Potential Influence of ADAM9 Genetic Variants and Expression Levels on the EGFR Mutation Status and Disease Progression in Patients with Lung Adenocarcinoma
Abstract
1. Introduction
2. Results
2.1. Demographic and Clinical Profiles of LUAD Patients with WT or Mutant EGFR
2.2. Distribution of ADAM9 Candidate SNPs (rs6474526, rs7006414, rs10105311, and rs78451751) Among LUAD Patients and Their Correlations with the EGFR Mutation Status
2.3. Associations Between ADAM9 Polymorphic Genotypes and Clinicopathological Features of LUAD Patients with WT or Mutant EGFR
2.4. Associations Between ADAM9 Polymorphic Genotypes and Clinicopathological Characteristics in Female LUAD Patients and Non-Smoking LUAD Patients
2.5. Potential Effects of ADAM9 Genetic Variants on ADAM9 Expression
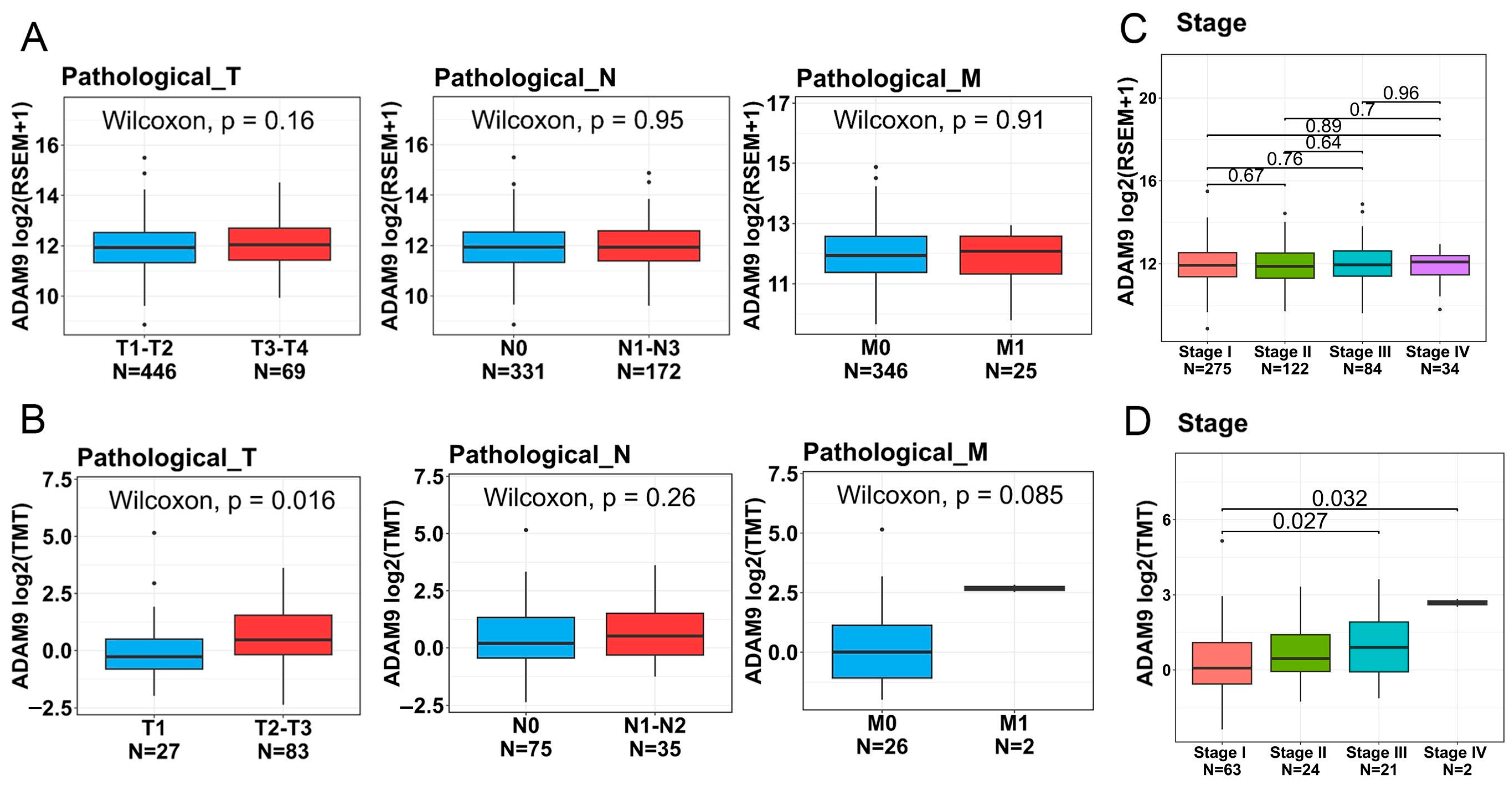
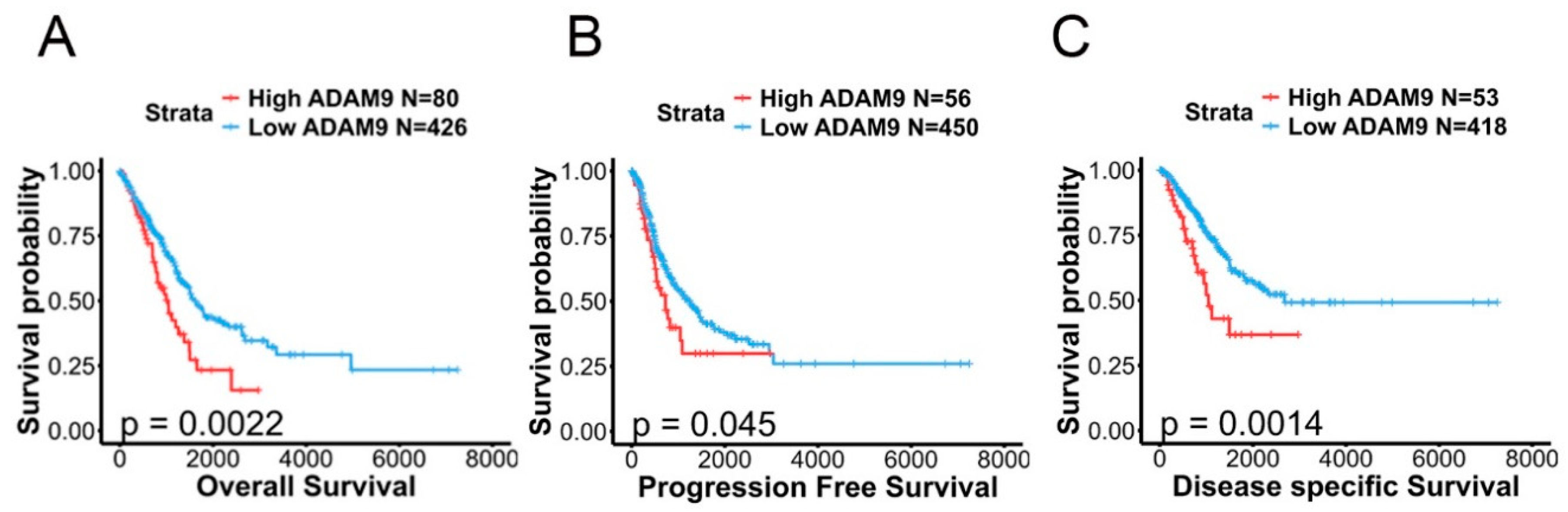
2.6. Associations Between ADAM9 Expression Levels and Clinicopathological Characteristics as Well as Prognoses of LUAD Patients
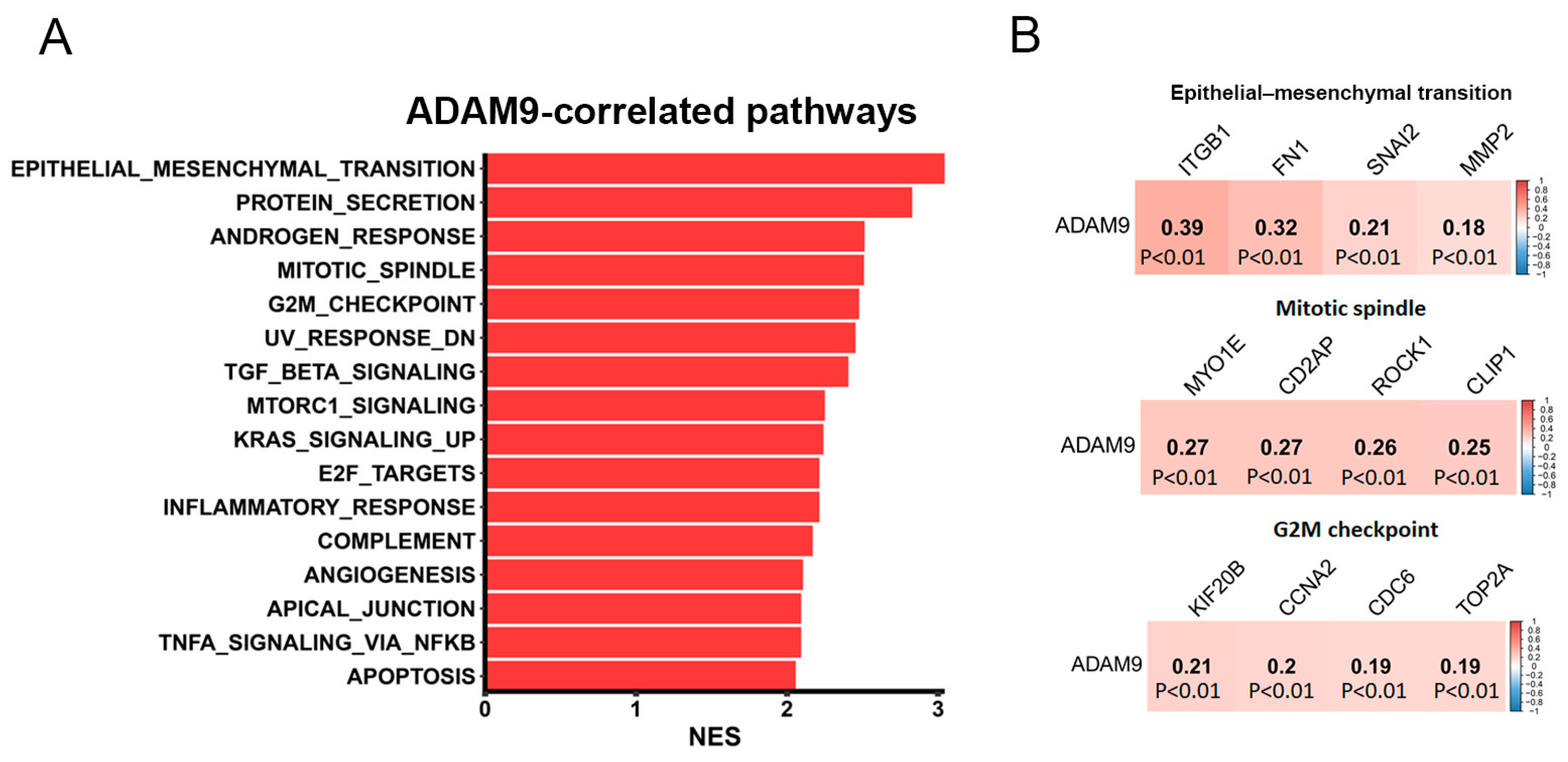
2.7. Investigation of Potential Molecular Mechanisms Regulated by ADAM9 in LUAD Progression
3. Discussion
4. Materials and Methods
4.1. Study Populations and Ethics
4.2. Sequencing of EGFR from Tumor Tissues
4.3. Genotyping of ADAM9 SNPs from Whole-Blood Samples
4.4. Bioinformatics Analysis
4.5. Western Blot Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Dearden, S.; Stevens, J.; Wu, Y.L.; Blowers, D. Mutation incidence and coincidence in non small-cell lung cancer: Meta-analyses by ethnicity and histology (mutMap). Ann. Oncol. 2013, 24, 2371–2376. [Google Scholar] [CrossRef]
- Midha, A.; Dearden, S.; McCormack, R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar] [PubMed]
- Hendriks, L.E.L.; Remon, J.; Faivre-Finn, C.; Garassino, M.C.; Heymach, J.V.; Kerr, K.M.; Tan, D.S.W.; Veronesi, G.; Reck, M. Non-small-cell lung cancer. Nat. Rev. Dis. Primers 2024, 10, 71. [Google Scholar] [CrossRef] [PubMed]
- Benusiglio, P.R.; Fallet, V.; Sanchis-Borja, M.; Coulet, F.; Cadranel, J. Lung cancer is also a hereditary disease. Eur. Respir. Rev. 2021, 30, 210045. [Google Scholar] [CrossRef]
- Siegelin, M.D.; Borczuk, A.C. Epidermal growth factor receptor mutations in lung adenocarcinoma. Lab. Investig. 2014, 94, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.Y. Next-generation sequencing methods: Impact of sequencing accuracy on SNP discovery. Methods Mol. Biol. 2009, 578, 95–111. [Google Scholar]
- Hunt, R.; Sauna, Z.E.; Ambudkar, S.V.; Gottesman, M.M.; Kimchi-Sarfaty, C. Silent (synonymous) SNPs: Should we care about them? Methods Mol. Biol. 2009, 578, 23–39. [Google Scholar]
- Shi, J.; Shiraishi, K.; Choi, J.; Matsuo, K.; Chen, T.Y.; Dai, J.; Hung, R.J.; Chen, K.; Shu, X.O.; Kim, Y.T.; et al. Genome-wide association study of lung adenocarcinoma in East Asia and comparison with a European population. Nat. Commun. 2023, 14, 3043. [Google Scholar] [CrossRef]
- Seow, W.J.; Matsuo, K.; Hsiung, C.A.; Shiraishi, K.; Song, M.; Kim, H.N.; Wong, M.P.; Hong, Y.C.; Hosgood, H.D.; Wang, Z., 3rd; et al. Association between GWAS-identified lung adenocarcinoma susceptibility loci and EGFR mutations in never-smoking Asian women, and comparison with findings from Western populations. Hum. Mol. Genet. 2017, 26, 454–465. [Google Scholar]
- Haoyuan, M.A.; Yanshu, L.I. Structure, regulatory factors and cancer-related physiological effects of ADAM9. Cell Adh. Migr. 2020, 14, 165–181. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.W.; Huang, Y.K.; Kuo, T.T.; Liu, J.P.; Sher, Y.P. An Overview of ADAM9: Structure, Activation, and Regulation in Human Diseases. Int. J. Mol. Sci. 2020, 21, 7790. [Google Scholar] [CrossRef]
- Kossmann, C.M.; Annereau, M.; Thomas-Schoemann, A.; Nicco-Overney, C.; Chéreau, C.; Batteux, F.; Alexandre, J.; Lemare, F. ADAM9 expression promotes an aggressive lung adenocarcinoma phenotype. Tumour Biol. 2017, 39, 1010428317716077. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Cho, C.F.; Bai, S.T.; Liu, J.P.; Kuo, T.T.; Wang, L.J.; Lin, Y.S.; Lin, C.C.; Lai, L.C.; Lu, T.P.; et al. ADAM9 promotes lung cancer progression through vascular remodeling by VEGFA, ANGPT2, and PLAT. Sci. Rep. 2017, 7, 15108. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, J.; Chen, N.; Fu, W.; Zhou, B.; He, A. High expression of a disintegrin and metalloproteinase-9 predicts a shortened survival time in completely resected stage I non-small cell lung cancer. Oncol. Lett. 2013, 5, 1461–1466. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Hirata, M.; Hasuwa, H.; Iwamoto, R.; Umata, T.; Miyado, K.; Tamai, Y.; Kurisaki, T.; Sehara-Fujisawa, A.; Ohno, S.; et al. A metalloprotease-disintegrin, MDC9/meltrin-gamma/ADAM9 and PKCdelta are involved in TPA-induced ectodomain shedding of membrane-anchored heparin-binding EGF-like growth factor. Embo J. 1998, 17, 7260–7272. [Google Scholar] [CrossRef]
- Ongusaha, P.P.; Kwak, J.C.; Zwible, A.J.; Macip, S.; Higashiyama, S.; Taniguchi, N.; Fang, L.; Lee, S.W. HB-EGF is a potent inducer of tumor growth and angiogenesis. Cancer Res. 2004, 64, 5283–5290. [Google Scholar] [CrossRef]
- Chiu, K.L.; Lin, Y.S.; Kuo, T.T.; Lo, C.C.; Huang, Y.K.; Chang, H.F.; Chuang, E.Y.; Lin, C.C.; Cheng, W.C.; Liu, Y.N.; et al. ADAM9 enhances CDCP1 by inhibiting miR-1 through EGFR signaling activation in lung cancer metastasis. Oncotarget 2017, 8, 47365–47378. [Google Scholar] [CrossRef]
- Lin, C.Y.; Chen, H.J.; Huang, C.C.; Lai, L.C.; Lu, T.P.; Tseng, G.C.; Kuo, T.T.; Kuok, Q.Y.; Hsu, J.L.; Sung, S.Y.; et al. ADAM9 promotes lung cancer metastases to brain by a plasminogen activator-based pathway. Cancer Res. 2014, 74, 5229–5243. [Google Scholar] [CrossRef]
- Choi, W.I.; Jeong, J.; Lee, C.W. Association between EGFR mutation and ageing, history of pneumonia and gastroesophageal reflux disease among patients with advanced lung cancer. Eur. J. Cancer 2019, 122, 101–108. [Google Scholar] [CrossRef]
- Dreymueller, D.; Uhlig, S.; Ludwig, A. ADAM-family metalloproteinases in lung inflammation: Potential therapeutic targets. Am. J. Physiol. Lung Cell Mol. Physiol. 2015, 308, L325–L343. [Google Scholar] [CrossRef]
- Lin, Y.W.; Wen, Y.C.; Lin, C.Y.; Hsiao, C.H.; Ho, K.H.; Huang, H.C.; Chang, L.C.; Wang, S.S.; Yang, S.F.; Chien, M.H. Genetic variants of ADAM9 as potential predictors for biochemical recurrence in prostate cancer patients after receiving a radical prostatectomy. Int. J. Med. Sci. 2024, 21, 2934–2942. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Jia, J. Promoter polymorphisms which regulate ADAM9 transcription are protective against sporadic Alzheimer’s disease. Neurobiol. Aging 2011, 32, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, K.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.Y.; Choi, S.J.; Cho, J.H.; Choi, H.J.; Lee, J.; Jung, K.; Irwin, D.; Liu, X.; Lira, M.E.; Mao, M.; et al. Lung cancer in never-smoker Asian females is driven by oncogenic mutations, most often involving EGFR. Oncotarget 2015, 6, 5465–5474. [Google Scholar] [CrossRef]
- Liu, R.; Gu, J.; Jiang, P.; Zheng, Y.; Liu, X.; Jiang, X.; Huang, E.; Xiong, S.; Xu, F.; Liu, G.; et al. DNMT1-microRNA126 epigenetic circuit contributes to esophageal squamous cell carcinoma growth via ADAM9-EGFR-AKT signaling. Clin. Cancer Res. 2015, 21, 854–863. [Google Scholar] [CrossRef]
- Fu, Q.; Cheng, J.; Zhang, J.; Zhang, Y.; Chen, X.; Luo, S.; Xie, J. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol. Rep. 2017, 37, 123–130. [Google Scholar] [CrossRef]
- Deng, N.; Zhou, H.; Fan, H.; Yuan, Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget 2017, 8, 110635–110649. [Google Scholar] [CrossRef]
- Gorlova, O.; Fedorov, A.; Logothetis, C.; Amos, C.; Gorlov, I. Genes with a large intronic burden show greater evolutionary conservation on the protein level. BMC Evol. Biol. 2014, 14, 50. [Google Scholar] [CrossRef]
- Kalsotra, A.; Cooper, T.A. Functional consequences of developmentally regulated alternative splicing. Nat. Rev. Genet. 2011, 12, 715–729. [Google Scholar] [CrossRef]
- Do, T.N.; Ucisik-Akkaya, E.; Davis, C.F.; Morrison, B.A.; Dorak, M.T. An intronic polymorphism of IRF4 gene influences gene transcription in vitro and shows a risk association with childhood acute lymphoblastic leukemia in males. Biochim. Biophys. Acta 2010, 1802, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Campbell, T.M.; Castro, M.A.A.; de Santiago, I.; Fletcher, M.N.C.; Halim, S.; Prathalingam, R.; Ponder, B.A.J.; Meyer, K.B. FGFR2 risk SNPs confer breast cancer risk by augmenting oestrogen responsiveness. Carcinogenesis 2016, 37, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Yao, X.Y.; Wang, H.Y.; Li, Y.J.; Zhang, X.X.; Sun, C. Breast cancer-associated SNP rs72755295 is a cis-regulatory variation for human EXO1. Genet. Mol. Biol. 2022, 45, e20210420. [Google Scholar] [CrossRef] [PubMed]
- Ahrendt, S.A.; Decker, P.A.; Alawi, E.A.; Zhu, Y.R.; Sanchez-Cespedes, M.; Yang, S.C.; Haasler, G.B.; Kajdacsy-Balla, A.; Demeure, M.J.; Sidransky, D. Cigarette smoking is strongly associated with mutation of the K-ras gene in patients with primary adenocarcinoma of the lung. Cancer 2001, 92, 1525–1530. [Google Scholar] [CrossRef]
- Li, D.; Wang, T.; Ma, Q.; Zhou, L.; Le, Y.; Rao, Y.; Jin, L.; Pei, Y.; Cheng, Y.; Huang, C.; et al. IL-17A Promotes Epithelial ADAM9 Expression in Cigarette Smoke-Related COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2022, 17, 2589–2602. [Google Scholar] [CrossRef]
- Huang, Y.K.; Cheng, W.C.; Kuo, T.T.; Yang, J.C.; Wu, Y.C.; Wu, H.H.; Lo, C.C.; Hsieh, C.Y.; Wong, S.C.; Lu, C.H.; et al. Inhibition of ADAM9 promotes the selective degradation of KRAS and sensitizes pancreatic cancers to chemotherapy. Nat. Cancer 2024, 5, 400–419. [Google Scholar] [CrossRef]
- Menju, T.; Date, H. Lung cancer and epithelial-mesenchymal transition. Gen. Thorac. Cardiovasc. Surg. 2021, 69, 781–789. [Google Scholar] [CrossRef]
- Cui, Y.; Wang, X.; Zhang, L.; Liu, W.; Ning, J.; Gu, R.; Cui, Y.; Cai, L.; Xing, Y. A novel epithelial-mesenchymal transition (EMT)-related gene signature of predictive value for the survival outcomes in lung adenocarcinoma. Front. Oncol. 2022, 12, 974614. [Google Scholar] [CrossRef]
- Lo, H.W.; Hsu, S.C.; Xia, W.; Cao, X.; Shih, J.Y.; Wei, Y.; Abbruzzese, J.L.; Hortobagyi, G.N.; Hung, M.C. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007, 67, 9066–9076. [Google Scholar] [CrossRef]
- Pallier, K.; Cessot, A.; Côté, J.F.; Just, P.A.; Cazes, A.; Fabre, E.; Danel, C.; Riquet, M.; Devouassoux-Shisheboran, M.; Ansieau, S.; et al. TWIST1 a new determinant of epithelial to mesenchymal transition in EGFR mutated lung adenocarcinoma. PLoS ONE 2012, 7, e29954. [Google Scholar] [CrossRef]
- Sher, Y.P.; Wang, L.J.; Chuang, L.L.; Tsai, M.H.; Kuo, T.T.; Huang, C.C.; Chuang, E.Y.; Lai, L.C. ADAM9 up-regulates N-cadherin via miR-218 suppression in lung adenocarcinoma cells. PLoS ONE 2014, 9, e94065. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Tsao, S.M.; Li, Y.T.; Lee, C.Y.; Tsao, T.C.; Hsieh, M.J.; Yang, S.F. The Relationship between Long Noncoding RNA H19 Polymorphism and the Epidermal Growth Factor Receptor Phenotypes on the Clinicopathological Characteristics of Lung Adenocarcinoma. Int. J. Environ. Res. Public Health 2021, 18, 2862. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.W.; Wang, S.S.; Wen, Y.C.; Tung, M.C.; Lee, L.M.; Yang, S.F.; Chien, M.H. Genetic Variations of Melatonin Receptor Type 1A are Associated with the Clinicopathologic Development of Urothelial Cell Carcinoma. Int. J. Med. Sci. 2017, 14, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.W.; Wen, Y.C.; Chu, C.Y.; Tung, M.C.; Yang, Y.C.; Hua, K.T.; Pan, K.F.; Hsiao, M.; Lee, W.J.; Chien, M.H. Stabilization of ADAM9 by N-α-acetyltransferase 10 protein contributes to promoting progression of androgen-independent prostate cancer. Cell Death. Dis. 2020, 11, 591. [Google Scholar] [CrossRef]


| Variable | EGFR Wild-Type (N = 219) n (%) | EGFR Mutation (N = 316) n (%) | p-Value |
|---|---|---|---|
| Age (years) | |||
| Mean ± SD | 64.63 ± 12.23 | 65.36 ± 12.51 | p = 0.523 |
| Gender | |||
| Male | 134 (61.2%) | 105 (33.2%) | p < 0.001 |
| Female | 85 (38.8%) | 211 (66.8%) | |
| Cigarette smoking status | |||
| Never-smoker | 103 (47.0%) | 255 (80.7%) | p < 0.001 |
| Ever-smoker | 116 (53.0%) | 61 (19.3%) | |
| Stage | |||
| I or II | 48 (21.9%) | 76 (24.1%) | p = 0.565 |
| III or IV | 171 (78.1%) | 240 (75.9%) | |
| Tumor T status | |||
| T1 or T2 | 108 (49.3%) | 171 (54.1%) | p = 0.275 |
| T3 or T4 | 111 (50.7%) | 145 (45.9%) | |
| Lymph node status | |||
| Negative | 59 (26.9%) | 89 (28.2%) | p = 0.756 |
| Positive | 160 (73.1%) | 227 (71.8%) | |
| Distant metastasis | |||
| Negative | 97 (44.3%) | 126 (39.9%) | p = 0.308 |
| Positive | 122 (55.7%) | 190 (60.1%) | |
| Cell differentiation | |||
| Well | 18 (8.2%) | 35 (11.1%) | p < 0.001 |
| Moderately | 136 (62.1%) | 248 (78.5%) | |
| Poorly | 65 (29.7%) | 33 (10.4%) |
| Genotypes | EGFR Wild-Type (N = 219) | EGFR Mutation (N = 316) | AOR (95% CI) | p-Value |
|---|---|---|---|---|
| rs6474526 | ||||
| TT | 192 (87.7%) | 296 (93.7%) | 1.000 | |
| TG | 27 (12.3%) | 20 (6.3%) | 0.477 (0.260~0.875) | p = 0.017 * |
| GG | 0 (0.0%) | 0 (0.0%) | --- | --- |
| TG + GG | 27 (12.3%) | 20 (6.3%) | 0.477 (0.260~0.875) | p = 0.017 * |
| rs7006414 | ||||
| TT | 136 (62.1%) | 198 (62.7%) | 1.000 | |
| TC | 67 (30.6%) | 102 (32.3%) | 1.156 (0.769~1.738) | p = 0.485 |
| CC | 16 (7.3%) | 16 (5.0%) | 0.652 (0.302~1.412) | p = 0.278 |
| TC + CC | 83 (37.9%) | 118 (37.3%) | 1.051 (0.717~1.540) | p = 0.799 |
| rs10105311 | ||||
| CC | 155 (70.8%) | 219 (69.3%) | 1.000 | |
| CT | 56 (25.6%) | 83 (26.3%) | 1.073 (0.700~1.644) | p = 0.746 |
| TT | 8 (3.6%) | 14 (4.4%) | 1.017 (0.397~2.607) | p = 0.972 |
| CT + TT | 64 (29.2%) | 97 (30.7%) | 1.065 (0.711~1.595) | p = 0.759 |
| rs78451751 | ||||
| TT | 216 (98.6%) | 314 (99.4%) | 1.000 | |
| TC | 3 (1.4%) | 2 (0.6%) | 0.372 (0.058~2.408) | p = 0.300 |
| CC | 0 (0.0%) | 0 (0.0%) | --- | --- |
| TC + CC | 3 (1.4%) | 2 (0.6%) | 0.372 (0.058~2.408) | p = 0.300 |
| Variable | All (N = 535) | EGFR Wild-Type (N = 219) | EGFR Mutation (N = 316) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TT (N = 488) | TG + GG (N = 47) | p-Value | TT (N = 192) | TG + GG (N = 27) | p-Value | TT (N = 296) | TG + GG (N = 20) | p-Value | |
| Stage | |||||||||
| I or II | 116 (23.8%) | 8 (22.8%) | p = 0.295 | 42 (21.9%) | 6 (22.2%) | p = 0.967 | 74 (25.0%) | 2 (10.0%) | p = 0.129 |
| III or IV | 372 (76.2%) | 39 (83.0%) | 150 (78.1%) | 21 (77.8%) | 222 (75.0%) | 18 (90.0%) | |||
| Tumor T status | |||||||||
| T1 or T2 | 262 (53.7%) | 17 (36.2%) | p = 0.022 a | 97 (50.5%) | 11 (40.7%) | p = 0.341 | 165 (55.7%) | 6 (30.0%) | p = 0.025 b |
| T3 or T4 | 226 (46.3%) | 30 (63.8%) | 95 (49.5%) | 16 (59.3%) | 131 (44.3%) | 14 (70.0%) | |||
| Lymph node status | |||||||||
| Negative | 138 (28.3%) | 10 (21.3%) | p = 0.305 | 52 (27.1%) | 7 (25.9%) | p = 0.899 | 86 (29.1%) | 3 (15.0%) | p = 0.176 |
| Positive | 350 (71.7%) | 37 (78.7%) | 140 (72.9%) | 20 (74.1%) | 210 (70.9%) | 17 (85.0%) | |||
| Distant metastasis | |||||||||
| Negative | 208 (42.6%) | 15 (31.9%) | p = 0.155 | 87 (45.3%) | 10 (37.0%) | p = 0.418 | 121 (40.9%) | 5 (25.0%) | p = 0.160 |
| Positive | 280 (57.4%) | 32 (68.1%) | 105 (54.7%) | 17 (63.0%) | 175 (59.1%) | 15 (75.0%) | |||
| Cell differentiation | |||||||||
| Well/Moderately | 403 (82.6%) | 34 (72.3%) | p = 0.083 | 137 (71.4%) | 17 (63.0%) | p = 0.372 | 266 (89.9%) | 17 (85.0%) | p = 0.491 |
| Poorly | 85 (17.4%) | 13 (27.7%) | 55 (28.6%) | 10 (37.0%) | 30 (10.1%) | 3 (15.0%) | |||
| Variable | All (N = 535) | EGFR Wild-Type (N = 219) | EGFR Mutation (N = 316) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CC (N = 374) | CT + TT (N = 161) | p-Value | CC (N = 155) | CT + TT (N = 64) | p-Value | CC (N = 219) | CT + TT (N = 97) | p-Value | |
| Stage | |||||||||
| I or II | 82 (21.9%) | 42 (26.1%) | p = 0.295 | 28 (18.1%) | 20 (31.2%) | p = 0.032 a | 54 (24.7%) | 22 (22.7%) | p = 0.704 |
| III or IV | 292 (78.1%) | 119 (73.9%) | 127 (81.9%) | 44 (68.8%) | 165 (75.3%) | 75 (77.3%) | |||
| Tumor T status | |||||||||
| T1 or T2 | 190 (50.8%) | 89 (55.3%) | p = 0.342 | 74 (47.7%) | 34 (53.1%) | p = 0.469 | 116 (53.0%) | 55 (56.7%) | p = 0.539 |
| T3 or T4 | 184 (49.2%) | 72 (44.7%) | 81 (52.3%) | 30 (46.9%) | 103 (47.0%) | 42 (43.3%) | |||
| Lymph node status | |||||||||
| Negative | 102 (27.3%) | 46 (28.6%) | p = 0.758 | 36 (23.2%) | 23 (35.9%) | p = 0.054 | 66 (30.1%) | 23 (23.7%) | p = 0.241 |
| Positive | 272 (72.7%) | 115 (71.4%) | 119 (76.8%) | 41 (64.1%) | 153 (69.9%) | 74 (76.3%) | |||
| Distant metastasis | |||||||||
| Negative | 149 (39.8%) | 74 (46.0%) | p = 0.188 | 63 (40.6%) | 34 (53.1%) | p = 0.091 | 86 (39.3%) | 40 (41.2%) | p = 0.742 |
| Positive | 225 (60.2%) | 87 (54.0%) | 92 (59.4%) | 30 (46.9%) | 133 (60.7%) | 57 (58.8%) | |||
| Cell differentiation | |||||||||
| Well/Moderately | 306 (81.8%) | 131 (81.4%) | p = 0.901 | 113 (72.9%) | 41 (64.1%) | p = 0.193 | 193 (88.1%) | 90 (92.8%) | p = 0.212 |
| Poorly | 68 (18.2%) | 30 (18.6%) | 42 (27.1%) | 23 (35.9%) | 26 (11.9%) | 7 (7.2%) | |||
| Variable | Female (N = 296) | Non-Smoker (N = 358) | ||||||
|---|---|---|---|---|---|---|---|---|
| TT (N = 278) | TG + GG (N = 18) | OR (95% CI) | p-Value | TT (N = 336) | TG + GG (N = 22) | OR (95% CI) | p-Value | |
| Stage | ||||||||
| I or II | 73 (26.3%) | 2 (11.1%) | 1.000 | p = 0.152 | 87 (25.9%) | 2 (9.1%) | 1.000 | p = 0.077 |
| III or IV | 205 (73.7%) | 16 (88.9%) | 2.849 (0.639~12.692) | 249 (74.1%) | 20 (90.9%) | 3.494 (0.800~15.256) | ||
| Tumor T status | ||||||||
| T1 or T2 | 156 (56.1%) | 4 (22.2%) | 1.000 | p = 0.005 | 185 (55.1%) | 4 (18.2%) | 1.000 | p = 0.001 |
| T3 or T4 | 122 (43.9%) | 14 (77.8%) | 4.475 (1.437~13.940) | 151 (44.9%) | 18 (81.8%) | 5.513 (1.827~16.638) | ||
| Lymph node status | ||||||||
| Negative | 89 (32.0%) | 3 (16.7%) | 1.000 | p = 0.173 | 101 (30.1%) | 4 (18.2%) | 1.000 | p = 0.236 |
| Positive | 189 (68.0%) | 15 (83.3%) | 2.354 (0.665~8.342) | 235 (69.9%) | 18 (81.8%) | 1.934 (0.639~5.858) | ||
| Distant metastasis | ||||||||
| Negative | 118 (42.4%) | 3 (16.7%) | 1.000 | p = 0.031 | 140 (41.7%) | 3 (13.6%) | 1.000 | p = 0.009 |
| Positive | 160 (57.6%) | 15 (83.3%) | 3.688 (1.044~13.029) | 196 (58.3%) | 19 (86.4%) | 4.524 (1.313~15.583) | ||
| Cell differentiation | ||||||||
| Well/ Moderately | 243 (87.4%) | 14 (77.8%) | 1.000 | p = 0.242 | 288 (85.7%) | 16 (72.7%) | 1.000 | p = 0.099 |
| Poorly | 35 (12.6%) | 4 (22.2%) | 1.984 (0.618~6.368) | 48 (14.3%) | 6 (27.3%) | 2.250 (0.839~6.036) | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, J.-H.; Lai, T.-C.; Ho, K.-H.; Tsao, T.C.-Y.; Chang, L.-C.; Yang, S.-F.; Chien, M.-H. Potential Influence of ADAM9 Genetic Variants and Expression Levels on the EGFR Mutation Status and Disease Progression in Patients with Lung Adenocarcinoma. Int. J. Mol. Sci. 2025, 26, 4606. https://doi.org/10.3390/ijms26104606
Chang J-H, Lai T-C, Ho K-H, Tsao TC-Y, Chang L-C, Yang S-F, Chien M-H. Potential Influence of ADAM9 Genetic Variants and Expression Levels on the EGFR Mutation Status and Disease Progression in Patients with Lung Adenocarcinoma. International Journal of Molecular Sciences. 2025; 26(10):4606. https://doi.org/10.3390/ijms26104606
Chicago/Turabian StyleChang, Jer-Hwa, Tsung-Ching Lai, Kuo-Hao Ho, Thomas Chang-Yao Tsao, Lun-Ching Chang, Shun-Fa Yang, and Ming-Hsien Chien. 2025. "Potential Influence of ADAM9 Genetic Variants and Expression Levels on the EGFR Mutation Status and Disease Progression in Patients with Lung Adenocarcinoma" International Journal of Molecular Sciences 26, no. 10: 4606. https://doi.org/10.3390/ijms26104606
APA StyleChang, J.-H., Lai, T.-C., Ho, K.-H., Tsao, T. C.-Y., Chang, L.-C., Yang, S.-F., & Chien, M.-H. (2025). Potential Influence of ADAM9 Genetic Variants and Expression Levels on the EGFR Mutation Status and Disease Progression in Patients with Lung Adenocarcinoma. International Journal of Molecular Sciences, 26(10), 4606. https://doi.org/10.3390/ijms26104606








