Reduced Sulfur Diet Reshapes the Microbiome and Metabolome in Mild–Moderate Ulcerative Colitis
Abstract
1. Introduction
2. Results
2.1. Participant Information
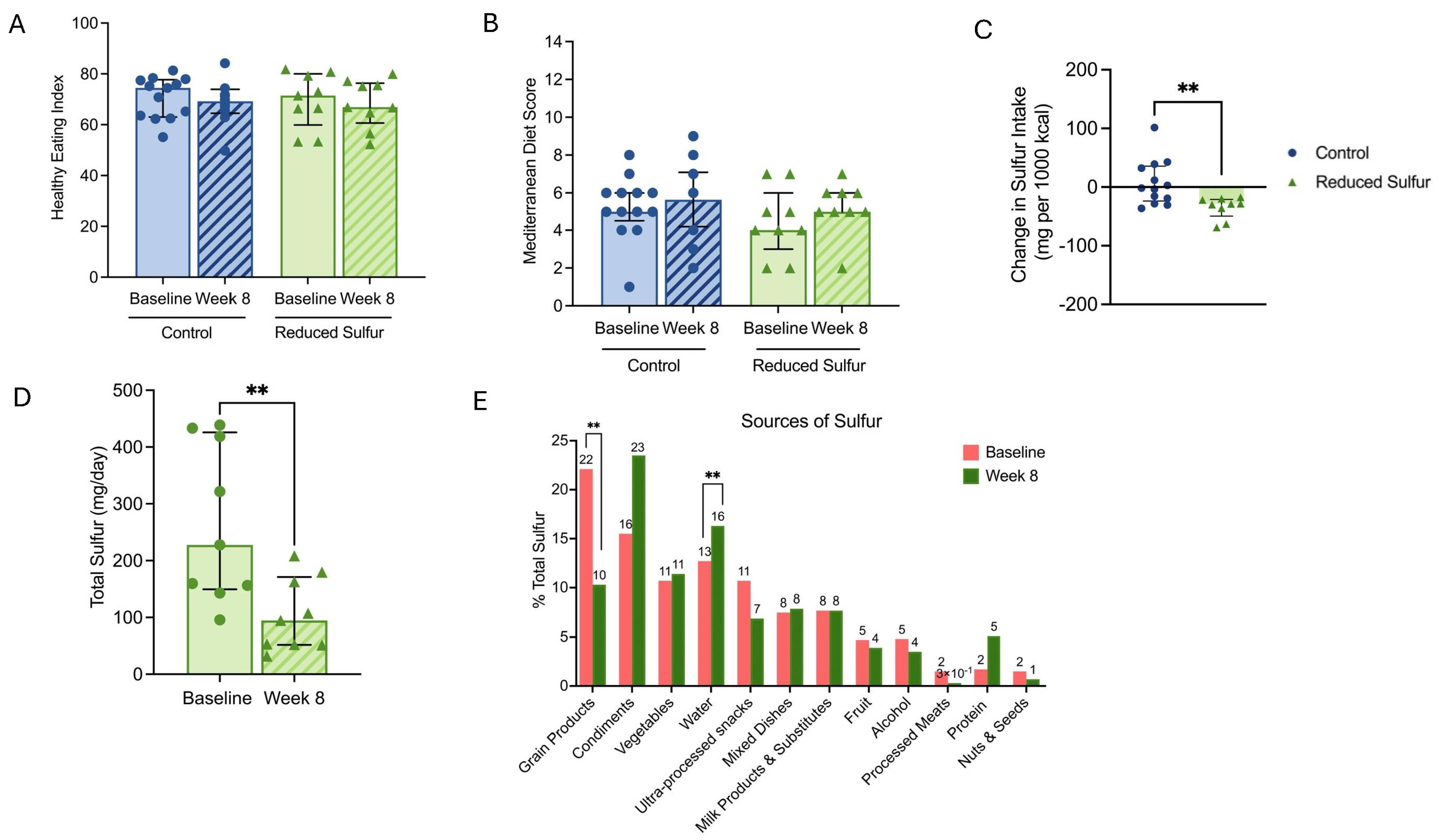
2.2. Diet Characteristics of the Study Participants
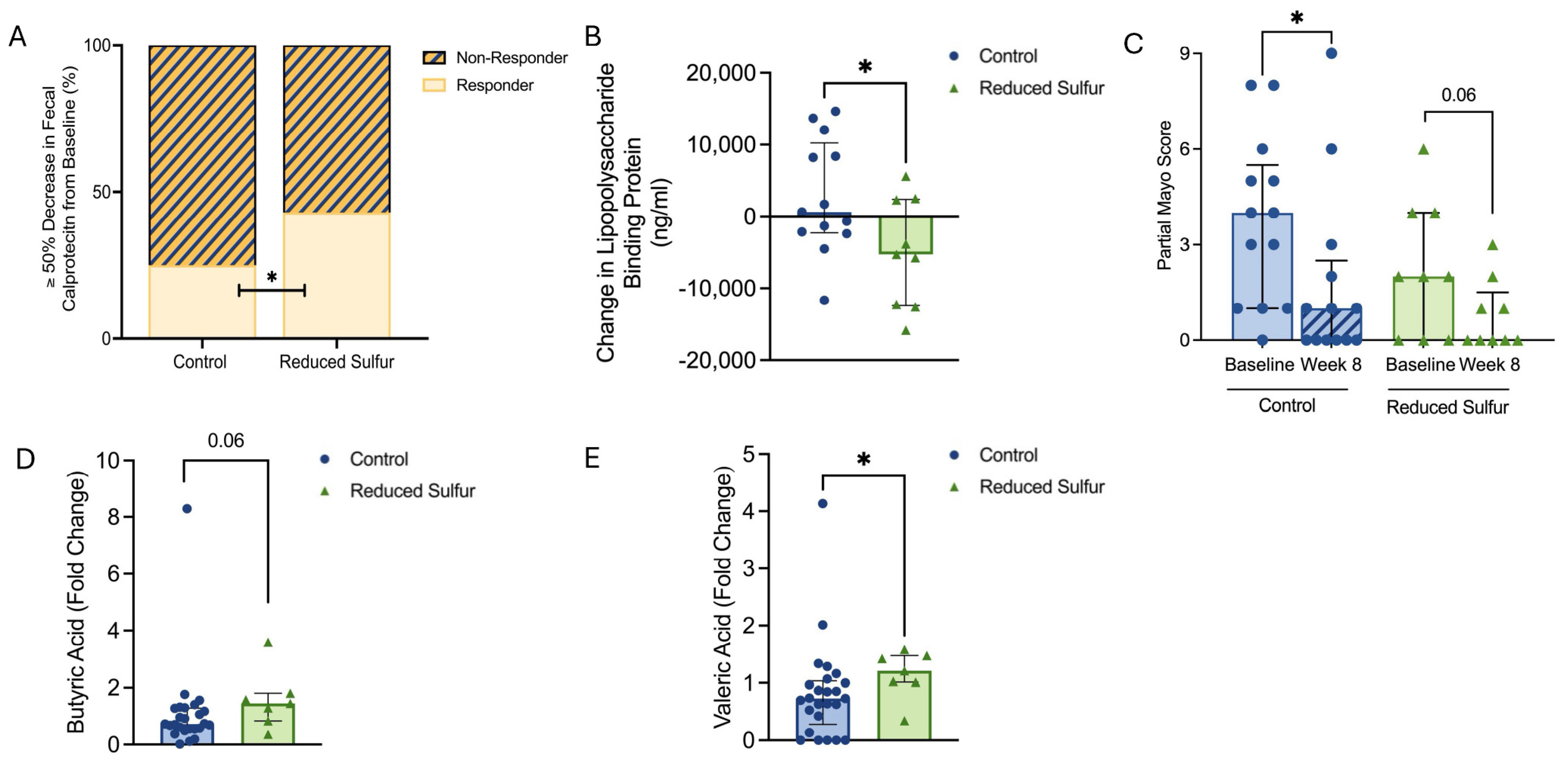
2.3. Reduced Dietary Sulfur Intake Improved Disease Activity and Influenced Short-Chain Fatty Acid Production
2.4. A Reduction in the Dietary-Sulfur-Induced Metabolic Shifts
2.5. Reduced Dietary Sulfur Induced Microbiome Shifts and Enhanced Diversity
2.6. Eggerthella Lenta and Collinsella Stercoris Played Opposing Roles in Gut Metabolism and Inflammation
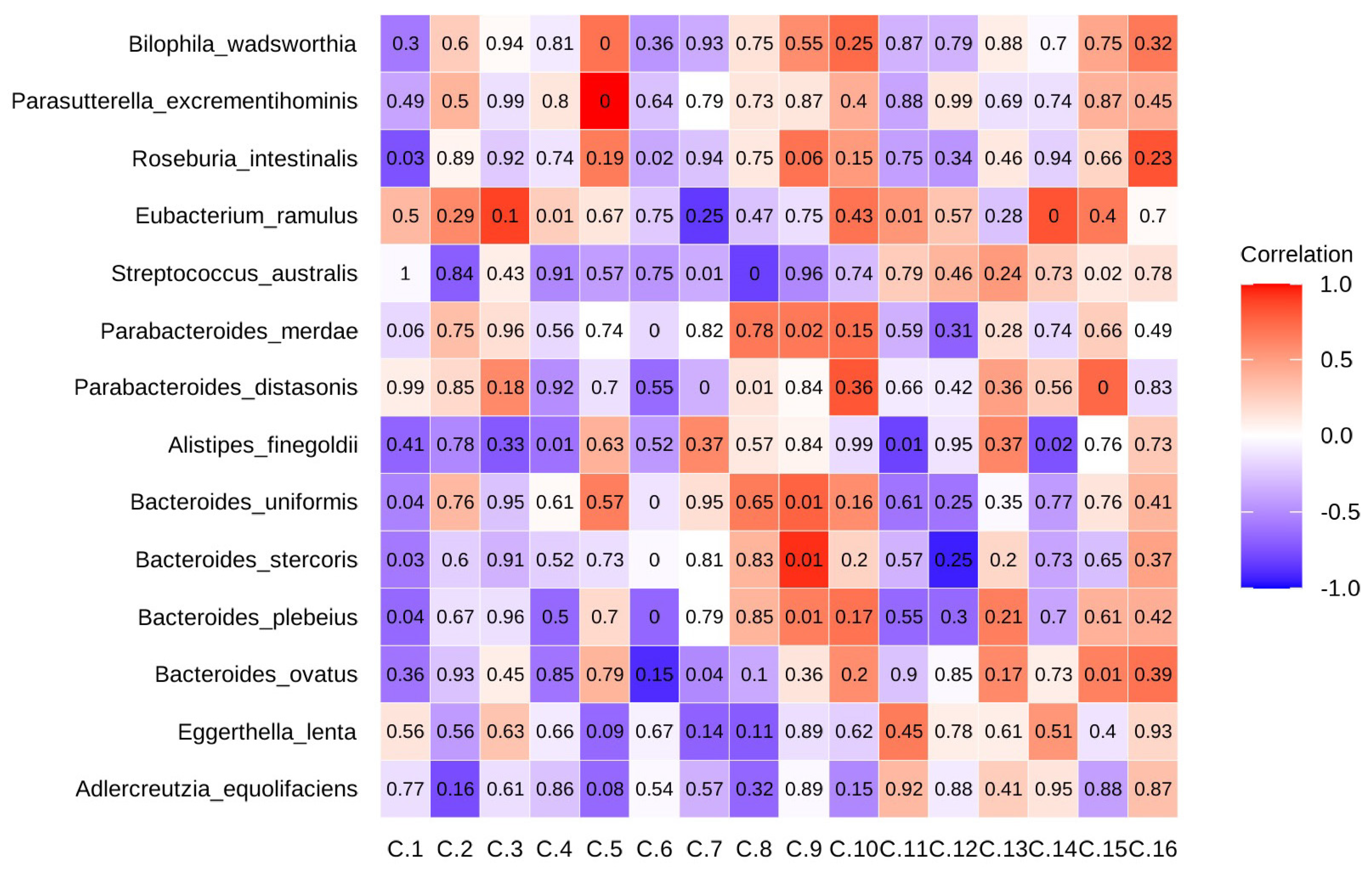
2.7. Dietary Choices Influenced Sulfur-Metabolizing Bacteria
3. Discussion
4. Materials and Methods
4.1. Study Design and Participants
4.2. Inclusion and Exclusion Criteria
4.3. Reduced Sulfur Intervention
4.4. Fecal Microbiome Analysis
4.5. Fecal Metabolome Analysis
4.6. Metabolomic Data Processing and Statistical Analyses
4.7. Short-Chain Fatty Acids
4.8. Statistical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansen, A.W.; Venkatachalam, K.V. Sulfur-Element containing metabolic pathways in human health and crosstalk with the microbiome. Biochem. Biophys. Rep. 2023, 35, 101529. [Google Scholar] [CrossRef] [PubMed]
- Cirino, G.; Szabo, C.; Papapetropoulos, A. Physiological roles of hydrogen sulfide in mammalian cells, tissues, and organs. Physiol. Rev. 2023, 103, 31–276. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, A.I.; Bernardino, R.M.; Pereira, I.A.C. Chapter Four—Role of sulfidogenic members of the gut microbiota in human disease. Adv. Microb. Physiol. 2024, 85, 145–200. [Google Scholar] [CrossRef]
- Dordevic, D.; Capikova, J.; Dordevic, S.; Tremlová, B.; Gajdács, M.; Kushkevych, I. Sulfur content in foods and beverages and its role in human and animal metabolism: A scoping review of recent studies. Heliyon 2023, 9, e15452. [Google Scholar] [CrossRef]
- Carbonero, F.; Benefiel, A.C.; Alizadeh-Ghamsari, A.H.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3, 448. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Distrutti, E.; Cirino, G.; Wallace, J.L. The Emerging Roles of Hydrogen Sulfide in the Gastrointestinal Tract and Liver. Gastroenterology 2006, 131, 259–271. [Google Scholar] [CrossRef]
- Guo, F.F.; Yu, T.C.; Hong, J.; Fang, J.Y. Emerging roles of hydrogen sulfide in inflammatory and neoplastic colonic diseases. Front. Physiol. 2016, 7, 156. [Google Scholar] [CrossRef]
- Medani, M.; Collins, D.; Docherty, N.G.; Baird, A.W.; O’Connell, P.R.; Winter, D.C. Emerging role of hydrogen sulfide in colonic physiology and pathophysiology. Inflamm. Bowel Dis. 2011, 17, 1620–1625. [Google Scholar] [CrossRef]
- Teigen, L.M.; Geng, Z.; Sadowsky, M.J.; Vaughn, B.P.; Hamilton, M.J.; Khoruts, A. Dietary factors in sulfur metabolism and pathogenesis of ulcerative colitis. Nutrients 2019, 11, 931. [Google Scholar] [CrossRef]
- Jørgensen, J.; Mortensen, P.B. Hydrogen sulfide and colonic epithelial metabolism: Implications for ulcerative colitis. Dig. Dis. Sci. 2001, 46, 1722–1732. [Google Scholar] [CrossRef]
- Pitcher, M.C.L.; Beatty, E.R.; Cummings, J.H. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 2000, 46, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.; Ellis, C.J.; Furne, J.K.; Springfield, J.; Levitt, M.D. Fecal hydrogen sulfide production in ulcerative colitis. Am. J. Gastroenterol. 1998, 93, 83–87. [Google Scholar] [CrossRef]
- Kushkevych, I.; Sangrador, J.C.; Dordević, D.; Rozehnalová, M.; Černý, M.; Fafula, R.; Vítězová, M.; Rittmann, S.K.-M.R. Evaluation of physiological parameters of intestinal sulfate-reducing bacteria isolated from patients suffering from ibd and healthy people. J. Clin. Med. 2020, 9, 1920. [Google Scholar] [CrossRef]
- Kushkevych, I.; Leščanová, O.; Dordević, D.; Jančíková, S.; Hošek, J.; Vítězová, M.; Buňková, L.; Drago, L. The sulfate-reducing microbial communities and meta-analysis of their occurrence during diseases of small–large intestine axis. J. Clin. Med. 2019, 8, 1656. [Google Scholar] [CrossRef]
- Khalil, N.A.; Walton, G.E.; Gibson, G.R.; Tuohy, K.M.; Andrews, S.C. In vitro batch cultures of gut microbiota from healthy and ulcerative colitis (UC) subjects suggest that sulphate-reducing bacteria levels are raised in UC and by a protein-rich diet. Int. J. Food Sci. Nutr. 2014, 65, 79–88. [Google Scholar] [CrossRef]
- Chen, J.; Wellens, J.; Kalla, R.; Fu, T.; Deng, M.; Zhang, H.; Yuan, S.; Wang, X.; Theodoratou, E.; Li, X.; et al. Intake of Ultra-processed Foods Is Associated with an Increased Risk of Crohn’s Disease: A Cross-sectional and Prospective Analysis of 187,154 Participants in the UK Biobank. J. Crohn’s Colitis 2023, 17, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Chiba, M.; Nakane, K.; Tsuji, T.; Tsuda, S.; Ishii, H.; Ohno, H.; Watanabe, K.; Obara, Y.; Komatsu, M.; Sugawara, T. Relapse Prevention by Plant-Based Diet Incorporated into Induction Therapy for Ulcerative Colitis: A Single-Group Trial. Perm. J. 2019, 23. [Google Scholar] [CrossRef] [PubMed]
- Day, A.S.; Yao, C.K.; Costello, S.P.; Ruszkiewicz, A.; Andrews, J.M.; Gibson, P.R.; Bryant, R.V. Therapeutic Potential of the 4 Strategies to SUlfide-REduction (4-SURE) Diet in Adults with Mild to Moderately Active Ulcerative Colitis: An Open-Label Feasibility Study. J. Nutr. 2022, 152, 1690–1701. [Google Scholar] [CrossRef]
- Chiba, M.; Tsuji, T.; Nakane, K.; Tsuda, S.; Ishii, H.; Ohno, H.; Obara, Y.; Komatsu, M.; Tozawa, H. High Remission Rate with Infliximab and Plant-Based Diet as First-Line (IPF) Therapy for Severe Ulcerative Colitis: Single-Group Trial. Perm. J. 2020, 24, 116. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Shumard, T.; Xie, H.; Dodda, A.; Varady, K.A.; Feferman, L.; Halline, A.G.; Goldstein, J.L.; Hanauer, S.B.; Tobacman, J.K. A randomized trial of the effects of the no-carrageenan diet on ulcerative colitis disease activity. Nutr. Health Aging 2017, 4, 181–192. [Google Scholar] [CrossRef]
- Roediger, W.E.W. Decreased sulphur aminoacid intake in ulcerative colitis. Lancet 1998, 351, 1555. [Google Scholar] [CrossRef] [PubMed]
- Papadaki, A.; Johnson, L.; Toumpakari, Z.; England, C.; Rai, M.; Toms, S.; Penfold, C.; Zazpe, I.; Martínez-González, M.A.; Feder, G. Validation of the English version of the 14-item mediterranean diet adherence screener of the PREDIMED study, in people at high cardiovascular risk in the UK. Nutrients 2018, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Shams-White, M.M.; Pannucci, T.E.; Lerman, J.L.; Herrick, K.A.; Zimmer, M.; Mathieu, K.M.; Stoody, E.E.; Reedy, J. Healthy Eating Index-2020: Review and Update Process to Reflect the Dietary Guidelines for Americans, 2020–2025. J. Acad. Nutr. Diet. 2023, 123, 1280–1288. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.H.; Ma, W.; Wang, D.D.; Cao, Y.; Mallick, H.; Gerbaba, T.K.; Lloyd-Price, J.; Abu-Ali, G.; Hall, A.B.; Sikavi, D.; et al. Association Between Sulfur-Metabolizing Bacterial Communities in Stool and Risk of Distal Colorectal Cancer in Men. Gastroenterology 2020, 158, 1313–1325. [Google Scholar] [CrossRef]
- Luo, W.; Zhao, M.; Dwidar, M.; Gao, Y.; Xiang, L.; Wu, X.; Medema, M.H.; Xu, S.; Li, X.; Schäfer, H.; et al. Microbial assimilatory sulfate reduction-mediated H2S: An overlooked role in Crohn’s disease development. Microbiome 2024, 12, 152. [Google Scholar] [CrossRef]
- Fusco, W.; Lorenzo, M.B.; Cintoni, M.; Porcari, S.; Rinninella, E.; Kaitsas, F.; Lener, E.; Mele, M.C.; Gasbarrini, A.; Collado, M.C.; et al. Short-Chain Fatty-Acid-Producing Bacteria: Key Components of the Human Gut Microbiota. Nutrients 2023, 15, 2211. [Google Scholar] [CrossRef] [PubMed]
- Oñate, F.P.; Chamignon, C.; Burz, S.D.; Lapaque, N.; Monnoye, M.; Philippe, C.; Bredel, M.; Chêne, L.; Farin, W.; Paillarse, J.-M.; et al. Adlercreutzia equolifaciens Is an Anti-Inflammatory Commensal Bacterium with Decreased Abundance in Gut Microbiota of Patients with Metabolic Liver Disease. Int. J. Mol. Sci. 2023, 24, 12232. [Google Scholar] [CrossRef]
- Gasaly, N.; de Vos, P.; Hermoso, M.A. Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation. Front. Immunol. 2021, 12, 658354. [Google Scholar] [CrossRef]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef]
- Monteleone, I.; Rizzo, A.; Sarra, M.; Sica, G.; Sileri, P.; Biancone, L.; Macdonald, T.T.; Pallone, F.; Monteleone, G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 2011, 141, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.; Vaughan, C.W.; Vandenberg, R.J. N-Acyl amino acids and N-acyl neurotransmitter conjugates: Neuromodulators and probes for new drug targets. Br. J. Pharmacol. 2010, 160, 1857–1871. [Google Scholar] [CrossRef]
- Crittenden, S.; Cheyne, A.; Adams, A.; Forster, T.; Robb, C.T.; Felton, J.; Ho, G.; Ruckerl, D.; Rossi, A.G.; Anderton, S.M.; et al. Purine metabolism controls innate lymphoid cell function and protects against intestinal injury. Immunol. Cell Biol. 2018, 96, 1049–1059. [Google Scholar] [CrossRef]
- Worledge, C.S.; Kostelecky, R.E.; Zhou, L.; Bhagavatula, G.; Colgan, S.P.; Lee, J.S. Allopurinol Disrupts Purine Metabolism to Increase Damage in Experimental Colitis. Cells 2024, 13, 373. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Oppedisano, F.; Gratteri, S.; Muscoli, C.; Mollace, V. Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 2016, 213, 8–14. [Google Scholar] [CrossRef]
- Hosomi, A.; Nakanishi, T.; Fujita, T.; Tamai, I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS ONE 2012, 7, e30456. [Google Scholar] [CrossRef]
- Dubinsky, M.C. Azathioprine, 6-mercaptopurine in inflammatory bowel disease: Pharmacology, efficacy, and safety. Clin. Gastroenterol. Hepatol. 2004, 2, 731–743. [Google Scholar] [CrossRef]
- Gearry, R.B.; Barclay, M.L. Azathioprine and 6-mercaptopurine pharmacogenetics and metabolite monitoring in inflammatory bowel disease. J. Gastroenterol. Hepatol. 2005, 20, 1149–1157. [Google Scholar] [CrossRef]
- Caldovic, L.; Ah Mew, N.; Shi, D.; Morizono, H.; Yudkoff, M.; Tuchman, M. N-acetylglutamate synthase: Structure, function and defects. Mol. Genet. Metab. 2010, 100, S13–S19. [Google Scholar] [CrossRef]
- Yokoo, K.; Yamamoto, Y.; Suzuki, T. Ammonia impairs tight junction barriers by inducing mitochondrial dysfunction in Caco-2 cells. FASEB J. 2021, 35, e21854. [Google Scholar] [CrossRef] [PubMed]
- Seethaler, B.; Basrai, M.; Neyrinck, A.M.; Nazare, J.-A.; Walter, J.; Delzenne, N.M.; Bischoff, S.C. Biomarkers for assessment of intestinal permeability in clinical practice. Am. J. Physiol. Liver Physiol. 2021, 321, G11–G17. [Google Scholar] [CrossRef]
- Dong, Z.; Gao, X.; Chinchilli, V.M.; Sinha, R.; Muscat, J.; Winkels, R.M.; Richie, J.P. Association of sulfur amino acid consumption with cardiometabolic risk factors: Cross-sectional findings from NHANES III. eClinicalMedicine 2020, 19, 100248. [Google Scholar] [CrossRef] [PubMed]
- Zizzo, M.G.; Caldara, G.; Bellanca, A.; Nuzzo, D.; Di Carlo, M.; Serio, R. Preventive effects of guanosine on intestinal inflammation in 2, 4-dinitrobenzene sulfonic acid (DNBS)-induced colitis in rats. Inflammopharmacology 2019, 27, 349–359. [Google Scholar] [CrossRef]
- Li, M.J.; Liu, B.W.; Li, R.; Yang, P.; Leng, P.; Huang, Y. Exploration of the link between gut microbiota and purinergic signalling. Purinergic Signal. 2023, 19, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.C.; Haskey, N.; Ramay, H.R.; Ghosh, T.S.; Taylor, L.M.; Yousuf, M.; Ohland, C.; McCoy, K.D.; Ingram, R.J.M.; Ghosh, S.; et al. Weighted Gene Co-Expression Network Analysis Identifies a Functional Guild and Metabolite Cluster Mediating the Relationship between Mucosal Inflammation and Adherence to the Mediterranean Diet in Ulcerative Colitis. Int. J. Mol. Sci. 2023, 24, 7323. [Google Scholar] [CrossRef]
- Magee, E.A.; Richardson, C.J.; Hughes, R.; Cummings, J.H. Contribution of dietary protein to sulfide production in the large intestine: An in vitro and a controlled feeding study in humans. Am. J. Clin. Nutr. 2000, 72, 1488–1494. [Google Scholar] [CrossRef]
- Jowett, S.L.; Seal, C.J.; Pearce, M.S.; Phillips, E.; Gregory, W.; Barton, J.R.; Welfare, M.R. Influence of dietary factors on the clinical course of ulcerative colitis: A prospective cohort study. Gut 2004, 53, 1479–1484. [Google Scholar] [CrossRef]
- Health Canada. Guidelines for Canadian Drinking Water Quality: Guideline Technical Document—Sulphate. Available online: https://www.canada.ca/en/health-canada/programs/guidelines-canadian-drinking-water-quality-operational-parameters.html (accessed on 9 May 2025).
- Rey, F.E.; Gonzalez, M.D.; Cheng, J.; Wu, M.; Ahern, P.P.; Gordon, J.I. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. USA 2013, 110, 13582–13587. [Google Scholar] [CrossRef]
- National Cancer Institute. Automated Self-Administered 24-Hour (ASA24) Dietary Assessment Tool. n.d. Available online: https://epi.grants.cancer.gov/asa24/ (accessed on 31 July 2021).
- National Cancer Institute: Reviewing and Cleaning ASA24® Data. 2018. Available online: https://epi.grants.cancer.gov/asa24/resources/cleaning.html (accessed on 9 May 2025).
- 53. Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef]
- Bishop, S.L.; Drikic, M.; Wacker, S.; Chen, Y.Y.; Kozyrskyj, A.L.; Lewis, I.A. Moving beyond descriptive studies: Harnessing metabolomics to elucidate the molecular mechanisms underpinning host-microbiome phenotypes. Mucosal Immunol. 2022, 15, 1071–1084. [Google Scholar] [CrossRef] [PubMed]
- Groves, R.A.; Mapar, M.; Aburashed, R.; Ponce, L.F.; Bishop, S.L.; Rydzak, T.; Drikic, M.; Bihan, D.G.; Benediktsson, H.; Clement, F.; et al. Methods for Quantifying the Metabolic Boundary Fluxes of Cell Cultures in Large Cohorts by High-Resolution Hydrophilic Liquid Chromatography Mass Spectrometry. Anal. Chem. 2022, 94, 8874–8882. [Google Scholar] [CrossRef] [PubMed]
- Rydzak, T.; Groves, R.A.; Zhang, R.; Aburashed, R.; Pushpker, R.; Mapar, M.; Lewis, I.A. Metabolic preference assay for rapid diagnosis of bloodstream infections. Nat. Commun. 2022, 13, 2332. [Google Scholar] [CrossRef]
- Melamud, E.; Vastag, L.; Rabinowitz, J.D. Metabolomic analysis and visualization engine for LC–MS data. Anal. Chem. 2010, 82, 9818–9826. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed]
- Rainer, J.; Vicini, A.; Salzer, L.; Stanstrup, J.; Badia, J.M.; Neumann, S.; Stravs, M.A.; Hernandes, V.V.; Gatto, L.; Gibb, S.; et al. A Modular and Expandable Ecosystem for Metabolomics Data Annotation in R. Metabolites 2022, 12, 173. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 10 May 2025).
- Bihan, D.G.; Rydzak, T.; Wyss, M.; Pittman, K.; McCoy, K.D.; Lewis, I.A. Method for absolute quantification of short chain fatty acids via reverse phase chromatography mass spectrometry. PLoS ONE 2022, 17, e0267093. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. Available online: https://www.r-project.org/ (accessed on 10 May 2025).


| Participant Characteristics | Control (n = 13) | Reduced Sulfur Diet (n = 9) |
|---|---|---|
| Sex, female | 5 (38%) * | 7 (78%) * |
| Age (years) | 47 (29–50) | 41 (32–46) |
| Body mass index (kg/m2) | 26 (25–31) | 24 (22–28) |
| Partial Mayo score (n, %) | ||
| Remission (<2) | 4 (31%) | 3 (33%) |
| Mild (2–4) | 4 (31%) | 5 (56%) |
| Moderate (5–7) | 3 (23%) | 1 (11%) |
| Severe (>7) | 2 (15%) | 0 (0%) |
| Fecal calprotectin (mcg/g) | 593 (92–1789) | 249 (44–1728) |
| Medications † | ||
| Steroids | 5 (38%) | 2 (22%) |
| 5-ASA | 9 (69%) | 6 (67%) |
| Immunomodulator | 1 (7%) | 2 (22%) |
| Biologic | 1 (7%) | 2 (22%) |
| Taxon | P52 | P45 | P44 | P42 | P30 | P29 | P28 |
|---|---|---|---|---|---|---|---|
| Unknown | 1.0 | 1.0 | 1.2 | 0.9 | 1.0 | 0.9 | 0.9 |
| Adlercreutzia_equolifaciens | 1.5 | 0.5 | 0.4 | 0.4 | 0.1 | 0.6 | 0.4 |
| Asaccharobacter_celatus | 1.2 | 0.5 | 0.5 | 0.2 | 0.3 | 0.5 | 0.4 |
| Eggerthella_lenta | 0.5 | 0.4 | 0.3 | 0.4 | 0.0 | 0.3 | 0.6 |
| Enterorhabdus_caecimuris | 1.0 | 0.5 | 0.5 | 0.2 | 0.0 | 1.0 | 0.5 |
| Gordonibacter_pamelaeae | 1.4 | 0.5 | 0.7 | 0.1 | 3.7 | 0.6 | 0.6 |
| Bacteroides_ovatus | 2.5 | 2.6 | 0.3 | 0.4 | 0.0 | 3.2 | 30.9 |
| Parabacteroides_distasonis | 2.2 | 1.2 | 0.1 | 8.7 | 0.1 | 11.7 | 54.5 |
| Streptococcus_salivarius | 1.7 | 1.1 | 0.8 | 21.4 | 2.1 | 1.2 | 81.2 |
| Blautia_wexlerae | 1.8 | 1.9 | 2.2 | 0.1 | 0.5 | 0.1 | 1.5 |
| Agathobaculum_butyriciproducens | 2.2 | 0.3 | 7.4 | 4.3 | 4.3 | 0.3 | 0.2 |
| Faecalibacterium_prausnitzii | 1.0 | 2.3 | 1.8 | 11.5 | 0.8 | 4.4 | 0.4 |
| Flavonifractor_plautii | 3.7 | 0.0 | 0.1 | 4.5 | 0.2 | 0.0 | 1.5 |
| Ruthenibacterium_lactatiformans | 0.8 | 0.2 | 0.2 | 0.3 | 2.5 | 0.2 | 1.0 |
| Clostridium_spiroforme | 0.7 | 1.2 | 1.4 | 0.0 | 2.0 | 0.9 | 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, J.; Raman, M.; Taylor, L.M.; Yousuf, M.; Panaccione, R.; Turbide, C.; Sinha, S.R.; Haskey, N. Reduced Sulfur Diet Reshapes the Microbiome and Metabolome in Mild–Moderate Ulcerative Colitis. Int. J. Mol. Sci. 2025, 26, 4596. https://doi.org/10.3390/ijms26104596
Ye J, Raman M, Taylor LM, Yousuf M, Panaccione R, Turbide C, Sinha SR, Haskey N. Reduced Sulfur Diet Reshapes the Microbiome and Metabolome in Mild–Moderate Ulcerative Colitis. International Journal of Molecular Sciences. 2025; 26(10):4596. https://doi.org/10.3390/ijms26104596
Chicago/Turabian StyleYe, Jiayu, Maitreyi Raman, Lorian M. Taylor, Munazza Yousuf, Remo Panaccione, Christian Turbide, Sidhartha R. Sinha, and Natasha Haskey. 2025. "Reduced Sulfur Diet Reshapes the Microbiome and Metabolome in Mild–Moderate Ulcerative Colitis" International Journal of Molecular Sciences 26, no. 10: 4596. https://doi.org/10.3390/ijms26104596
APA StyleYe, J., Raman, M., Taylor, L. M., Yousuf, M., Panaccione, R., Turbide, C., Sinha, S. R., & Haskey, N. (2025). Reduced Sulfur Diet Reshapes the Microbiome and Metabolome in Mild–Moderate Ulcerative Colitis. International Journal of Molecular Sciences, 26(10), 4596. https://doi.org/10.3390/ijms26104596







