Integrative ATAC-Seq and RNA-Seq Analysis Reveals Key Transcription Factors Mediating Low Salinity Adaptation in Penaeid Shrimp
Abstract
1. Introduction
2. Results
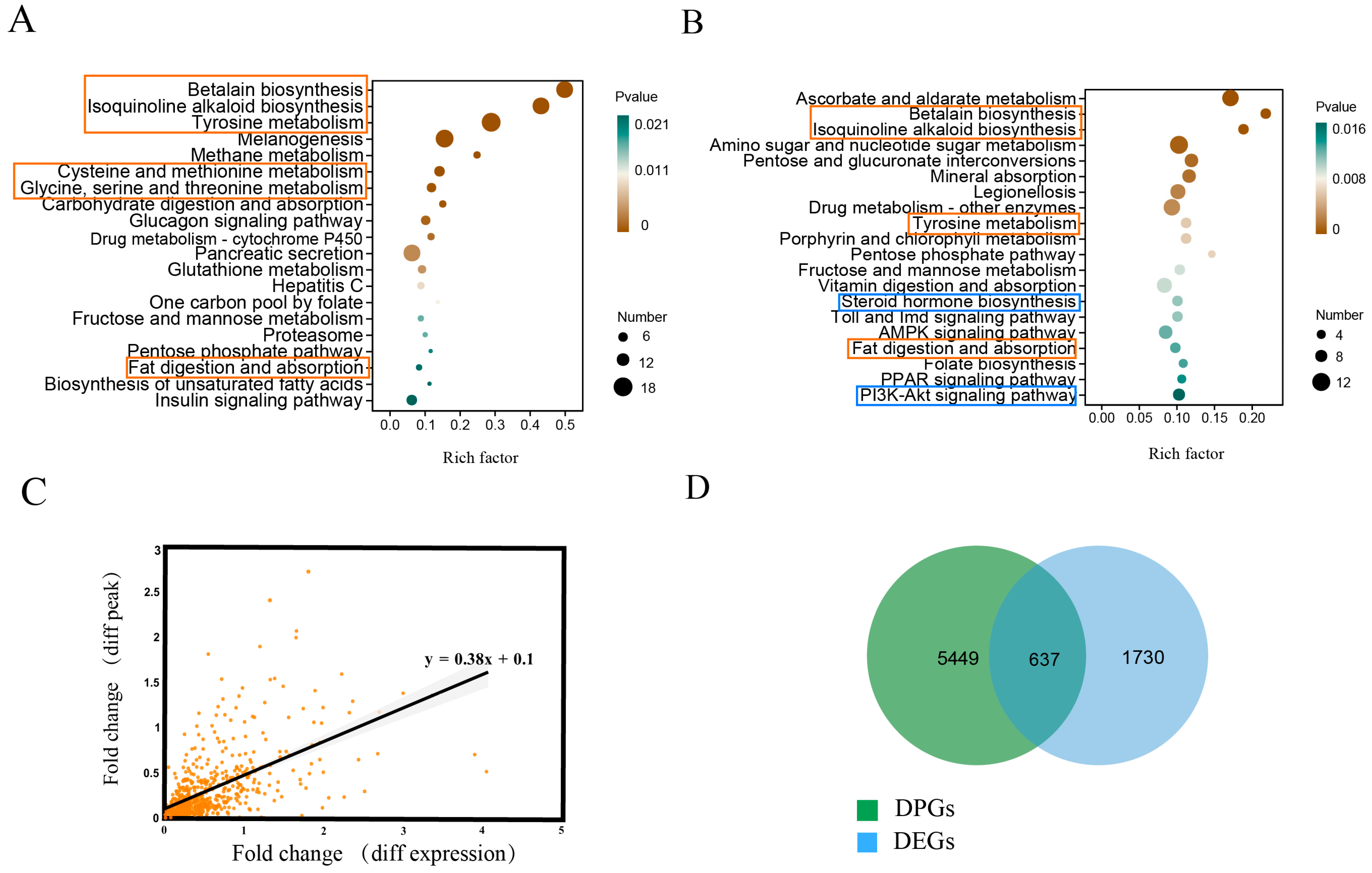
2.1. Differentially Expressed Genes in Shrimp in Response to Low Salinity Stress
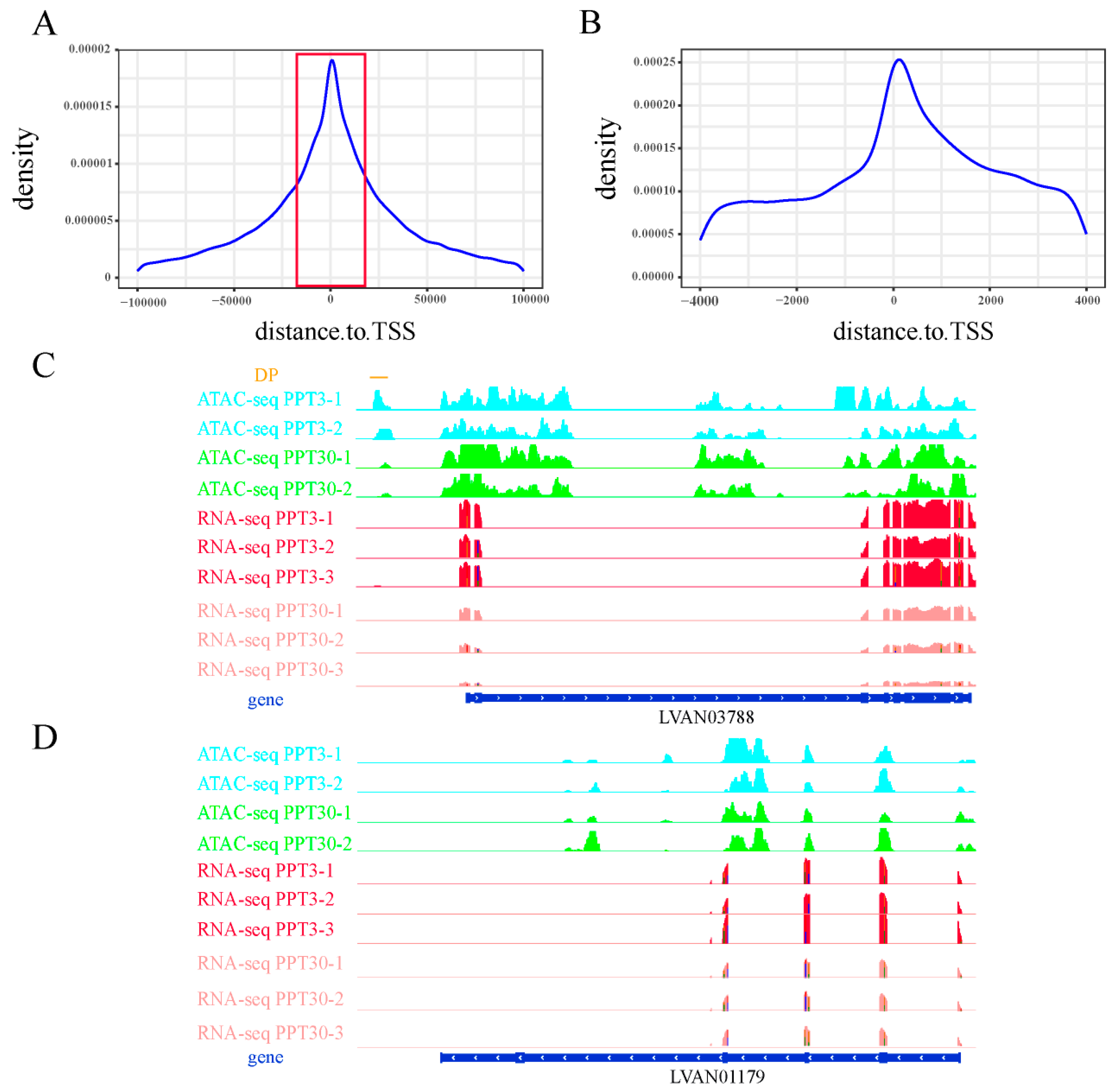
2.2. ATAC-Seq Analysis in Shrimp Under Low Salinity Stress
2.3. Integration of ATAC-Seq and RNA-Seq Reveals Unique Cis-Regulatory Mechanisms of Shrimp
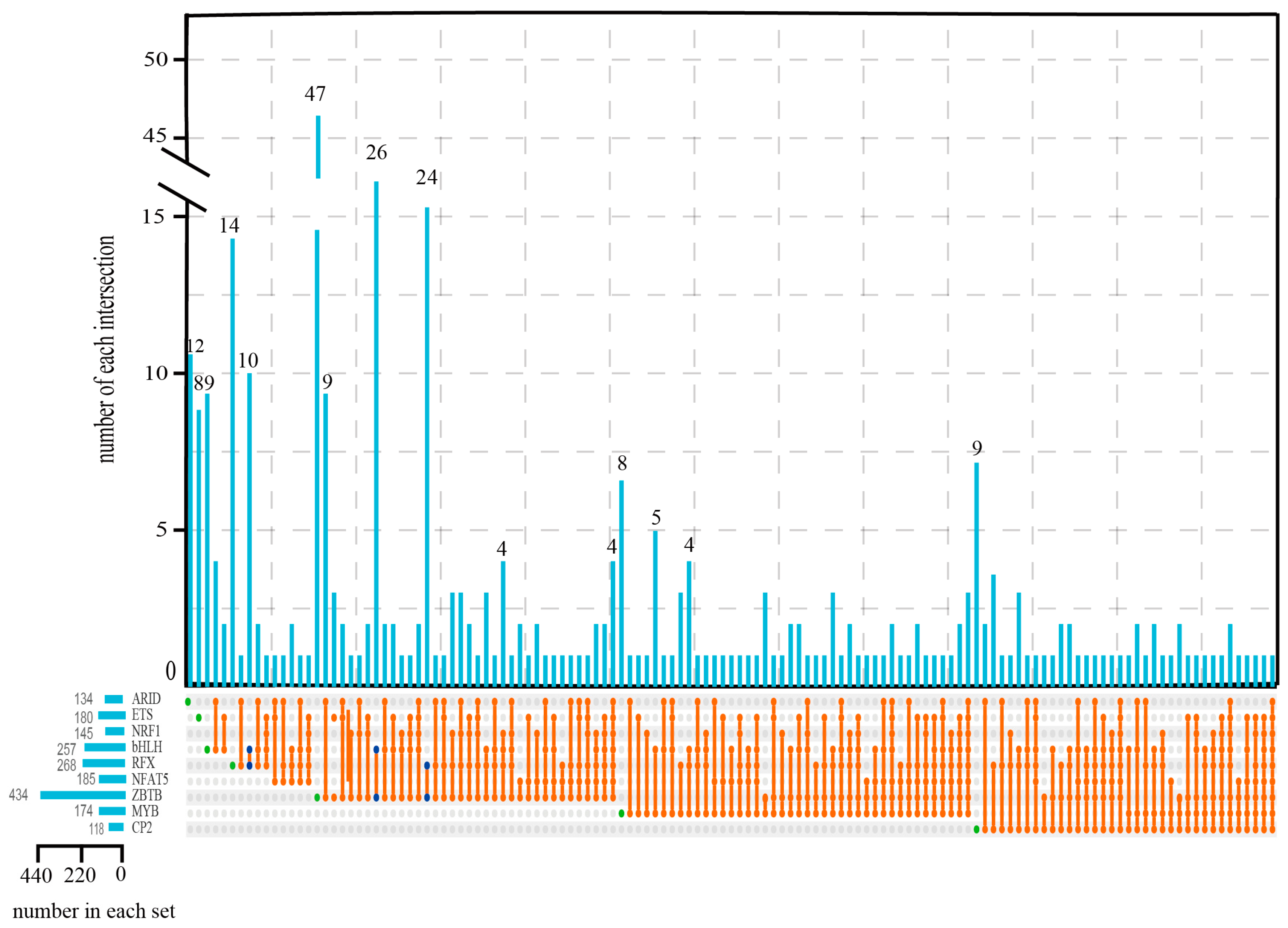
2.4. Identification of Salinity Adaptation-Related TFs in Shrimp
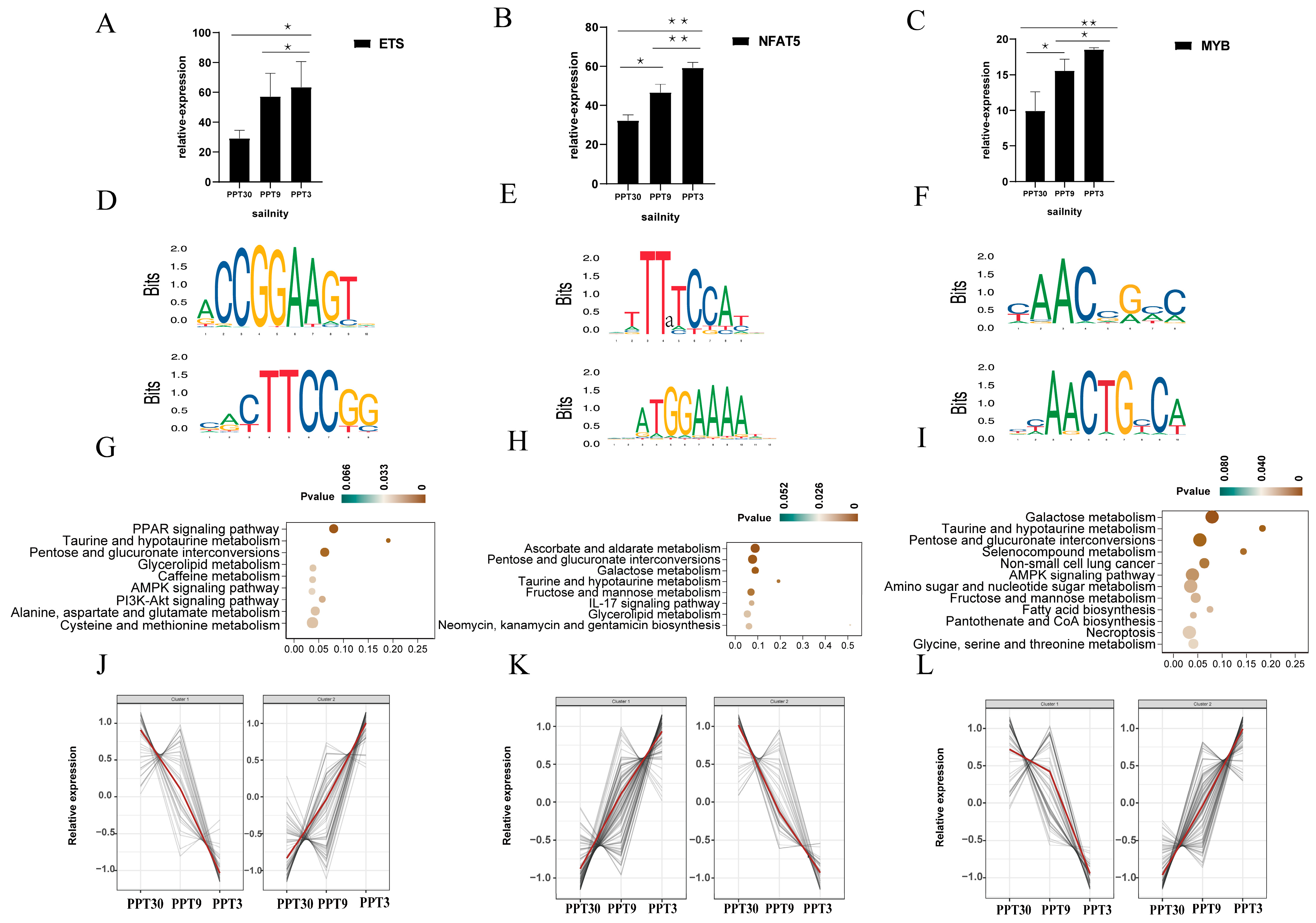
2.5. Characteristics and Targeted Genes of the Nine TFs
2.5.1. ZBTB, bHLH, and RFX
2.5.2. ETS, NFAT5, and MYB
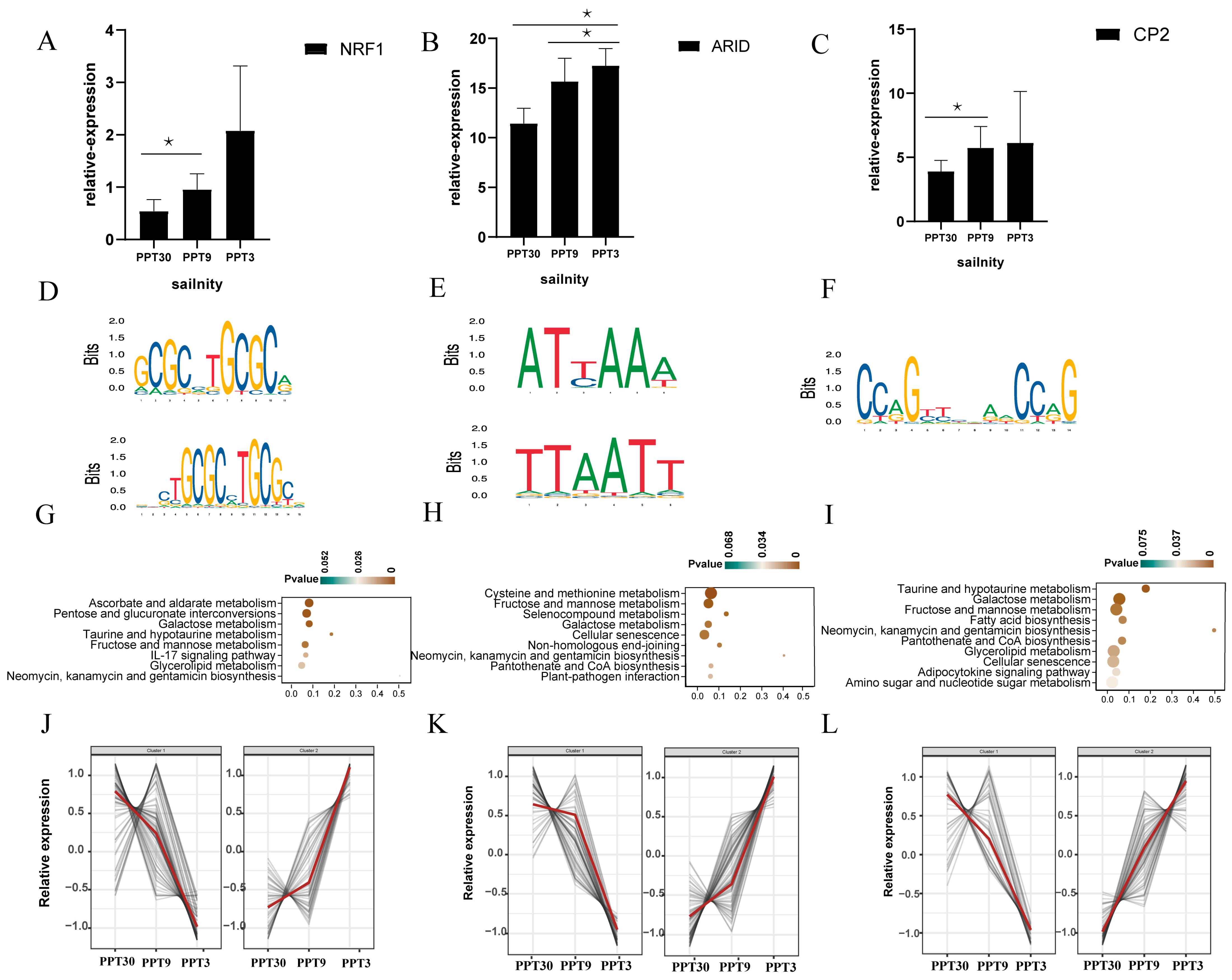
2.5.3. NRF1, ARID, and CP2
3. Discussion
3.1. Cis-Regulation Mechanisms of Salinity Adaptation in Shrimp
3.2. The Hepatopancreas: A Central Hub for Metabolic and Osmotic Adaptation to Low Salinity Stress
3.3. Novel Osmoregulation-Related TFs Identified in Shrimp
4. Materials and Methods
4.1. Shrimp Sampling for ATAC-Seq and RNA-Seq
4.2. RNA-Seq and Analyses
4.3. ATAC-Seq and Analyses
4.4. Integrative Analysis of ATAC-Seq and RNA-Seq
4.5. TF and TFBS Prediction
4.6. TFBS Prediction by AlphaFold3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perazzolo, L.M.; Gargioni, R.; Ogliari, P.; Barracco, M.A.A. Evaluation of some hemato-immunological parameters in the shrimp Farfantepenaeus paulensis submitted to environmental and physiological stress. Aquaculture 2002, 214, 19–33. [Google Scholar] [CrossRef]
- Ponce-Palafox, J.; Martinez-Palacios, C.A.; Ross, L.G. The effects of salinity and temperature on the growth and survival rates of juvenile white shrimp, Penaeus vannamei, Boone, 1931. Aquaculture 1997, 157, 107–115. [Google Scholar] [CrossRef]
- Perez-Velazquez, M.; Gonzalez-Felix, M.L.; Jaimes-Bustamente, F.; Martinez-Cordova, L.R.; Trujillo-Villalba, D.A.; Davis, D.A. Investigation of the effects of salinity and dietary protein level on growth and survival of Pacific white shrimp, Litopenaeus vannamei. J. World Aquac. Soc. 2007, 38, 475–485. [Google Scholar] [CrossRef]
- Alberto Valentino-Alvarez, J.; Nunez-Nogueira, G.; Fernandez-Bringas, L. Acute Toxicity of Arsenic under Different Temperatures and Salinity Conditions on the White Shrimp Litopenaeus vannamei. Biol. Trace Elem. Res. 2013, 152, 350–357. [Google Scholar] [CrossRef]
- Yang, Z.; Zhu, L.; Zhao, X.; Cheng, Y. Effects of salinity stress on osmotic pressure, free amino acids, and immune-associated parameters of the juvenile Chinese mitten crab, Eriocheir sinensis. Aquaculture 2022, 549, 737776. [Google Scholar] [CrossRef]
- Vieira, L.R.; Gravato, C.; Soares, A.M.V.M.; Morgado, F.; Guilhermino, L. Acute effects of copper and mercury on the estuarine fish Pomatoschistus microps: Linking biomarkers to behaviour. Chemosphere 2009, 76, 1416–1427. [Google Scholar] [CrossRef]
- Racotta, I.S.; Palacios, E. Hemolymph metabolic variables in response to experimental manipulation stress and serotonin injection in Penaeus vannamei. J. World Aquac. Soc. 1998, 29, 351–356. [Google Scholar] [CrossRef]
- Dzinyela, R.; Alhassan, A.R.; Suglo, P.; Movahedi, A. Advanced study of functional proteins involved in salt stress regulatory pathways in plants. S. Afr. J. Bot. 2023, 159, 425–438. [Google Scholar] [CrossRef]
- Minnoye, L.; Marinov, G.K.; Krausgruber, T.; Pan, L.; Marand, A.P.; Secchia, S.; Greenleaf, W.J.; Furlong, E.E.M.; Zhao, K.; Schmitz, R.J.; et al. Chromatin accessibility profiling methods. Nat. Rev. Methods Primers 2021, 1, 10. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, Z. Nrf2 is involved in osmoregulation, antioxidation and immunopotentiation in Coilia nasus under salinity stress. Biotechnol. Biotechnol. Equip. 2019, 33, 1453–1463. [Google Scholar] [CrossRef]
- Miller, W.A.; Terwilliger, N.B. Effects of molting and salinity stress on the expression of HIF, molting, and immune response genes in juvenile Cancer magister. Integr. Comp. Biol. 2009, 49, E273. [Google Scholar]
- Pan, L.; Si, L.; Hu, D. Ion Transport Signal Pathways Mediated by Neurotransmitter (Biogenic Amines) of Litopenaeus vannamei Under Low Salinity Challenge. J. Ocean Univ. China 2019, 18, 210–218. [Google Scholar] [CrossRef]
- Wang, X.; Chen, C.; He, C.; Chen, D.; Yan, W. Mapping open chromatin by ATAC-seq in bread wheat. Front. Plant Sci. 2022, 13, 1074873. [Google Scholar] [CrossRef]
- Merrill, C.B.; Montgomery, A.B.; Pabon, M.A.; Shabalin, A.A.; Rodan, A.R.; Rothenfluh, A. Harnessing changes in open chromatin determined by ATAC-seq to generate insulin-responsive reporter constructs. BMC Genom. 2022, 23, 399. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Liu, H.; Wang, T.; Zhang, T.; Kuang, J.; Luo, Y.; Chung, S.S.M.; Yuan, L.; Yang, J.Y. Tonicity-responsive microRNAs contribute to the maximal induction of osmoregulatory transcription factor OREBP in response to high-NaCl hypertonicity. Nucleic Acids Res. 2010, 39, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front. Plant Sci. 2021, 12, 677611. [Google Scholar] [CrossRef]
- Cheng, Z.-Y.; He, T.-T.; Gao, X.-M.; Zhao, Y.; Wang, J. ZBTB Transcription Factors: Key Regulators of the Development, Differentiation and Effector Function of T Cells. Front. Immunol. 2021, 12, 713294. [Google Scholar] [CrossRef]
- Qiu, J.-R.; Huang, Z.; Xiang, X.-Y.; Xu, W.-X.; Wang, J.-T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Sugiaman-Trapman, D.; Vitezic, M.; Jouhilahti, E.-M.; Mathelier, A.; Lauter, G.; Misra, S.; Daub, C.O.; Kere, J.; Swoboda, P. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genom. 2018, 19, 181. [Google Scholar] [CrossRef]
- Jaffer, Y.D.; Bhat, I.A.; Mir, I.N.; Bhat, R.A.H.; Sidiq, M.J.; Jana, P. Adaptation of cultured decapod crustaceans to changing salinities: Physiological responses, molecular mechanisms and disease implications. Rev. Aquacult. 2024, 16, 1520–1543. [Google Scholar] [CrossRef]
- Li, P.; Zheng, T.; Li, L.; Liu, W.; Qiu, L.; Ahmad, S.; Wang, J.; Cheng, T.; Zhang, Q. Integration of chromatin accessibility and gene expression reveals new regulators of cold hardening to enhance freezing tolerance in Prunus mume. J. Exp. Bot. 2023, 74, 2173–2187. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Zhou, Q.; Wong, W.H. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21521–21526. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses (May, 10.1038/s41580-022-00479-6, 2022). Nat. Rev. Mol. Cell Biol. 2022, 23, 516. [Google Scholar] [CrossRef]
- Vilarrasa-Blasi, J.; Vellosillo, T.; Jinkerson, R.E.; Fauser, F.; Xiang, T.; Minkoff, B.B.; Wang, L.; Kniazev, K.; Guzman, M.; Osaki, J.; et al. Multi-omics analysis of green lineage osmotic stress pathways unveils crucial roles of different cellular compartments. Nat. Commun. 2024, 15, 5988. [Google Scholar] [CrossRef]
- Meister, A. GLUTATHIONE METABOLISM AND ITS SELECTIVE MODIFICATION. J. Biol. Chem. 1988, 263, 17205–17208. [Google Scholar] [CrossRef]
- Sardella, B.A.; Kueltz, D. Osmo- and ionoregulatory responses of green sturgeon (Acipenser medirostris) to salinity acclimation. J. Comp. Physiol. B-Biochem. Syst. Environ. Physiol. 2009, 179, 383–390. [Google Scholar] [CrossRef]
- Yuan, J.B.; Zhang, X.J.; Liu, C.Z.; Duan, H.; Li, F.H.; Xiang, J.H. Convergent Evolution of the Osmoregulation System in Decapod Shrimps. Mar. Biotechnol. 2017, 19, 76–88. [Google Scholar] [CrossRef]
- Zhang, R.; Shi, X.; Guo, J.; Mao, X.; Fan, B. Acute stress response in hepatopancreas of Pacific white shrimp Litopenaeus vannamei to high alkalinity. Aquac. Rep. 2024, 35. [Google Scholar] [CrossRef]
- Cui, W.X.; Ma, A.J.; Wang, X.N. Response of the PI3K-AKT signalling pathway to low salinity and the effect of its inhibition mediated by wortmannin on ion channels in turbot. Aquac. Res. 2020, 51, 2676–2686. [Google Scholar] [CrossRef]
- Gagandeep, J. Functional Role of Betalains in Disphyma Australe Under Salinty Stress. Ph.D. Thesis, Victoria University of Wellington, Wellington, New Zealand, 2016. [Google Scholar]
- Lu, H.; Chen, W.; Peng, K.; Huang, M.; Zhao, J.; Chen, X.; Sun, Y.; Ruan, Z.; Li, C.; Liu, D.; et al. Rapid adaptive and acute stimulatory responses to low salinity stress in Pacific white shrimp (Litopenaeus vannamei): Insights from integrative transcriptomic and proteomic analysis. Comp. Biochem. Physiol. D-Genom. Proteom. 2023, 48, 101149. [Google Scholar] [CrossRef]
- Pequeux, A. OSMOTIC REGULATION IN CRUSTACEANS. J. Crustac. Biol. 1995, 15, 1–60. [Google Scholar] [CrossRef]
- Lignot, J.H.; Spanings-Pierrot, C.; Charmantier, G. Osmoregulatory capacity as a tool in monitoring the physiological condition and the effect of stress in crustaceans. Aquaculture 2000, 191, 209–245. [Google Scholar] [CrossRef]
- Hoffmann, E.K.; Pedersen, S.F. Sensors and signal transduction pathways in vertebrate cell volume regulation. In Mechanisms and Significance of Cell Volume Regulation; Lang, F., Ed.; Contributions to Nephrology; Karger Publishers: Basel, Switzerland, 2006; Volume 152, pp. 54–104. [Google Scholar]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.-X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, L.; Ying, Y.; Wang, J.; Shao, J.F.; Yamaji, N.; Whelan, J.; Ma, J.F.; Shou, H. A transcription factor OsbHLH156 regulates Strategy II iron acquisition through localising IRO2 to the nucleus in rice. New Phytol. 2020, 225, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Gavello, D.; Carbone, E.; Carabelli, V. Leptin-mediated ion channel regulation: PI3K pathways, physiological role, and therapeutic potential. Channels 2016, 10, 282–296. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.; Seri, L.; Levine, A. Induction of phosphatidylinositol 3-kinase-mediated endocytosis by salt stress leads to intracellular production of reactive oxygen species and salt tolerance. Plant J. 2007, 51, 185–197. [Google Scholar] [CrossRef]
- Yao, H.; Li, X.; Tang, L.; Wang, H.; Wang, C.; Mu, C.; Shi, C. Metabolic mechanism of the mud crab (Scylla paramamosain) adapting to salinity sudden drop based on GC-MS technology. Aquacult. Rep. 2020, 18, 100533. [Google Scholar] [CrossRef]
- Panda, A.; Rangani, J.; Parida, A.K. Unraveling salt responsive metabolites and metabolic pathways using non-targeted metabolomics approach and elucidation of salt tolerance mechanisms in the xero-halophyte Haloxylon salicornicum. Plant Physiol. Biochem. PPB 2021, 158, 284–296. [Google Scholar] [CrossRef]
- Yang, Y.; Han, X.; Sun, L.; Shao, F.; Yin, Y.; Zhang, W. ETS Transcription Factors in Immune Cells and Immune-Related Diseases. Int. J. Mol. Sci. 2024, 25, 10004. [Google Scholar] [CrossRef]
- Musa, J.; Aynaud, M.-M.; Mirabeau, O.; Delattre, O.; Gruenewald, T.G.P. MYBL2 (B-Myb): A central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis. 2017, 8, e2895. [Google Scholar] [CrossRef]
- Luo, C.T.; Osmanbeyoglu, H.U.; Do, M.H.; Bivona, M.R.; Toure, A.; Kang, D.; Xie, Y.; Leslie, C.S.; Li, M.O. Ets transcription factor GABP controls T cell homeostasis and immunity. Nat. Commun. 2017, 8, 1062. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, N.; Jaiswal, A.; Bhartiya, P.; Choi, E.H.; Kaushik, N.K. TFCP2 as a therapeutic nexus: Unveiling molecular signatures in cancer. Cancer Metastasis Rev. 2024, 43, 959–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Wu, B.; Chang, H.Y.; Greenleaf, W.J. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nussbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Cui, X.; Wang, T.; Chen, H.S.; Busov, V.; Wei, H. TF-finder: A software package for identifying transcription factors involved in biological processes using microarray data and existing knowledge base. BMC Bioinform. 2010, 11, 425. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]

| TF | Full Name | Encoding Gene Number (DEGs) | Score * | p-Value * | Expression Profile | TFBS Number | TFBSs in Promoter | Targeted Gene Number |
|---|---|---|---|---|---|---|---|---|
| ZBTB | Zinc finger and BTB domain | 2 | 21.2273 | 2.99 × 10−9 | up | 63,247 | 6064 | 434 |
| CP2 | Transcription factor CP2 | 1 | 18.202 | 3.14 × 10−8 | up | 11,537 | 1561 | 118 |
| MYB | Myeloblastosis | 2 | 22.3279 | 6.21 × 10−9 | up | 15,068 | 2817 | 174 |
| NFAT5 | Nuclear factor of activated T-cells 5 | 1 | 16.0845 | 1.85 × 10−7 | up | 16,368 | 2814 | 185 |
| RFX | Regulatory Factor X | 1 | 25.4138 | 7.25 × 10−11 | up | 54,216 | 3725 | 268 |
| bHLH | Basic helix-loop-helix | 23 | 17.4545 | 1.02 × 10−7 | up | 59,169 | 4008 | 257 |
| NRF1 | Nuclear factor E2-related factor 1 | 2 | 22.4634 | 2.24 × 10−9 | up | 26,183 | 2700 | 145 |
| ETS | Erythroblast transformation-specific | 11 | 20.1897 | 1.82 × 10−9 | up | 18,086 | 3025 | 180 |
| ARID | AT-rich interactive domain | 2 | 15.6071 | 2.40 × 10−8 | up | 11,149 | 2331 | 134 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Yuan, J.; Li, R.; Yang, Z.; Luo, M.; Zhong, X.; Hu, J.; Si, S.; Zhang, X.; Li, F. Integrative ATAC-Seq and RNA-Seq Analysis Reveals Key Transcription Factors Mediating Low Salinity Adaptation in Penaeid Shrimp. Int. J. Mol. Sci. 2025, 26, 4605. https://doi.org/10.3390/ijms26104605
Zhang C, Yuan J, Li R, Yang Z, Luo M, Zhong X, Hu J, Si S, Zhang X, Li F. Integrative ATAC-Seq and RNA-Seq Analysis Reveals Key Transcription Factors Mediating Low Salinity Adaptation in Penaeid Shrimp. International Journal of Molecular Sciences. 2025; 26(10):4605. https://doi.org/10.3390/ijms26104605
Chicago/Turabian StyleZhang, Chuntao, Jianbo Yuan, Roujing Li, Zhanyuan Yang, Man Luo, Xiaoyun Zhong, Jie Hu, Shuqing Si, Xiaojun Zhang, and Fuhua Li. 2025. "Integrative ATAC-Seq and RNA-Seq Analysis Reveals Key Transcription Factors Mediating Low Salinity Adaptation in Penaeid Shrimp" International Journal of Molecular Sciences 26, no. 10: 4605. https://doi.org/10.3390/ijms26104605
APA StyleZhang, C., Yuan, J., Li, R., Yang, Z., Luo, M., Zhong, X., Hu, J., Si, S., Zhang, X., & Li, F. (2025). Integrative ATAC-Seq and RNA-Seq Analysis Reveals Key Transcription Factors Mediating Low Salinity Adaptation in Penaeid Shrimp. International Journal of Molecular Sciences, 26(10), 4605. https://doi.org/10.3390/ijms26104605





