Genome-Wide Identification and Expression Profiling of Phosphatidylethanolamine-Binding Protein (PEBP) Genes in Helianthus annuus L.
Abstract
1. Introduction
2. Results
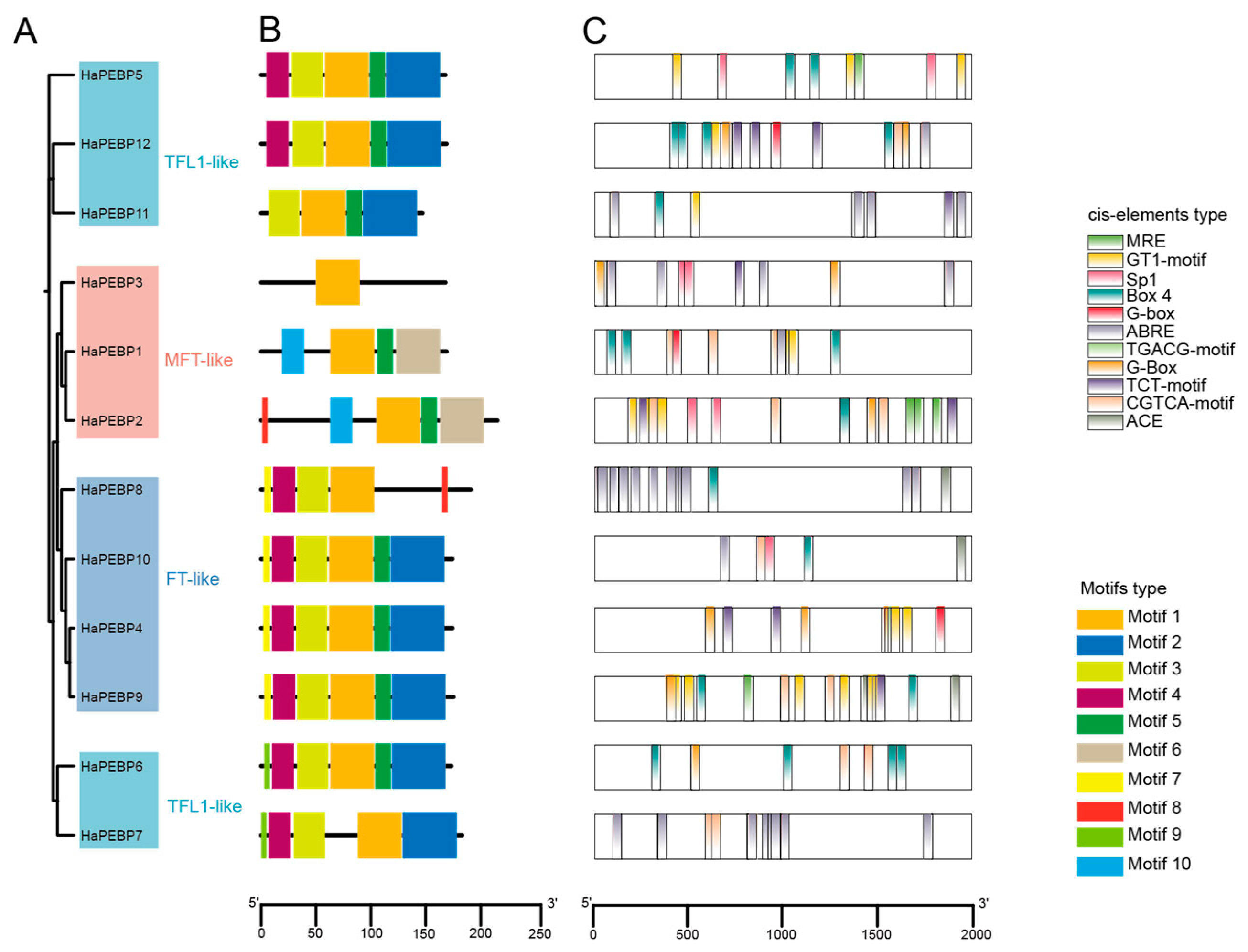
2.1. Genome-Wide Identification and Phylogenetic Relationship of HaPEBP Members in Sunflower
2.2. Conserved Motifs and Cis-Acting Elements of HaPEBP Genes
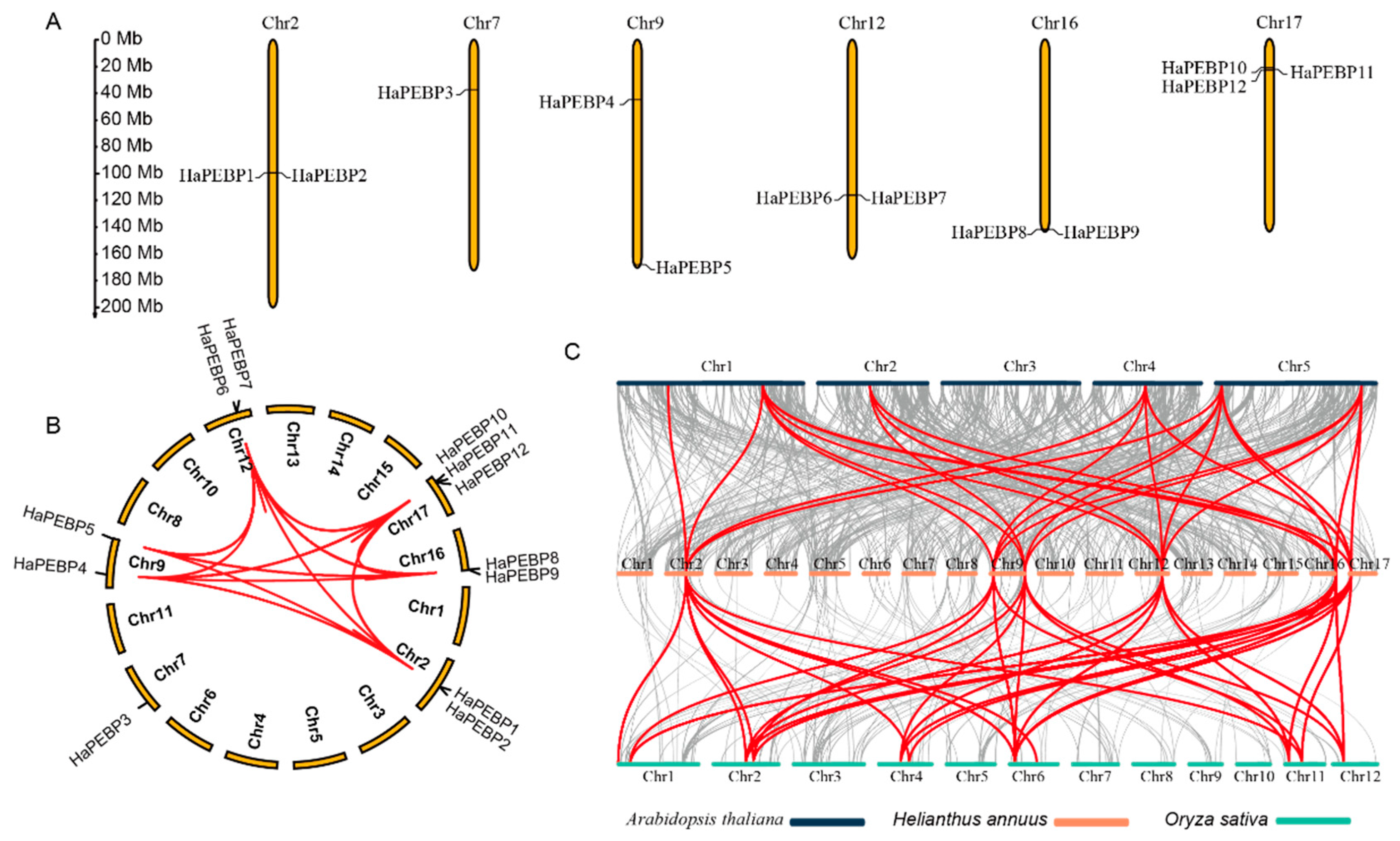
2.3. Chromosomal Localization and Gene Ontology Analysis of HaPEBP Genes
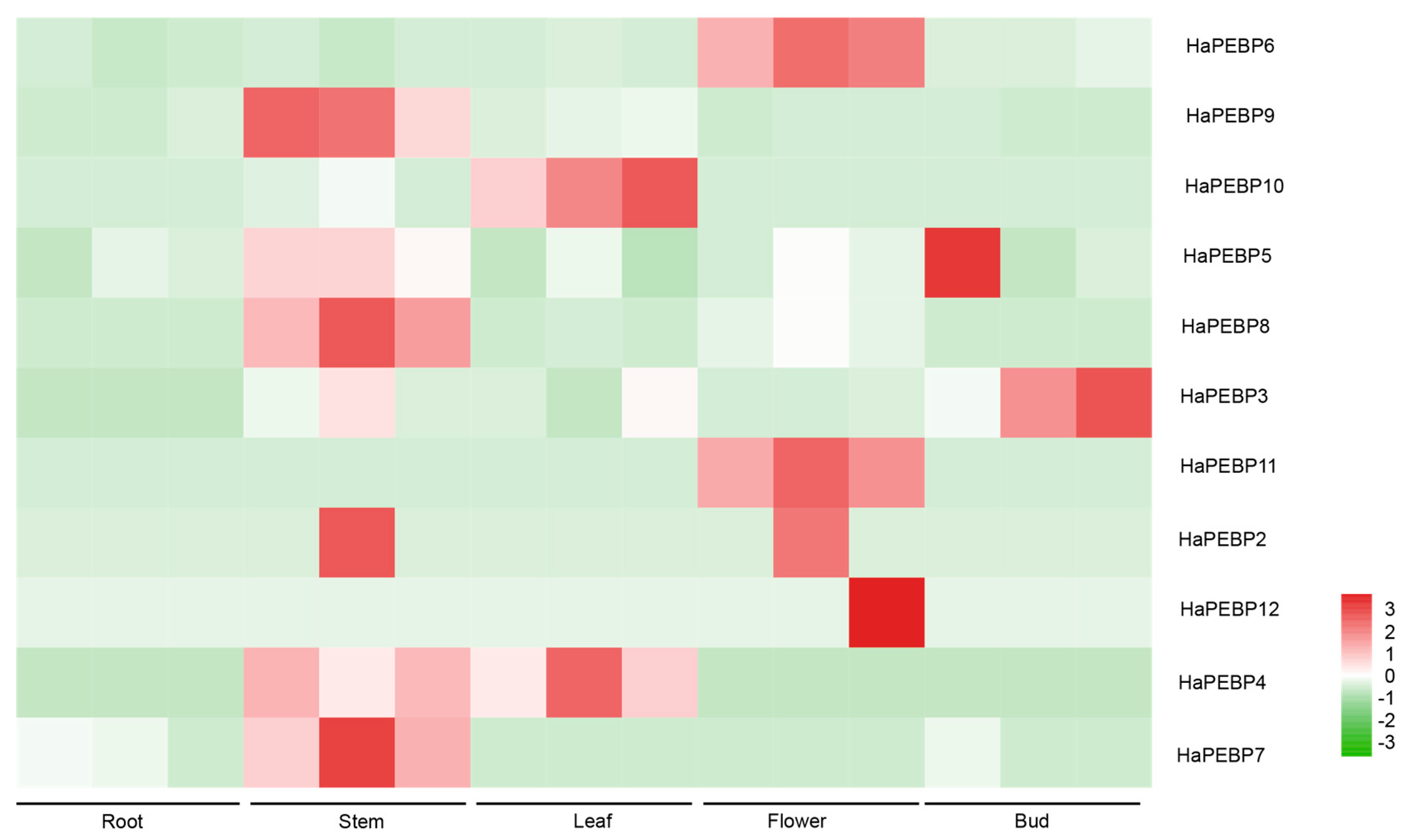
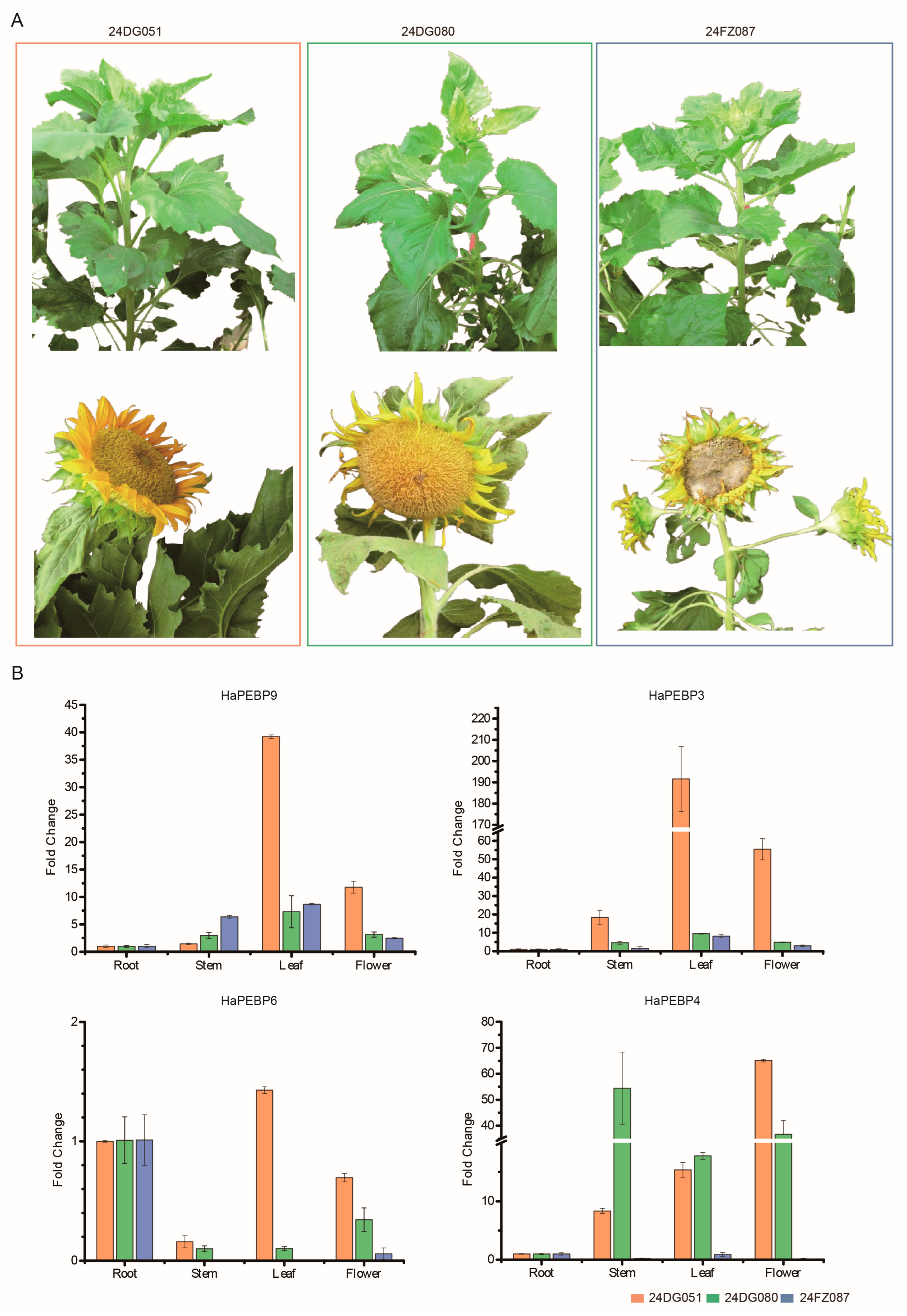
2.4. Expression Characterization Analysis of HaPEBP Genes in Different Sunflower Tissues
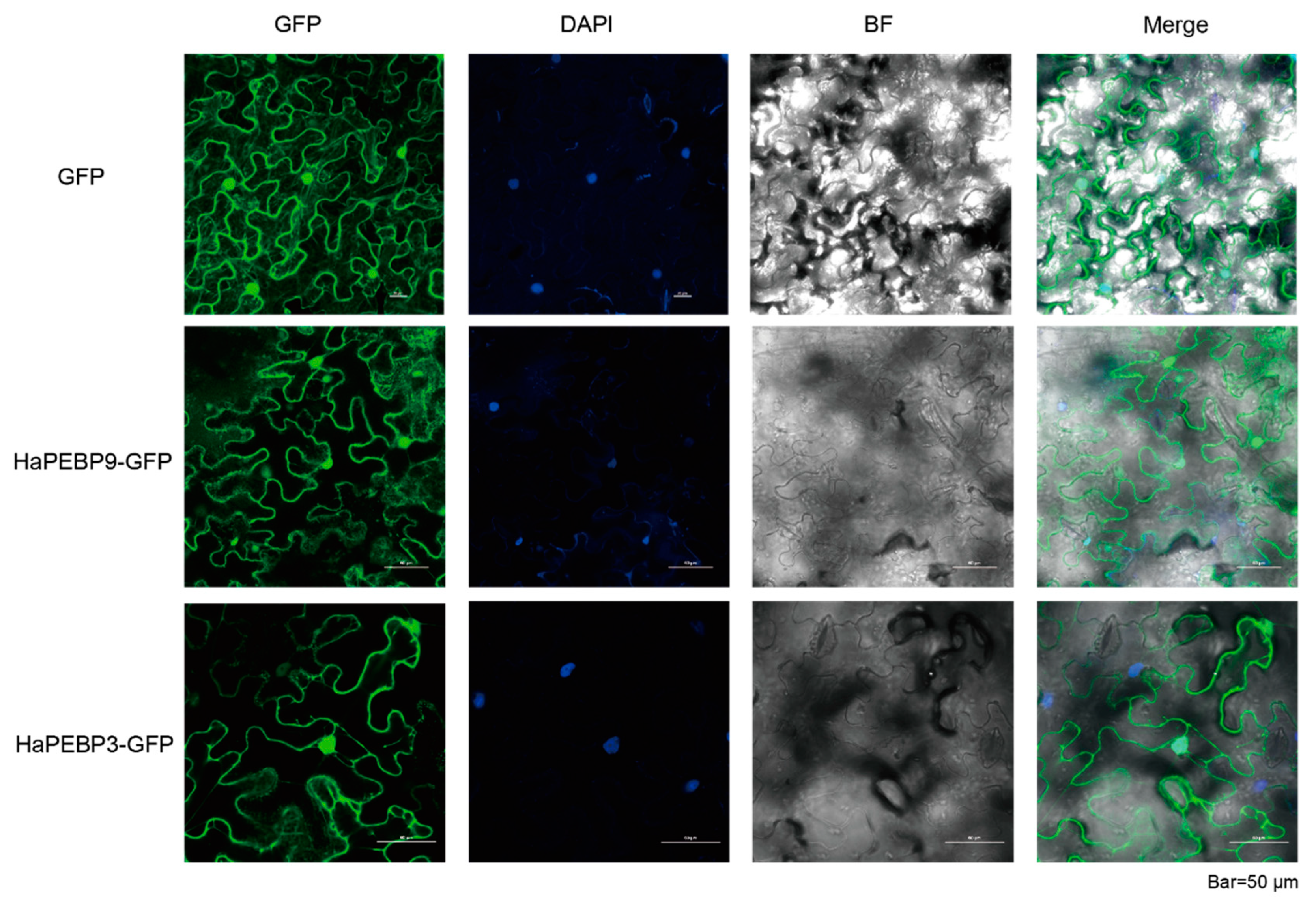
2.5. Subcellular Localization Analysis of HaPEBPs
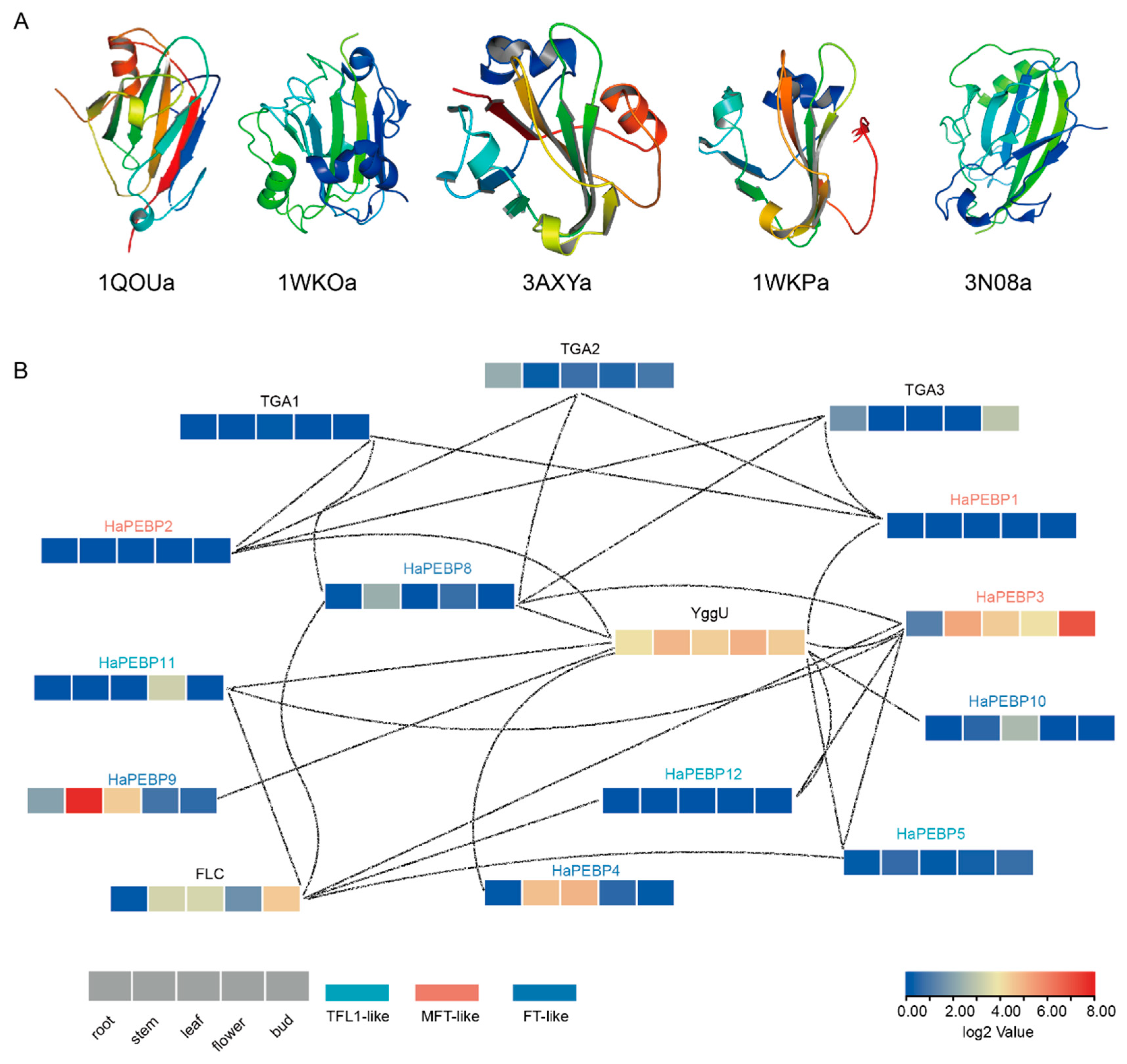
2.6. Three-Dimensional Structure and Protein–Protein Interaction Analysis of HaPEBPs
3. Discussion
3.1. Evolution Characteristics of HaPEBP Genes
3.2. Structure Variation of HaPEBP Genes
3.3. Biological Functions of HaPEBP Genes
4. Methods and Materials
4.1. Plant Materials
4.2. Identification of the PEBP Gene Family in Sunflower
4.3. Multiple Sequence Alignment and Phylogenetic Analysis of HaPEBP
4.4. Motifs and Gene Structure Analysis
4.5. Identification and Analysis of Cis-Acting Elements in Gene Promoters
4.6. Characterization, Chromosomal Localization, and Synteny Analysis of HaPEBP Genes
4.7. Analysis of Selective Pressures on HaPEBP Genes
4.8. Subcellular Localization
4.9. Expression Profile Assay of HaPEBP Gene and RNA-Seq Data Analysis
4.10. Analysis of the Three-Dimensional Structure and Protein–Protein Interactions of HaPEBP Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wickland, D.P.; Hanzawa, Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: Functional evolution and molecular mechanisms. Mol. Plant 2015, 8, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Trakul, N.; Rosner, M.R. Modulation of the MAP kinase signaling cascade by Raf kinase inhibitory protein. Cell Res. 2005, 15, 19–23. [Google Scholar] [CrossRef]
- Karlgren, A.; Gyllenstrand, N.; Källman, T.; Sundström, J.F.; Moore, D.; Lascoux, M.; Lagercrantz, U. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution. Plant Physiol. 2011, 156, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Kawahigashi, H.; Ando, T.; Tonooka, T.; Handa, H. Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 2009, 149, 1341–1353. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jia, X.; Yang, Z.; Fu, Q.; Yang, H.; Xu, X. Genome-wide identification of PEBP gene family in Solanum lycopersicum. Int. J. Mol. Sci. 2023, 24, 9185. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, Z.; Liu, Y.; Liu, T.; Li, Q.; Ji, Y.; Li, C.; Fang, C.; Wang, M.; Wu, M. Functional evolution of phosphatidylethanolamine binding proteins in soybean and Arabidopsis. Plant Cell 2015, 27, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhu, M.; Guo, Y.; Sun, J.; Ma, W.; Wang, X. Genomic survey of PEBP gene family in rice: Identification, phylogenetic analysis, and expression profiles in organs and under abiotic stresses. Plants 2022, 11, 1576. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Gan, Z.; Xu, F.; Qian, J.; Qian, M.; Ai, H.; Feng, T.; Lu, X.; Li, R.; Huang, X. Molecular characterization of pepper PEBP genes reveals the diverse functions of CaFTs in flowering and plant architecture. Sci. Hortic. 2024, 335, 113345. [Google Scholar] [CrossRef]
- Putterill, J.; Varkonyi-Gasic, E. FT and florigen long-distance flowering control in plants. Curr. Opin. Plant Biol. 2016, 33, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Yang, K.Z.; Wei, X.X.; Wang, X.Q. Revisiting the phosphatidylethanolamine-binding protein (PEBP) gene family reveals cryptic FLOWERING LOCUS T gene homologs in gymnosperms and sheds new light on functional evolution. New Phytol. 2016, 212, 730–744. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Kobayashi, Y.; Goto, K.; Abe, M.; Araki, T. TWIN SISTER OF FT (TSF) Acts as a Floral Pathway Integrator Redundantly with FT. Plant Cell Physiol. 2005, 46, 1175–1189. [Google Scholar] [CrossRef]
- Yoo, S.J.; Chung, K.S.; Jung, S.H.; Yoo, S.Y.; Lee, J.S.; Ahn, J.H. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. Plant J. 2010, 63, 241–253. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Lee, H.-J.; Seo, P.J.; Jung, J.-H.; Ahn, J.H.; Park, C.-M. The Arabidopsis Floral Repressor BFT Delays Flowering by Competing with FT for FD Binding under High Salinity. Mol. Plant 2014, 7, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xian, P.; Cheng, Y.; Zhong, Y.; Yang, Y.; Zhou, Q.; Lian, T.; Ma, Q.; Nian, H.; Ge, L. MOTHER-OF-FT-AND-TFL1 regulates the seed oil and protein content in soybean. New Phytol. 2023, 239, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.; Källman, T.; Lagercrantz, U. Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 2009, 70, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Klasfeld, S.; Jeong, C.W.; Jin, R.; Goto, K.; Yamaguchi, N.; Wagner, D. TERMINAL FLOWER 1-FD complex target genes and competition with FLOWERING LOCUS T. Nat. Commun. 2020, 11, 5118. [Google Scholar] [CrossRef]
- Su, T.; Wu, Y.; Fang, C.; Liu, B.; Lu, S.; Kong, F.; Liu, H. The Critical Roles of Phosphatidylethanolamine-Binding Proteins in Legumes. Plant Cell Environ. 2024. [Google Scholar] [CrossRef]
- Zhao, H.; Huang, X.; Yang, Z.; Li, F.; Ge, X. Synergistic optimization of crops by combining early maturation with other agronomic traits. Trends Plant Sci. 2023, 28, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef]
- Miao, H.; Wang, L.; Qu, L.; Liu, H.; Sun, Y.; Le, M.; Wang, Q.; Wei, S.; Zheng, Y.; Lin, W. Genomic evolution and insights into agronomic trait innovations of Sesamum species. Plant Commun. 2024, 5, 100729. [Google Scholar] [CrossRef]
- Mandel, J.R.; Dikow, R.B.; Siniscalchi, C.M.; Thapa, R.; Watson, L.E.; Funk, V.A. A fully resolved backbone phylogeny reveals numerous dispersals and explosive diversifications throughout the history of Asteraceae. Proc. Natl. Acad. Sci. USA 2019, 116, 14083–14088. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S. Chapter 7—Helianthus annuus L. In Edible Flowers; Gupta, A.K., Kumar, V., Naik, B., Mishra, P., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 105–126. [Google Scholar]
- Puttha, R.; Venkatachalam, K.; Hanpakdeesakul, S.; Wongsa, J.; Parametthanuwat, T.; Srean, P.; Pakeechai, K.; Charoenphun, N. Exploring the Potential of Sunflowers: Agronomy, Applications, and Opportunities within Bio-Circular-Green Economy. Horticulturae 2023, 9, 1079. [Google Scholar] [CrossRef]
- Blackman, B.K. Interacting duplications, fluctuating selection, and convergence: The complex dynamics of flowering time evolution during sunflower domestication. J. Exp. Bot. 2013, 64, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Blackman, B.K.; Michaels, S.D.; Rieseberg, L.H. Connecting the sun to flowering in sunflower adaptation. Mol. Ecol. 2011, 20, 3503–3512. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Li, P.; Li, L.; Zhang, Q. Research advances in and prospects of ornamental plant genomics. Hortic. Res. 2021, 8, 65. [Google Scholar] [CrossRef]
- Jin, H.; Tang, X.; Xing, M.; Zhu, H.; Sui, J.; Cai, C.; Li, S. Molecular and transcriptional characterization of phosphatidyl ethanolamine-binding proteins in wild peanuts Arachis duranensis and Arachis ipaensis. BMC Plant Biol. 2019, 19, 484. [Google Scholar] [CrossRef]
- Martignago, D.; da Silveira Falavigna, V.; Lombardi, A.; Gao, H.; Korwin Kurkowski, P.; Galbiati, M.; Tonelli, C.; Coupland, G.; Conti, L. The bZIP transcription factor AREB3 mediates FT signalling and floral transition at the Arabidopsis shoot apical meristem. PLoS Genet. 2023, 19, e1010766. [Google Scholar]
- Niwa, M.; Daimon, Y.; Kurotani, K.-I.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 Interacts with FLOWERING LOCUS T to Repress the Floral Transition of the Axillary Meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef]
- Jaeger, K.E.; Pullen, N.; Lamzin, S.; Morris, R.J.; Wigge, P.A. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell 2013, 25, 820–833. [Google Scholar] [CrossRef]
- Kaneko-Suzuki, M.; Kurihara-Ishikawa, R.; Okushita-Terakawa, C.; Kojima, C.; Nagano-Fujiwara, M.; Ohki, I.; Tsuji, H.; Shimamoto, K.; Taoka, K.-I. TFL1-Like Proteins in Rice Antagonize Rice FT-Like Protein in Inflorescence Development by Competition for Complex Formation with 14-3-3 and FD. Plant Cell Physiol. 2018, 59, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, M.F.; Mostafa, K.; Kavas, M. CRISPR/Cas9-mediated mutagenesis of FT/TFL1 in petunia improves plant architecture and early flowering. Plant Mol. Biol. 2024, 114, 69. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Damerval, C. Phylogenomic analysis of the PEBP gene family in cereals. J. Mol. Evol. 2005, 61, 579–590. [Google Scholar] [CrossRef]
- Marand, A.P.; Eveland, A.L.; Kaufmann, K.; Springer, N.M. cis-Regulatory Elements in Plant Development, Adaptation, and Evolution. Annu. Rev. Plant Biol. 2023, 74, 111–137. [Google Scholar] [CrossRef]
- Zhao, S.; Wei, Y.; Pang, H.; Xu, J.; Li, Y.; Zhang, H.; Zhang, J.; Zhang, Y. Genome-wide identification of the PEBP genes in pears and the putative role of PbFT in flower bud differentiation. PeerJ 2020, 8, e8928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-M.; Zhao, X.; He, X.; Zheng, Q.; Huang, Y.; Li, Y.; Ke, S.; Liu, Z.-J.; Lan, S. Genome-wide identification of PEBP gene family in two Dendrobium species and expression patterns in Dendrobium chrysotoxum. Int. J. Mol. Sci. 2023, 24, 17463. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Rodgers-Melnick, E.; Mane, S.P.; Dharmawardhana, P.; Slavov, G.T.; Crasta, O.R.; Strauss, S.H.; Brunner, A.M.; DiFazio, S.P. Contrasting patterns of evolution following whole genome versus tandem duplication events in Populus. Genome Res. 2012, 22, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kaya, H.; Goto, K.; Iwabuchi, M.; Araki, T. A pair of related genes with antagonistic roles in mediating flowering signals. Science 1999, 286, 1960–1962. [Google Scholar] [CrossRef]
- Borello, U.; Ceccarelli, E.; Giuliano, G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different I box elements in plant light-regulated promoters. Plant J. 1993, 4, 611–619. [Google Scholar] [CrossRef]
- Kumar, G.M.; Mamidala, P.; Podile, A.R. Regulation of Polygalacturonase-inhibitory proteins in plants is highly dependent on stress and light responsive elements. Plant Omics 2009, 2, 238–249. [Google Scholar]
- Lifschitz, E.; Eshed, Y. Universal florigenic signals triggered by FT homologues regulate growth and flowering cycles in perennial day-neutral tomato. J. Exp. Bot. 2006, 57, 3405–3414. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Li, C.; Zhang, Y.; Zhang, K.; Guo, D.; Cui, B.; Wang, X.; Huang, X. Promoting flowering, lateral shoot outgrowth, leaf development, and flower abscission in tobacco plants overexpressing cotton FLOWERING LOCUS T (FT)-like gene GhFT1. Front. Plant Sci. 2015, 6, 454. [Google Scholar] [CrossRef] [PubMed]
- Moraes, T.S.; Dornelas, M.C.; Martinelli, A.P. FT/TFL1: Calibrating plant architecture. Front. Plant Sci. 2019, 10, 97. [Google Scholar] [CrossRef]
- Mendapara, I.; Modha, K.; Patel, S.; Parekh, V.; Patel, R.; Chauhan, D.; Bardhan, K.; Siddiqui, M.H.; Alamri, S.; Rahman, M.A. Characterization of CcTFL1 governing plant architecture in pigeon pea (Cajanus cajan (L.) Millsp.). Plants 2023, 12, 2168. [Google Scholar] [CrossRef] [PubMed]
- Sriboon, S.; Li, H.; Guo, C.; Senkhamwong, T.; Dai, C.; Liu, K. Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus. BMC Genet. 2020, 21, 52. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, J.; Li, C.; Deng, X.; Wang, T.; Dong, L. Genome-wide analysis of TCP transcription factor family in sunflower and identification of HaTCP1 involved in the regulation of shoot branching. BMC Plant Biol. 2023, 23, 222. [Google Scholar] [CrossRef] [PubMed]
- Taoka, K.-i.; Ohki, I.; Tsuji, H.; Furuita, K.; Hayashi, K.; Yanase, T.; Yamaguchi, M.; Nakashima, C.; Purwestri, Y.A.; Tamaki, S.; et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature 2011, 476, 332–335. [Google Scholar] [CrossRef]
- Banfield, M.J.; Brady, R.L. The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 2000, 297, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Ho, W.W.H.; Weigel, D. Structural Features Determining Flower-Promoting Activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 2014, 26, 552–564. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Miller, D.; Winter, V.J.; Banfield, M.J.; Lee, J.H.; Yoo, S.Y.; Henz, S.R.; Brady, R.L.; Weigel, D. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 2006, 25, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.H.; Choi, D.I. Roles of plant proteases in pathogen defense. Plant Pathol. J. 2008, 24, 367–374. [Google Scholar] [CrossRef]
- Li, D.; Shao, L.; Zhang, J.; Wang, X.; Zhang, D.; Horvath, D.P.; Zhang, L.; Zhang, J.; Xia, Y. MADS-box transcription factors determine the duration of temporary winter dormancy in closely related evergreen and deciduous Iris spp. J. Exp. Bot. 2022, 73, 1429–1449. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, J.; Yuan, C.; Hu, Y.; Liu, Q.; Chen, Q.; Zhang, P.; Shi, N.; Qin, C. Characterization of genes associated with TGA7 during the floral transition. BMC Plant Biol. 2021, 21, 367. [Google Scholar] [CrossRef]
- Koornneef, M.; Alonso-Blanco, C.; Vreugdenhil, D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu. Rev. Plant Biol. 2004, 55, 141–172. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Prakash, A.; Jeffryes, M.; Bateman, A.; Finn, R.D. The HMMER web server for protein sequence similarity search. Curr. Protoc. Bioinform. 2017, 60, 3.15.1–3.15.23. [Google Scholar] [CrossRef]
- Mistry, J.; Bateman, A.; Finn, R.D. Predicting active site residue annotations in the Pfam database. BMC Bioinform. 2007, 8, 298. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34 (Suppl. S2), W369–W373. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Chao, J.-T.; Kong, Y.-Z.; Wang, Q.; Sun, Y.-H.; Gong, D.-P.; Lv, J.; Liu, G.-S. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan Hered. 2015, 37, 91–97. [Google Scholar]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Zeng, X.; Xu, Z.; Taranto, A.; Lomas, J.S.; Zhang, Y.; Huang, Y.; Wang, Y.; Yim, W.C. JCVI: A versatile toolkit for comparative genomics analysis. iMeta 2024, 3, e211. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 19, 2206–2229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. KaKs_Calculator 3.0: Calculating selective pressure on coding and non-coding sequences. Genom. Proteom. Bioinform. 2022, 20, 536–540. [Google Scholar] [CrossRef]
- Thumuluri, V.; Almagro Armenteros, J.J.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chao, H.; Chen, L.; Craig, P.A.; Crichlow, G.V.; Dalenberg, K.; Duarte, J.M. RCSB Protein Data Bank (RCSB. org): Delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023, 51, D488–D508. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8.13.1–8.13.24. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Wang, Y.; Bai, J.; Guo, J.; Li, G.; Tian, Q.; Lv, S.; Cao, H.; Yang, X.; Liu, L. Genome-Wide Identification and Expression Profiling of Phosphatidylethanolamine-Binding Protein (PEBP) Genes in Helianthus annuus L. Int. J. Mol. Sci. 2025, 26, 4602. https://doi.org/10.3390/ijms26104602
Sun Y, Wang Y, Bai J, Guo J, Li G, Tian Q, Lv S, Cao H, Yang X, Liu L. Genome-Wide Identification and Expression Profiling of Phosphatidylethanolamine-Binding Protein (PEBP) Genes in Helianthus annuus L. International Journal of Molecular Sciences. 2025; 26(10):4602. https://doi.org/10.3390/ijms26104602
Chicago/Turabian StyleSun, Yiyi, Yanwen Wang, Jingyan Bai, Jiatong Guo, Guiting Li, Qiuzhen Tian, Shuping Lv, Hengchun Cao, Xiaojie Yang, and Lingyun Liu. 2025. "Genome-Wide Identification and Expression Profiling of Phosphatidylethanolamine-Binding Protein (PEBP) Genes in Helianthus annuus L." International Journal of Molecular Sciences 26, no. 10: 4602. https://doi.org/10.3390/ijms26104602
APA StyleSun, Y., Wang, Y., Bai, J., Guo, J., Li, G., Tian, Q., Lv, S., Cao, H., Yang, X., & Liu, L. (2025). Genome-Wide Identification and Expression Profiling of Phosphatidylethanolamine-Binding Protein (PEBP) Genes in Helianthus annuus L. International Journal of Molecular Sciences, 26(10), 4602. https://doi.org/10.3390/ijms26104602





