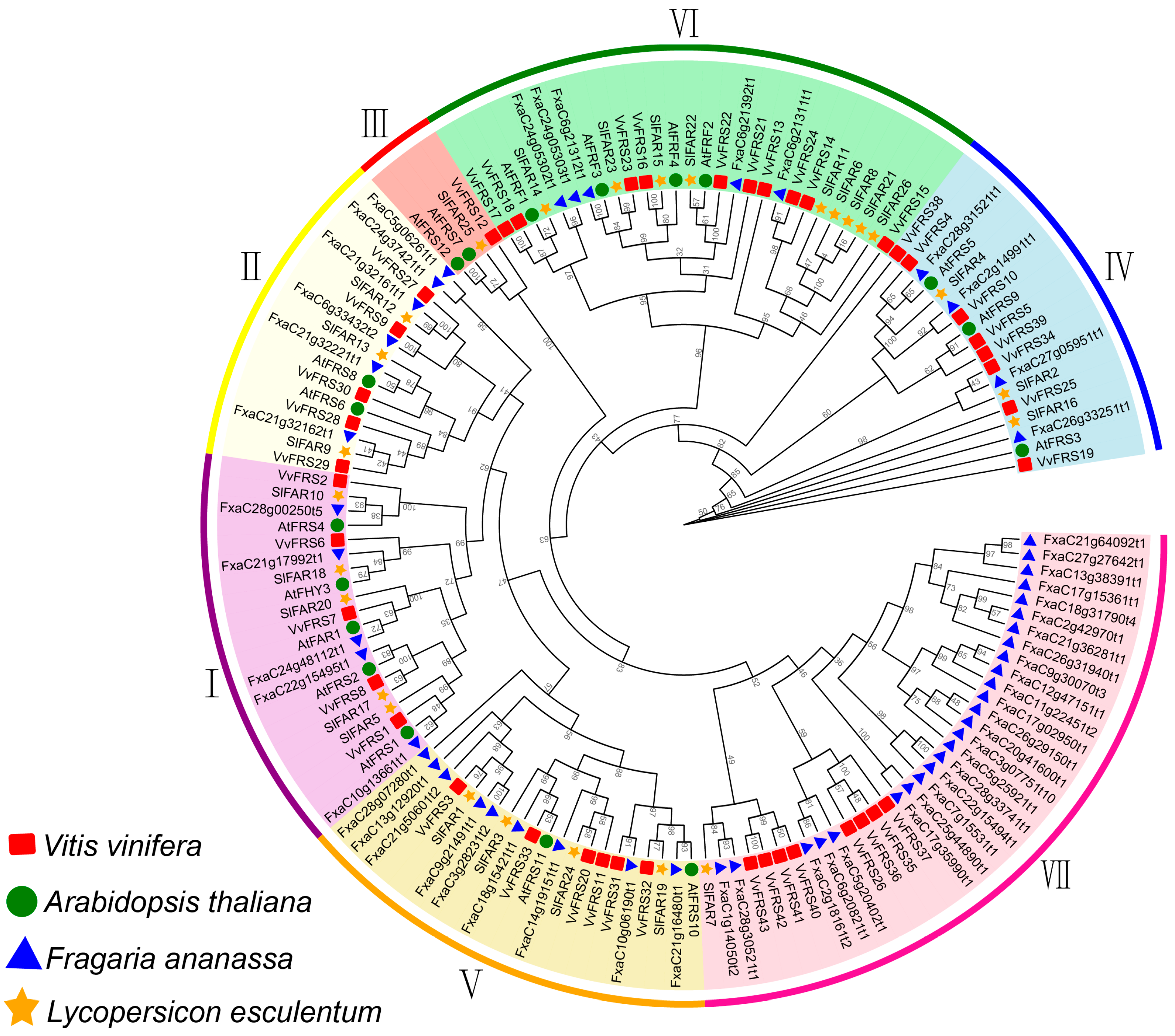
2.2. Phylogenetic Analysis of VvFRSs
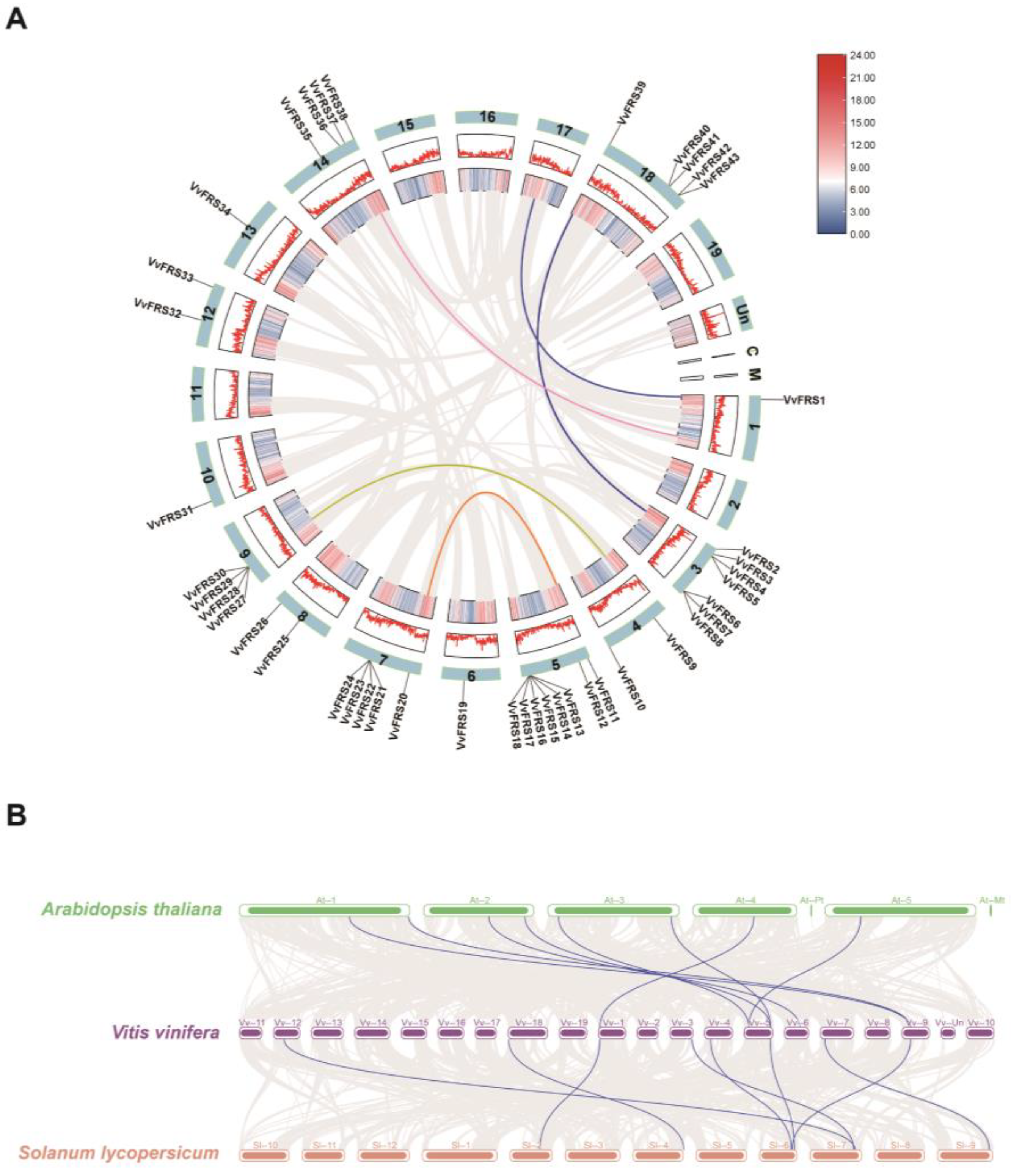
2.3. Synteny Analysis of VvFRSs in Grape
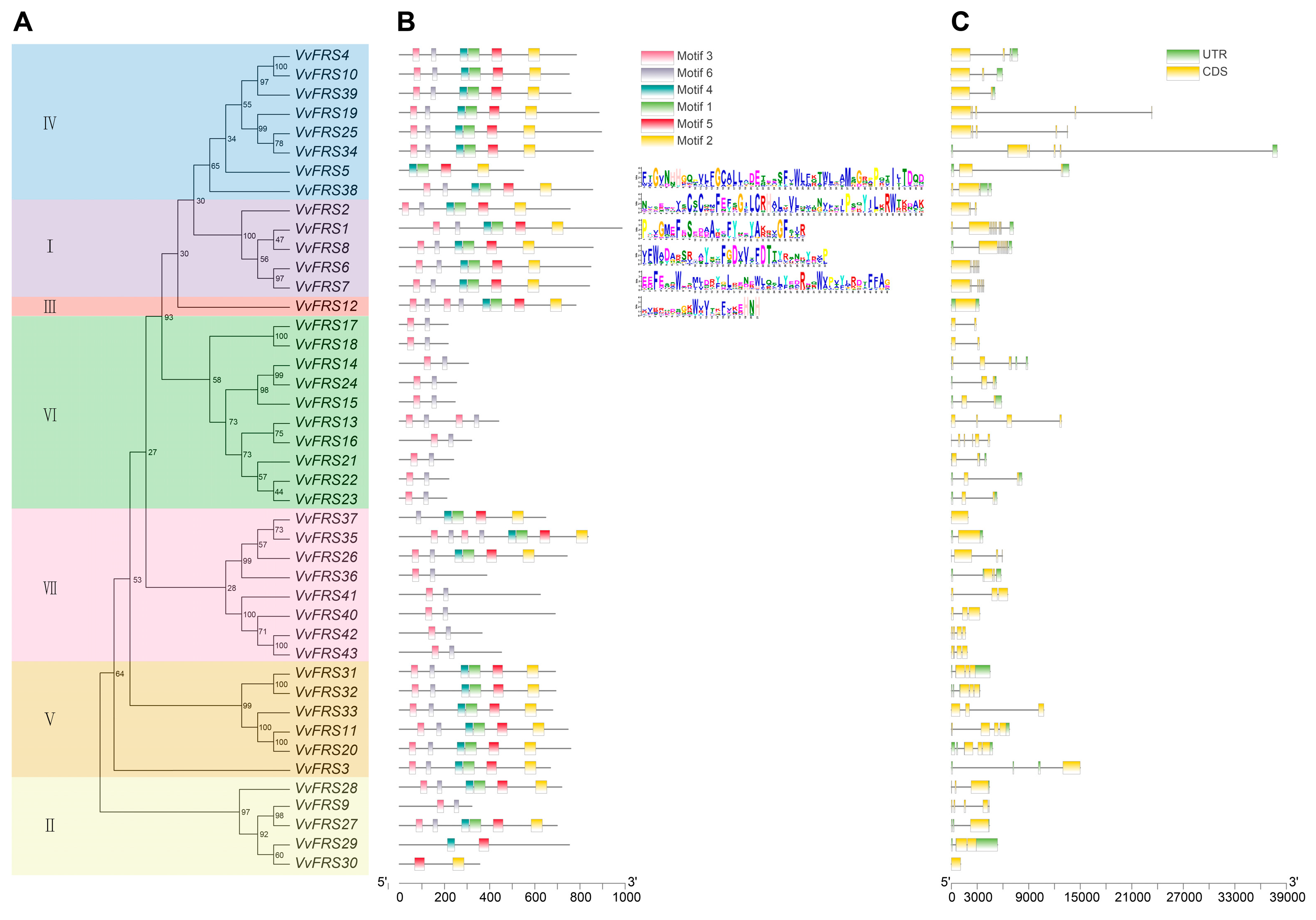
2.4. Conserved Motifs of Grape VvFRS Members
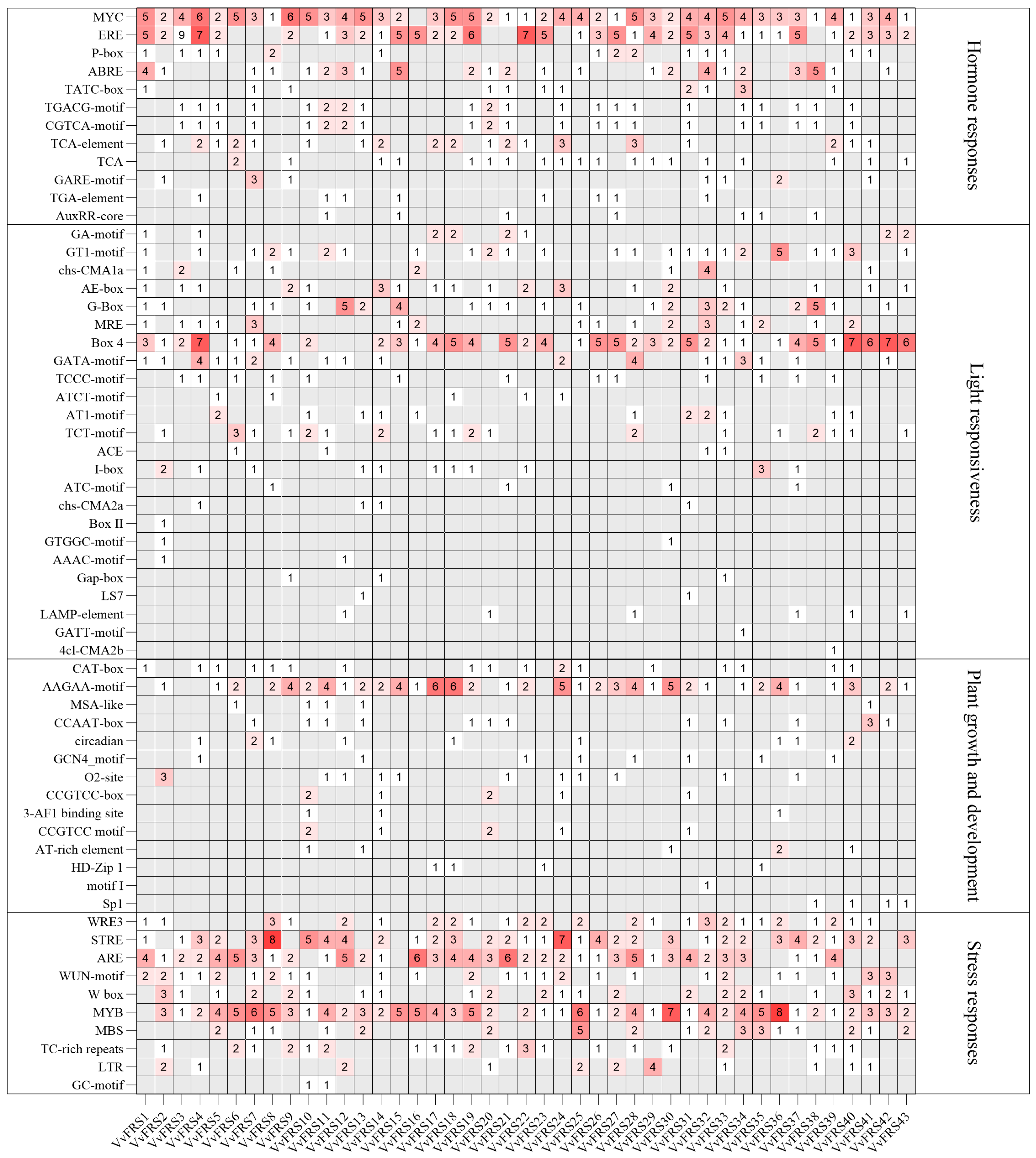
2.5. Cis-Elements in the Promoters of VvFRSs
2.6. Structural Features of VvFRS Proteins
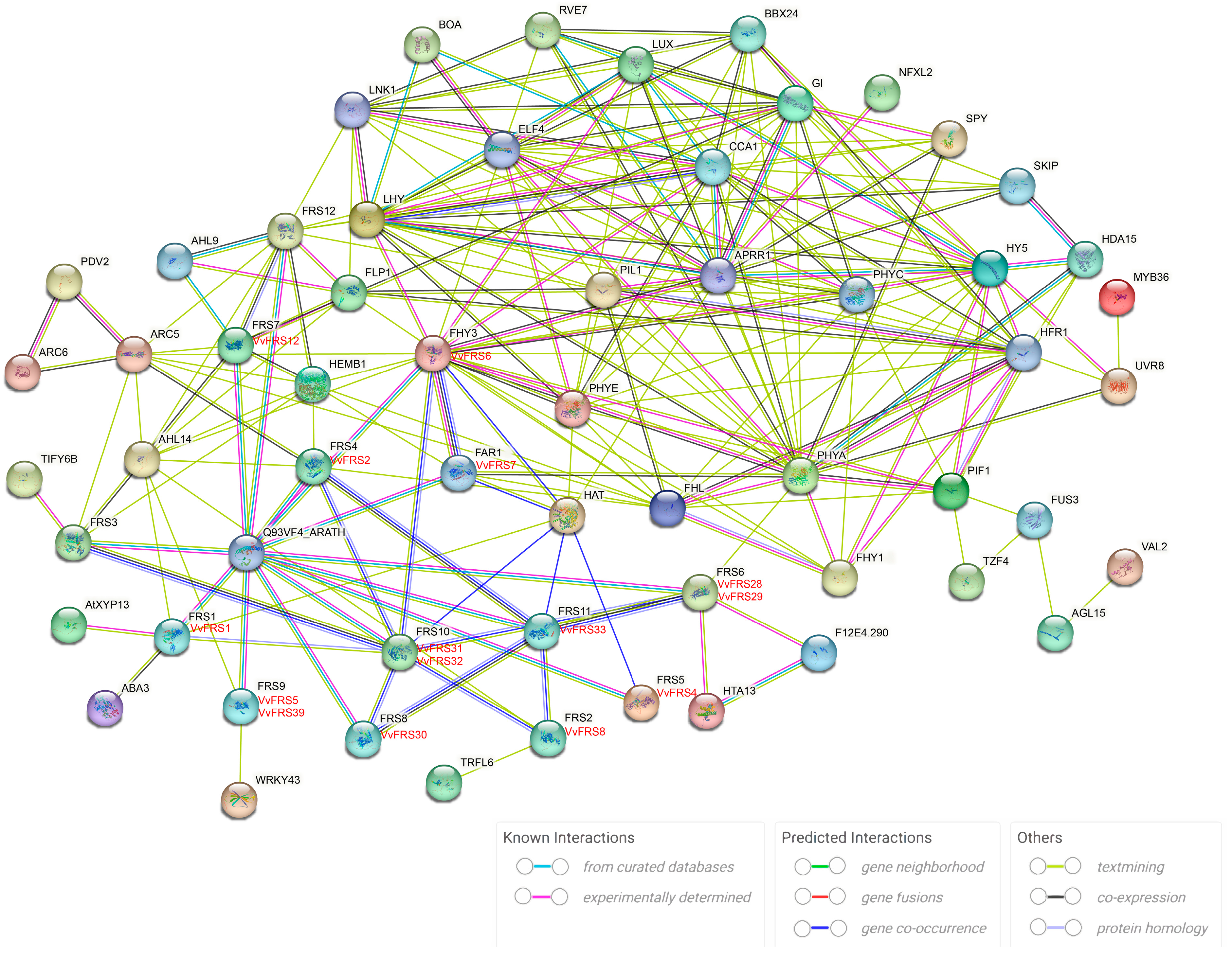
2.7. Protein Interaction Networks of VvFRS
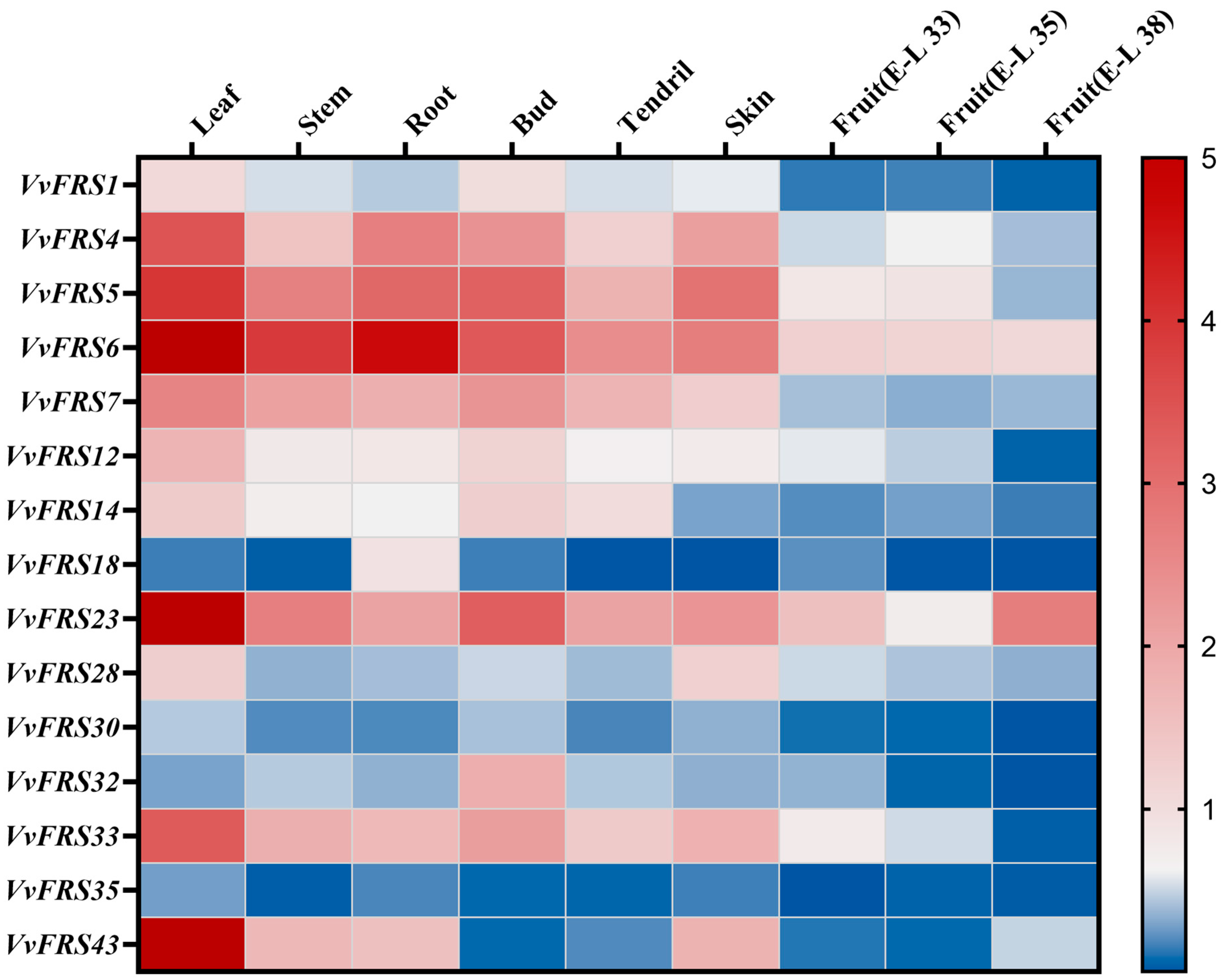
2.8. Expression Analysis of VvFRS in Different Tissues
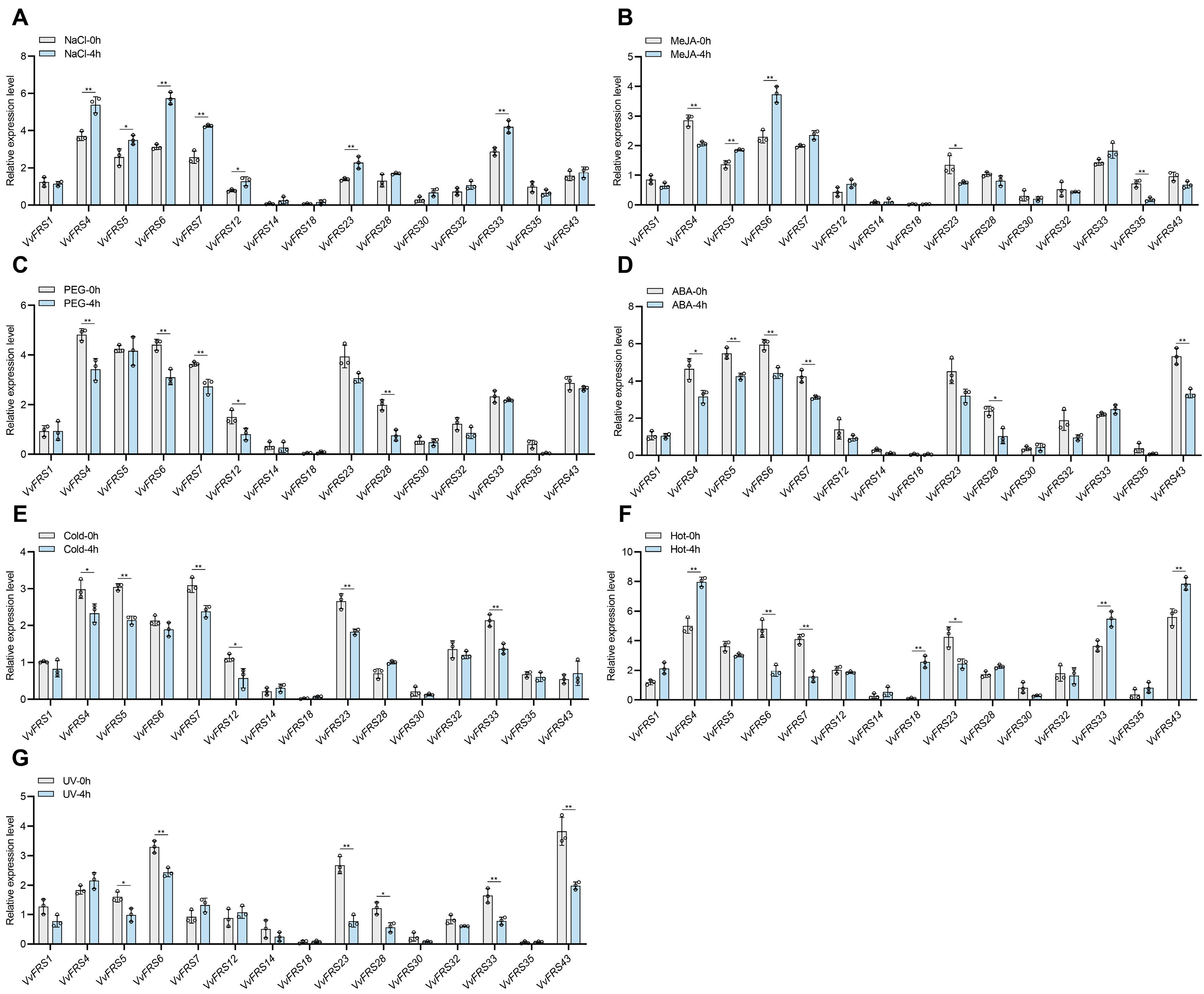
2.9. Expression Analysis of VvFRS Under Different Treatments
3. Discussion
3.1. The Structure, Evolutionary Characteristics, and Functions of VvFRSs
3.2. The Promoters of VvFRSs Contain Abundant Stress Response Elements
3.3. VvFRS Proteins Interaction Network
3.4. VvFRF6 Plays an Important Role in Stress Resistance
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Identification of VvFRS Family Genes in Grape
4.3. Phylogenetic Evolutionary Analysis of FRS Genes in Grape
4.4. Collinearity Analysis of Grape VvFRS
4.5. Gene Structure and Conserved Motifs Analysis of VvFRS
4.6. Analysis of Cis-Elements in Promoters
4.7. Structural Characteristics and Interaction Network of VvFRS Proteins
4.8. Quantitative Real-Time PCR Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pierik, R.; de Wit, M. Shade avoidance: Phytochrome signalling and other aboveground neighbour detection cues. J. Exp. Bot. 2014, 65, 2815–2824. [Google Scholar] [CrossRef]
- Jung, J.H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M.; Blackwood, L.; Petersen, J.; Sullivan, S. Plant flavoprotein photoreceptors. Plant Cell Physiol. 2015, 56, 401–413. [Google Scholar] [CrossRef]
- Wang, H.; Wang, H. Phytochrome signaling: Time to tighten up the loose ends. Mol. Plant 2015, 8, 540–551. [Google Scholar] [CrossRef]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome Signaling Networks. Annu. Rev. Plant Biol. 2021, 72, 217–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Deng, X.W. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 2003, 8, 172–178. [Google Scholar] [CrossRef]
- Lin, R.; Ding, L.; Casola, C.; Ripoll, D.R.; Feschotte, C.; Wang, H. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 2007, 318, 1302–1305. [Google Scholar] [CrossRef]
- Wang, H.; Deng, X.W. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. EMBO J. 2002, 21, 1339–1349. [Google Scholar] [CrossRef]
- Lin, R.; Wang, H. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol. 2004, 136, 4010–4022. [Google Scholar] [CrossRef]
- Huang, X.; Ouyang, X.; Yang, P.; Lau, O.S.; Li, G.; Li, J.; Chen, H.; Deng, X.W. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 2012, 24, 4590–4606. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Gao, Y.; Zhao, L.; Liu, X.; Gao, H. Arabidopsis FHY3/CPD45 regulates far-red light signaling and chloroplast division in parallel. Sci Rep. 2015, 5, 9612. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, H.; An, C.; Shi, Y.; Liu, X.; Yuan, W.; Zhang, B.; Yang, J.; Yu, C.; Gao, H. Arabidopsis FRS4/CPD25 and FHY3/CPD45 work cooperatively to promote the expression of the chloroplast division gene ARC5 and chloroplast division. Plant J. 2013, 75, 795–807. [Google Scholar] [CrossRef]
- Ouyang, X.; Li, J.; Li, G.; Li, B.; Chen, B.; Shen, H.; Huang, X.; Mo, X.; Wan, X.; Lin, R.; et al. Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 2011, 23, 2514–2535. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Wang, W.; Chen, D.; Ji, Q.; Jing, Y.; Wang, H.; Lin, R. Transposase-derived proteins FHY3/FAR1 interact with PHYTOCHROME-INTERACTING FACTOR1 to regulate chlorophyll biosynthesis by modulating HEMB1 during deetiolation in Arabidopsis. Plant Cell 2012, 24, 1984–2000. [Google Scholar] [CrossRef] [PubMed]
- McCormac, A.C.; Terry, M.J. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 2002, 32, 549–559. [Google Scholar] [CrossRef]
- Ma, L.; Xue, N.; Fu, X.; Zhang, H.; Li, G. Arabidopsis thaliana FAR-RED ELONGATED HYPOCOTYLS3 (FHY3) and FAR-RED-IMPAIRED RESPONSE1 (FAR1) modulate starch synthesis in response to light and sugar. New Phytol. 2017, 213, 1682–1696. [Google Scholar] [CrossRef]
- Allen, T.; Koustenis, A.; Theodorou, G.; Somers, D.E.; Kay, S.A.; Whitelam, G.C.; Devlin, P.F. Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell 2006, 18, 2506–2516. [Google Scholar] [CrossRef]
- Li, G.; Siddiqui, H.; Teng, Y.; Lin, R.; Wan, X.Y.; Li, J.; Lau, O.S.; Ouyang, X.; Dai, M.; Wan, J. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 2011, 13, 616–622. [Google Scholar] [CrossRef]
- Siddiqui, H.; Khan, S.; Rhodes, B.M.; Devlin, P.F. FHY3 and FAR1 act downstream of light stable phytochromes. Front. Plant Sci. 2016, 7, 175. [Google Scholar] [CrossRef]
- Ritter, A.; Iñigo, S.; Fernández-Calvo, P.; Heyndrickx, K.S.; Dhondt, S.; Shi, H.; De Milde, L.; Vanden Bossche, R.; De Clercq, R.; Eeckhout, D. The transcriptional repressor complex FRS7-FRS12 regulates flowering time and growth in Arabidopsis. Nat. Commun. 2017, 8, 15235. [Google Scholar] [CrossRef] [PubMed]
- Stirnberg, P.; Zhao, S.; Williamson, L.; Ward, S.; Leyser, O. FHY3 promotes shoot branching and stress tolerance in Arabidopsis in an AXR1-dependent manner. Plant J. 2012, 71, 907–920. [Google Scholar] [CrossRef]
- Li, D.; Fu, X.; Guo, L.; Huang, Z.; Li, Y.; Liu, Y.; He, Z.; Cao, X.; Ma, X.; Zhao, M. FAR-RED ELONGATED HYPOCOTYL3 activates SEPALLATA2 but inhibits CLAVATA3 to regulate meristem determinacy and maintenance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 9375–9380. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tian, T.; Lin, R.; Deng, X.W.; Wang, H.; Li, G. Arabidopsis FHY3 and FAR1 regulate light-induced myo-inositol biosynthesis and oxidative stress responses by transcriptional activation of MIPS1. Mol. Plant 2016, 9, 541–557. [Google Scholar] [CrossRef]
- Wang, W.; Tang, W.; Ma, T.; Niu, D.; Jin, J.B.; Wang, H.; Lin, R. A pair of light signaling factors FHY3 and FAR1 regulates plant immunity by modulating chlorophyll biosynthesis. J. Integr. Plant Biol. 2016, 58, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Ji, Q.; Huang, Y.; Jiang, Z.; Bao, M.; Wang, H.; Lin, R. FAR-RED ELONGATED HYPOCOTYL3 and FAR-RED IMPAIRED RESPONSE1 transcription factors integrate light and abscisic acid signaling in Arabidopsis. Plant Physiol. 2013, 163, 857–866. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 2017, 29, 2269–2284. [Google Scholar] [CrossRef]
- Dai, J.; Sun, J.; Peng, W.; Liao, W.; Zhou, Y.; Zhou, X.R.; Qin, Y.; Cheng, Y.; Cao, S. FAR1/FHY3 Transcription Factors Positively Regulate the Salt and Temperature Stress Responses in Eucalyptus grandis. Front Plant Sci. 2022, 13, 883654. [Google Scholar] [CrossRef]
- Liu, Z.; An, C.; Zhao, Y.; Xiao, Y.; Bao, L.; Gong, C.; Gao, Y. Genome-Wide Identification and Characterization of the CsFHY3/FAR1 Gene Family and Expression Analysis under Biotic and Abiotic Stresses in Tea Plants (Camellia sinensis). Plants 2021, 10, 570. [Google Scholar] [CrossRef]
- Chen, S.; Chen, Y.; Liang, M.; Qu, S.; Shen, L.; Zeng, Y.; Hou, N. Genome-wide identification and molecular expression profile analysis of FHY3/FAR1 gene family in walnut (Juglans sigillata L.) development. BMC Genom. 2023, 24, 673. [Google Scholar] [CrossRef]
- Chen, Q.; Song, Y.; Liu, K.; Su, C.; Yu, R.; Li, Y.; Yang, Y.; Zhou, B.; Wang, J.; Hu, G. Genome-Wide Identification and Functional Characterization of FAR1-RELATED SEQUENCE (FRS) Family Members in Potato (Solanum tuberosum). Plants 2023, 12, 2575. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Qiao, Y.; Lu, S.; Yao, K.; Wang, C.; Liao, W. Genome-Wide Identification and Expression Analysis of FAR1/FHY3 Gene Family in Cucumber (Cucumis sativus L.). Agronomy 2024, 14, 50. [Google Scholar] [CrossRef]
- Samuels, L.J.; Setati, M.E.; Blancquaert, E.H. Towards a Better Understanding of the Potential Benefits of Seaweed Based Biostimulants in Vitis vinifera L. Cultivars. Plants 2022, 11, 348. [Google Scholar] [CrossRef] [PubMed]
- Paiola, A.; Assandri, G.; Brambilla, M.; Zottini, M.; Pedrini, P.; Nascimbene, J. Exploring the potential of vineyards for biodiversity conservation and delivery of biodiversity-mediated ecosystem services: A global-scale systematic review. Sci. Total Environ. 2020, 706, 135839. [Google Scholar] [CrossRef]
- Shields, M.W.; Tompkins, J.M.; Saville, D.J.; Meurk, C.D.; Wratten, S. Potential ecosystem service delivery by endemic plants in New Zealand vineyards: Successes and prospects. PeerJ 2016, 4, e2042. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Monder, H.; Maillard, M.; Chérel, I.; Zimmermann, S.D.; Paris, N.; Cuéllar, T.; Gaillard, I. Adjustment of K+ Fluxes and Grapevine Defense in the Face of Climate Change. Int. J. Mol. Sci. 2021, 22, 10398. [Google Scholar] [CrossRef]
- Carvalho, L.C.; Ramos, M.J.N.; Faísca-Silva, D.; van der Kellen, D.; Fernandes, J.C.; Egipto, R.; Lopes, C.M.; Amâncio, S. Developmental Regulation of Transcription in Touriga Nacional Berries under Deficit Irrigation. Plants 2022, 11, 827. [Google Scholar] [CrossRef]
- De Rosa, V.; Vizzotto, G.; Falchi, R. Cold Hardiness Dynamics and Spring Phenology: Climate-Driven Changes and New Molecular Insights Into Grapevine Adaptive Potential. Front Plant Sci. 2021, 12, 644528. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.; Wang, Z.; Zhu, K.; Wu, W. Comparative metagenomic analysis reveals rhizosphere microbial community composition and functions help protect grapevines against salt stress. Front Microbiol. 2023, 14, 1102547. [Google Scholar] [CrossRef]
- Zhou, H.; Li, Q.; Gichuki, D.K.; Hou, Y.; Fan, P.; Gong, L.; Xin, H. Dynamics of starch degradation and expression of related genes during chilling stress in grapevine. Hortic. Adv. 2023, 1, 3. [Google Scholar] [CrossRef]
- Chen, Y.; Deng, J.; Chen, J.; Zeng, T.; Yu, T.; Huang, Q.; Chen, P.; Liu, Q.; Jian, W.; Yang, X. Genome-wide identification and expression analysis of FAR1/FHY3 transcription factor family in tomato. Plant Physiol. J. 2021, 57, 1983–1995. [Google Scholar]
- Aguilar-Martinez, J.A.; Uchida, N.; Townsley, B.; West, D.A.; Yanez, A.; Lynn, N.; Kimura, S.; Sinha, N. Transcriptional, posttranscriptional, and posttranslational regulation of SHOOT MERISTEMLESS gene expression in Arabidopsis determines gene function in the shoot apex. Plant Physiol. 2015, 167, 424–442. [Google Scholar] [CrossRef] [PubMed]
- Joly-Lopez, Z.; Hoen, D.R.; Blanchette, M.; Bureau, T.E. Phylogenetic and genomic analyses resolve the origin of important plant genes derived from transposable elements. Mol. Biol. Evol. 2016, 33, 1937–1956. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Tang, H.; Jing, D.; Liu, C.; Xie, X.; Zhang, L.; Chen, X.; Li, C. Genome-Wide Identification and Expression Analyses of the FAR1/FHY3 Gene Family Provide Insight into Inflorescence Development in Maize. Curr. Issues Mol. Biol. 2024, 46, 430–449. [Google Scholar] [CrossRef]
- Ma, L.; Li, G. FAR1-RELATED SEQUENCE (FRS) and FRS-RELATED FACTOR (FRF) Family Proteins in Arabidopsis Growth and Development. Front Plant Sci. 2018, 9, 692. [Google Scholar] [CrossRef]
- Vondras, A.M.; Commisso, M.; Guzzo, F.; Deluc, L.G. Metabolite Profiling Reveals Developmental Inequalities in Pinot Noir Berry Tissues Late in Ripening. Front Plant Sci. 2017, 8, 1108. [Google Scholar] [CrossRef]
- Ma, T.; Ma, H.; Zhao, H.; Qi, H.; Zhao, J. Identification, characterization, and transcription analysis of xylogen-like arabinogalactan proteins in rice (Oryza sativa L.). BMC Plant Biol. 2014, 14, 299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, J.; Wang, L.; Guo, H.; Liao, B.; King, G.; Zhang, X. Genome evolutionary dynamics followed by diversifying selection explains the complexity of the Sesamum indicum genome. BMC Genom. 2017, 18, 257. [Google Scholar] [CrossRef]
- Xiao, Y.; Wen, J.; Meng, R.; Meng, Y.; Zhou, Q.; Nie, Z. The expansion and diversity of the CYP75 gene family in Vitaceae. PeerJ 2021, 9, e12174. [Google Scholar] [CrossRef]
- Ahmad, B.; Yao, J.; Zhang, S.; Li, X.; Zhang, X.; Yadav, V.; Wang, X. Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development. Int. J. Mol. Sci. 2020, 21, 1088. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Q. Genome-wide characterization of the WRKY gene family in cultivated strawberry (Fragaria × ananassa Duch.) and the importance of several group III members in continuous cropping. Sci. Rep. 2019, 9, 8423. [Google Scholar] [CrossRef]
- Liu, J.; Wang, J.; Wang, M.; Zhao, J.; Zheng, Y.; Zhang, T.; Xue, L.; Lei, J. Genome-Wide Analysis of the R2R3-MYB Gene Family in Fragaria × ananassa and Its Function Identification During Anthocyanins Biosynthesis in Pink-Flowered Strawberry. Front. Plant Sci. 2021, 12, 702160. [Google Scholar] [CrossRef] [PubMed]
- Magris, G.; Jurman, I.; Fornasiero, A.; Paparelli, E.; Schwope, R.; Marroni, F.; Di Gaspero, G.; Morgante, M. The genomes of 204 Vitis vinifera accessions reveal the origin of European wine grapes. Nat. Commun. 2021, 12, 7240. [Google Scholar] [CrossRef]
- Xiong, J.; Zheng, D.; Zhu, H.; Chen, J.; Na, R.; Cheng, Z. Genome-wide identification and expression analysis of the SPL gene family in woodland strawberry Fragaria vesca. Genome 2018, 61, 675–683. [Google Scholar] [CrossRef]
- Cantu, D.; Massonnet, M.; Cochetel, N. The wild side of grape genomics. Trends Genet. 2024, 40, 601–612. [Google Scholar] [CrossRef]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Dinneny, J.R.; Long, T.A.; Wang, J.Y.; Jung, J.W.; Mace, D.; Pointer, S.; Barron, C.; Brady, S.M.; Schiefelbein, J.; Benfey, P.N. Cell Identity Mediates the response of Arabidopsis Roots to abiotic stress. Science 2008, 320, 942–945. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.E.; Lisch, D.R.; Quail, P.H. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003, 34, 453–471. [Google Scholar] [CrossRef]
- Cheng, C.; Wang, Y.; Chai, F.; Li, S.; Xin, H.; Liang, Z. Genome-wide identification and characterization of the 14-3-3 family in Vitis vinifera L. during berry development and cold- and heat-stress response. BMC Genom. 2018, 19, 579. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Zhang, C.; Qin, T.; Liu, Y.; Liu, Z.; Xie, X.; Bai, J.; Sun, C.; Bi, Z. Comprehensive Analysis of GH3 Gene Family in Potato and Functional Characterization of StGH3.3 under Drought Stress. Int. J. Mol. Sci. 2023, 24, 15122. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, R.; Ye, X.; Zheng, X.; Tan, B.; Wang, W.; Li, Z.; Li, J.; Cheng, J.; Feng, J. Overexpressing VvWRKY18 from grapevine reduces the drought tolerance in Arabidopsis by increasing leaf stomatal density. J. Plant Physiol. 2022, 275, 153741. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Sun, J.; Mao, X.; Wang, J.; Wang, J.; Liu, H.; Zheng, H.; Zhen, Z.; Zhao, H.; et al. Genome-wide characterization and identification of trihelix transcription factor and expression profiling in response to abiotic stresses in rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 251. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, X.; Hou, H.; Fan, Y.; Wang, L.; Zeng, H.; Chen, X.; Ding, Y.; Hu, X.; Yan, Q. Genome-wide characterization of the tomato UDP-glycosyltransferase gene family and functional identification of SlUDPGT52 in drought tolerance. Hortic. Adv. 2023, 1, 14. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simon-ovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Locus ID | Protein Length (aa) | Molecular Weight (Da) | Theoretical Isoelectric Point | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| VvFRS1 | Vitvi01g00116_t001 | 985 | 113,743.53 | 8.52 | 56.77 | 63.26 | −0.694 | Nucleus |
| VvFRS2 | Vitvi03g04004_t001 | 755 | 87,847.54 | 6.68 | 52.81 | 72.23 | −0.583 | Nucleus |
| VvFRS3 | Vitvi03g00193_t001 | 752 | 85,954.31 | 8.69 | 40.56 | 85.56 | −0.283 | Chloroplast |
| VvFRS4 | Vitvi03g01475_t001 | 783 | 89,339.54 | 6.16 | 47.37 | 70.59 | −0.528 | Chloroplast |
| VvFRS5 | Vitvi03g00266_t001 | 549 | 62,553.56 | 6.88 | 40.42 | 69.87 | −0.512 | Nucleus |
| VvFRS6 | Vitvi03g01239_t001 | 847 | 97,096.12 | 6.20 | 52.23 | 66.46 | −0.662 | Nucleus |
| VvFRS7 | Vitvi03g01243_t001 | 841 | 96,465.3 | 8.03 | 50.23 | 69.1 | −0.571 | Chloroplast |
| VvFRS8 | Vitvi03g01244_t001 | 857 | 98,181.2 | 6.24 | 47.67 | 72.67 | −0.54 | Chloroplast |
| VvFRS9 | Vitvi04g00504_t001 | 668 | 77,635.63 | 6.45 | 42.17 | 77.14 | −0.438 | Chloroplast |
| VvFRS10 | Vitvi04g01750_t001 | 751 | 85,598.73 | 6.41 | 45.31 | 73.87 | −0.45 | Chloroplast |
| VvFRS11 | Vitvi05g00081_t001 | 746 | 85,345.3 | 6.19 | 49.93 | 80.64 | −0.288 | Nucleus |
| VvFRS12 | Vitvi05g01873_t001 | 781 | 89,714.38 | 6.88 | 43.13 | 74.55 | −0.507 | Chloroplast |
| VvFRS13 | Vitvi05g01497_t001 | 439 | 51,066.38 | 6.51 | 58.27 | 69.75 | −0.868 | Nucleus |
| VvFRS14 | Vitvi05g01498_t001 | 305 | 35,433.14 | 6.65 | 52.65 | 78 | −0.666 | Nucleus |
| VvFRS15 | Vitvi05g01499_t001 | 246 | 28,349.96 | 5.23 | 50.88 | 71.71 | −0.694 | Chloroplast |
| VvFRS16 | Vitvi05g04445_t001 | 319 | 36,844.6 | 9.24 | 40.6 | 83.39 | −0.464 | Chloroplast |
| VvFRS17 | Vitvi05g01506_t001 | 215 | 25,134.58 | 7.06 | 45.71 | 68.56 | −0.771 | Chloroplast |
| VvFRS18 | Vitvi05g01510_t001 | 215 | 25,082.41 | 6.34 | 49.63 | 67.63 | −0.8 | Cytoplasm |
| VvFRS19 | Vitvi06g01048_t001 | 883 | 99,376.01 | 5.89 | 48.17 | 71.7 | −0.468 | Chloroplast |
| VvFRS20 | Vitvi07g00384_t001 | 758 | 86,535.53 | 6.46 | 49.54 | 76.15 | −0.346 | Nucleus |
| VvFRS21 | Vitvi07g04446_t001 | 239 | 27,624.01 | 5.27 | 39.56 | 70.5 | −0.878 | Nucleus |
| VvFRS22 | Vitvi07g01198_t001 | 218 | 25,372.89 | 8.94 | 51.09 | 70.6 | −0.843 | Nucleus |
| VvFRS23 | Vitvi07g01199_t001 | 209 | 24,052.29 | 8.56 | 57.83 | 68.42 | −0.722 | Nucleus |
| VvFRS24 | Vitvi07g02480_t001 | 252 | 29,153.62 | 6.32 | 48.73 | 68.81 | −0.923 | Nucleus |
| VvFRS25 | Vitvi08g00891_t001 | 894 | 100,926.01 | 6.18 | 41.1 | 71.88 | −0.49 | Chloroplast |
| VvFRS26 | Vitvi08g01733_t001 | 742 | 84,686.41 | 5.27 | 46.39 | 74.77 | −0.431 | Nucleus |
| VvFRS27 | Vitvi09g00649_t001 | 718 | 82,911.95 | 5.85 | 37.53 | 76.14 | −0.431 | Chloroplast |
| VvFRS28 | Vitvi09g04199_t001 | 355 | 41,623.75 | 8.63 | 45.54 | 85.41 | −0.402 | Nucleus |
| VvFRS29 | Vitvi09g04200_t001 | 320 | 36,041.05 | 9.20 | 44.62 | 70.94 | −0.55 | Chloroplast |
| VvFRS30 | Vitvi09g00650_t001 | 698 | 81,102.44 | 5.37 | 42.64 | 74 | −0.561 | Cytoplasm |
| VvFRS31 | Vitvi10g00230_t001 | 690 | 80,193.97 | 5.98 | 45.84 | 74.17 | −0.416 | Nucleus |
| VvFRS32 | Vitvi12g00713_t001 | 692 | 80,089.01 | 6.21 | 46.85 | 79.57 | −0.364 | Nucleus |
| VvFRS33 | Vitvi12g02073_t001 | 678 | 77,793.76 | 8.08 | 42.55 | 77.83 | −0.354 | Nucleus |
| VvFRS34 | Vitvi13g01139_t001 | 858 | 97,221.73 | 6.16 | 48.68 | 72.52 | −0.451 | Cell wall |
| VvFRS35 | Vitvi14g00948_t001 | 647 | 75,285.04 | 6.73 | 46.49 | 76.57 | −0.43 | Nucleus |
| VvFRS36 | Vitvi14g01432_t001 | 386 | 43,770.41 | 5.77 | 55.88 | 64.64 | −0.698 | Nucleus |
| VvFRS37 | Vitvi14g01622_t001 | 835 | 96,149.99 | 6.32 | 39.69 | 73.62 | −0.523 | Chloroplast |
| VvFRS38 | Vitvi14g02030_t001 | 855 | 97,537.38 | 6.00 | 49.11 | 68.78 | −0.483 | Nucleus |
| VvFRS39 | Vitvi18g00039_t001 | 759 | 86,033.65 | 6.14 | 46.26 | 71.71 | −0.511 | Chloroplast |
| VvFRS40 | Vitvi18g01884_t001 | 689 | 76,873.44 | 5.34 | 49.22 | 75.99 | −0.535 | Chloroplast |
| VvFRS41 | Vitvi18g03049_t001 | 623 | 69,497.33 | 5.73 | 47.02 | 69.52 | −0.722 | Nucleus |
| VvFRS42 | Vitvi18g04647_t001 | 365 | 40,838.6 | 6.07 | 30.46 | 68.14 | −0.646 | Nucleus |
| VvFRS43 | Vitvi18g03185_t001 | 451 | 50,798.04 | 5.60 | 42.97 | 76.32 | −0.644 | Nucleus |
| Name | Sequence Length | Amino Acid Number/Proportion | |||
|---|---|---|---|---|---|
| Alpha Helix (Hh) | Extended Strand (Ee) | Beta Turn (Tt) | Random Coil (Cc) | ||
| VvFRS1 | 985 | 339/34.42% | 89/9.04% | 30/3.05% | 527/53.50% |
| VvFRS2 | 755 | 309/40.93% | 81/10.73% | 26/3.44% | 339/44.90% |
| VvFRS3 | 752 | 346/46.01% | 102/13.56% | 23/3.06% | 281/37.37% |
| VvFRS4 | 783 | 322/41.12% | 88/11.24% | 25/3.19% | 348/44.44% |
| VvFRS5 | 549 | 275/50.09% | 51/9.29% | 14/2.55% | 209/38.07% |
| VvFRS6 | 847 | 319/37.66% | 85/10.04% | 28/3.31% | 415/49.00% |
| VvFRS7 | 841 | 328/39.00% | 81/9.63% | 25/2.97% | 407/48.39% |
| VvFRS8 | 857 | 305/35.59% | 84/9.80% | 23/2.68% | 445/51.93% |
| VvFRS9 | 668 | 310/46.41% | 83/12.43% | 27/4.04% | 248/37.13% |
| VvFRS10 | 751 | 325/43.28% | 83/11.05% | 27/3.60% | 316/42.08% |
| VvFRS11 | 746 | 311/41.69% | 82/10.99% | 24/3.22% | 329/44.10% |
| VvFRS12 | 781 | 332/42.51% | 113/14.47% | 27/3.46% | 309/39.56% |
| VvFRS13 | 439 | 140/31.89% | 61/13.90% | 18/4.10% | 220/50.11% |
| VvFRS14 | 305 | 110/36.07% | 39/12.79% | 8/2.62% | 148/48.52% |
| VvFRS15 | 246 | 81/32.93% | 28/11.38% | 10/4.07% | 127/51.63% |
| VvFRS16 | 319 | 113/35.42% | 36/11.29% | 11/3.45% | 159/49.84% |
| VvFRS17 | 215 | 86/40.00% | 29/13.49% | 11/5.12% | 89/41.40% |
| VvFRS18 | 215 | 85/39.53% | 30 /13.95% | 10/4.65% | 90/41.86% |
| VvFRS19 | 883 | 356/40.32% | 103/11.66% | 30/3.40% | 394/44.62% |
| VvFRS20 | 758 | 329/43.40% | 80/10.55% | 27/3.56% | 322/42.48% |
| VvFRS21 | 239 | 87/36.40% | 29/12.13% | 13/5.44% | 110/46.03% |
| VvFRS22 | 218 | 84/38.53% | 29/13.30% | 10/4.59% | 95/43.58% |
| VvFRS23 | 209 | 84/40.19% | 31/14.83% | 8/3.83% | 86/41.15% |
| VvFRS24 | 252 | 82/32.54% | 29/11.51% | 12/4.76% | 129/51.19% |
| VvFRS25 | 894 | 343/38.37% | 103/11.52% | 26/2.91% | 422/47.20% |
| VvFRS26 | 742 | 314/42.32% | 83/11.19% | 23/3.10% | 322/43.40% |
| VvFRS27 | 718 | 311/43.31% | 84/11.70% | 29/4.04% | 294/40.95% |
| VvFRS28 | 355 | 222/62.54% | 25/7.04% | 8/2.25% | 100/28.17% |
| VvFRS29 | 320 | 48/15.00% | 40/12.50% | 14/4.38% | 218/68.12% |
| VvFRS30 | 698 | 325/46.56% | 85/12.18% | 23/3.30% | 265/37.97% |
| VvFRS31 | 690 | 324/46.96% | 80/11.59% | 27/3.91% | 259/37.54% |
| VvFRS32 | 692 | 316/45.66% | 80/11.56% | 26/3.76% | 270/39.02% |
| VvFRS33 | 678 | 315/46.46% | 78/11.50% | 27/3.98% | 258/8.05% |
| VvFRS34 | 858 | 344/40.09% | 90/10.49% | 21/2.45% | 403/46.97% |
| VvFRS35 | 647 | 314/48.53% | 79/12.21% | 25/3.86% | 229/35.39% |
| VvFRS36 | 386 | 114/29.53% | 29/7.51% | 8/2.07% | 235/60.88% |
| VvFRS37 | 835 | 310/37.13% | 111/13.29% | 30/3.59% | 384/45.99% |
| VvFRS38 | 855 | 336/39.30% | 94/10.99% | 24/2.81% | 401/46.90% |
| VvFRS39 | 759 | 333/43.87% | 82/10.80% | 26/3.43% | 318/41.90% |
| VvFRS40 | 689 | 275/39.91% | 69/10.01% | 32/4.64% | 313/45.43% |
| VvFRS41 | 623 | 288/46.23% | 64/10.27% | 22/3.53% | 249/39.97% |
| VvFRS42 | 365 | 131/35.89% | 39/10.68% | 14/3.84% | 181/49.59% |
| VvFRS43 | 451 | 146/32.37% | 56/12.42% | 19/4.21% | 230/51.00% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, H.; Zheng, Y.; Sun, Y.; Liu, Y. Genome-Wide Identification and Expression Analysis of the FAR1-RELATED SEQUENCE (FRS) Gene Family in Grape (Vitis vinifera L.). Int. J. Mol. Sci. 2025, 26, 4675. https://doi.org/10.3390/ijms26104675
Yao H, Zheng Y, Sun Y, Liu Y. Genome-Wide Identification and Expression Analysis of the FAR1-RELATED SEQUENCE (FRS) Gene Family in Grape (Vitis vinifera L.). International Journal of Molecular Sciences. 2025; 26(10):4675. https://doi.org/10.3390/ijms26104675
Chicago/Turabian StyleYao, Heng, Yanyan Zheng, Yanzhao Sun, and Yang Liu. 2025. "Genome-Wide Identification and Expression Analysis of the FAR1-RELATED SEQUENCE (FRS) Gene Family in Grape (Vitis vinifera L.)" International Journal of Molecular Sciences 26, no. 10: 4675. https://doi.org/10.3390/ijms26104675
APA StyleYao, H., Zheng, Y., Sun, Y., & Liu, Y. (2025). Genome-Wide Identification and Expression Analysis of the FAR1-RELATED SEQUENCE (FRS) Gene Family in Grape (Vitis vinifera L.). International Journal of Molecular Sciences, 26(10), 4675. https://doi.org/10.3390/ijms26104675





