Novel Metabolites Genetically Linked to Salt Sensitivity of Blood Pressure: Evidence from mGWAS in Chinese Population
Abstract
1. Introduction
2. Results
2.1. Basic Characteristics of the Participants
2.2. Metabolite Genome-Wide Association Analysis
2.3. Identification of Independent mQTLs
2.4. Effector Genes of Genetically Influenced Metabolites
2.5. Potential Proteins Related to Genetically Influenced Metabolites
2.6. Associations Between GIMs and SSBP
2.7. Mendelian Randomization Analyses
2.8. Construction of SNP–Gene–Metabolite Association Network Related to SSBP
2.9. Replication Analyses
3. Discussion
4. Materials and Methods
4.1. Study Population and Samples
4.2. Determination of SS and SR
4.3. Untargeted Metabolomics Profiling and Data Processing
4.4. DNA Extraction, Genotyping, and Quality Control
4.5. mGWAS Analysis
4.6. Statistical Analysis
4.7. Replication Set
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GIM | Genetically influenced metabolite |
| mGWAS | Metabolome genome-wide association study |
| MR | Mendelian randomization |
| LD | Linear dichroism |
| SSBP | Salt sensitivity of blood pressure |
| SS | Salt-sensitive |
| SR | Salt-resistant |
| GWAS | Genome-wide association study |
| mQTLs | Metabolic quantitative trait loci |
| SBP | Systolic blood pressure |
| DBP | Diastolic blood pressure |
| MAP | Mean arterial pressure |
| uGRSs | Unweighted genetic risk scores |
| wGRSs | Weighted genetic risk scores |
| 2SLSs | Two-stage least squares |
References
- Manosroi, W.; Williams, G.H. Genetics of Human Primary Hypertension: Focus on Hormonal Mechanisms. Endocr. Rev. 2019, 40, 825–856. [Google Scholar] [CrossRef]
- Elijovich, F.; Weinberger, M.H.; Anderson, C.A.; Appel, L.J.; Bursztyn, M.; Cook, N.R.; Dart, R.A.; Newton-Cheh, C.H.; Sacks, F.M.; Laffer, C.L. Salt Sensitivity of Blood Pressure: A Scientific Statement From the American Heart Association. Hypertension 2016, 68, e7–e46. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.A.; Dhaun, N. Salt Sensitivity: Causes, Consequences, and Recent Advances. Hypertension 2024, 81, 476–489. [Google Scholar] [CrossRef]
- Afolabi, J.; Laffer, C.L.; Beasley, H.K.; Hinton, A.; Masenga, S.K.; Kirabo, A. Salt Sensitivity of Blood Pressure. Circ. Res. 2024, 134, 1234–1239. [Google Scholar] [CrossRef]
- Wright, J.T., Jr.; Rahman, M.; Scarpa, A.; Fatholahi, M.; Griffin, V.; Jean-Baptiste, R.; Islam, M.; Eissa, M.; White, S.; Douglas, J.G. Determinants of salt sensitivity in black and white normotensive and hypertensive women. Hypertension 2003, 42, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Parksook, W.W.; Williams, G.H. Challenges and Approach to Identifying Individuals with Salt Sensitivity of Blood Pressure. Am. J. Nephrol. 2022, 53, 847–855. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Dominiczak, A.F. Genomics of hypertension: The road to precision medicine. Nat. Rev. Cardiol. 2021, 18, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Tam, V.; Patel, N.; Turcotte, M.; Bossé, Y.; Paré, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Shin, S.Y.; Fauman, E.B.; Petersen, A.K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef]
- Adamski, J. Genome-wide association studies with metabolomics. Genome Med. 2012, 4, 34. [Google Scholar] [CrossRef]
- Luo, S.; Feofanova, E.V.; Tin, A.; Tung, S.; Rhee, E.P.; Coresh, J.; Arking, D.E.; Surapaneni, A.; Schlosser, P.; Li, Y.; et al. Genome-wide association study of serum metabolites in the African American Study of Kidney Disease and Hypertension. Kidney Int. 2021, 100, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, Q.; Liu, Y.; Chen, S.; Zhang, Y.; Ma, Q.; Chen, X.; Liu, C.; Lei, H.; Chen, H.; et al. Genome-wide association study of metabolites in patients with coronary artery disease identified novel metabolite quantitative trait loci. Clin. Transl. Med. 2021, 11, e290. [Google Scholar] [CrossRef]
- Feofanova, E.V.; Chen, H.; Dai, Y.; Jia, P.; Grove, M.L.; Morrison, A.C.; Qi, Q.; Daviglus, M.; Cai, J.; North, K.E.; et al. A Genome-wide Association Study Discovers 46 Loci of the Human Metabolome in the Hispanic Community Health Study/Study of Latinos. Am. J. Hum. Genet. 2020, 107, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Hartiala, J.A.; Tang, W.H.; Wang, Z.; Crow, A.L.; Stewart, A.F.; Roberts, R.; McPherson, R.; Erdmann, J.; Willenborg, C.; Hazen, S.L.; et al. Genome-wide association study and targeted metabolomics identifies sex-specific association of CPS1 with coronary artery disease. Nat. Commun. 2016, 7, 10558. [Google Scholar] [CrossRef]
- Lee, S.; Han, M.; Moon, S.; Kim, K.; An, W.J.; Ryu, H.; Oh, K.H.; Park, S.K. Identifying Genetic Variants and Metabolites Associated with Rapid Estimated Glomerular Filtration Rate Decline in Korea Based on Genome-Metabolomic Integrative Analysis. Metabolites 2022, 12, 1139. [Google Scholar] [CrossRef]
- Johnson, R.K.; Brunetti, T.; Quinn, K.; Doenges, K.; Campbell, M.; Arehart, C.; Taub, M.A.; Mathias, R.A.; Reisdorph, N.; Barnes, K.C.; et al. Discovering metabolite quantitative trait loci in asthma using an isolated population. J. Allergy Clin. Immunol. 2022, 149, 1807–1811.e16. [Google Scholar] [CrossRef]
- Hoff, S.; Halbritter, J.; Epting, D.; Frank, V.; Nguyen, T.M.; van Reeuwijk, J.; Boehlke, C.; Schell, C.; Yasunaga, T.; Helmstädter, M.; et al. ANKS6 is a central component of a nephronophthisis module linking NEK8 to INVS and NPHP3. Nat. Genet. 2013, 45, 951–956. [Google Scholar] [CrossRef]
- Sung, Y.J.; Pérusse, L.; Sarzynski, M.A.; Fornage, M.; Sidney, S.; Sternfeld, B.; Rice, T.; Terry, J.G.; Jacobs, D.R., Jr.; Katzmarzyk, P.; et al. Genome-wide association studies suggest sex-specific loci associated with abdominal and visceral fat. Int. J. Obes. 2016, 40, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, T.; Pettersson-Kymmer, U.; Stewart, I.D.; Butler-Laporte, G.; Nakanishi, T.; Cerani, A.; Liang, K.Y.H.; Yoshiji, S.; Willett, J.D.S.; et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 2023, 55, 44–53. [Google Scholar] [CrossRef]
- Feofanova, E.V.; Yu, B.; Metcalf, G.A.; Liu, X.; Muzny, D.; Below, J.E.; Wagenknecht, L.E.; Gibbs, R.A.; Morrison, A.C.; Boerwinkle, E. Sequence-Based Analysis of Lipid-Related Metabolites in a Multiethnic Study. Genetics 2018, 209, 607–616. [Google Scholar] [CrossRef]
- Lotta, L.A.; Pietzner, M.; Stewart, I.D.; Wittemans, L.B.L.; Li, C.; Bonelli, R.; Raffler, J.; Biggs, E.K.; Oliver-Williams, C.; Auyeung, V.P.W.; et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nat. Genet. 2021, 53, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Chan, L.S.; Bose, D.; Jackson, A.U.; VandeHaar, P.; Locke, A.E.; Fuchsberger, C.; Stringham, H.M.; Welch, R.; Yu, K.; et al. Genome-wide association studies of metabolites in Finnish men identify disease-relevant loci. Nat. Commun. 2022, 13, 1644. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Li, Q.; Sui, H.; Zhang, H. Role of Oxidation-Dependent CaMKII Activation in the Genesis of Abnormal Action Potentials in Atrial Cardiomyocytes: A Simulation Study. Biomed. Res. Int. 2020, 2020, 1597012. [Google Scholar] [CrossRef] [PubMed]
- Ben-Haim, Y.; Bird, M.; Johnson, D.; Mohiddin, S.; Favaloro, L.; Dyrberg Andersen, J.; Sheppard, M.N.; Pittman, A.; Futema, M.; Behr, E.R. RYR2 Variant and Sudden Death in Patients with Dilated Cardiomyopathy. J. Am. Coll. Cardiol. 2024, 83, 1105–1107. [Google Scholar] [CrossRef]
- He, K.Y.; Wang, H.; Cade, B.E.; Nandakumar, P.; Giri, A.; Ware, E.B.; Haessler, J.; Liang, J.; Smith, J.A.; Franceschini, N.; et al. Rare variants in fox-1 homolog A (RBFOX1) are associated with lower blood pressure. PLoS Genet. 2017, 13, e1006678. [Google Scholar] [CrossRef]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; Hrabé de Angelis, M.; Kronenberg, F.; Meitinger, T.; Mewes, H.W.; Wichmann, H.E.; Weinberger, K.M.; Adamski, J.; et al. Genetics meets metabolomics: A genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef]
- Cullell, N.; Soriano-Tárraga, C.; Gallego-Fábrega, C.; Cárcel-Márquez, J.; Muiño, E.; Llucià-Carol, L.; Lledós, M.; Esteller, M.; de Moura, M.C.; Montaner, J.; et al. Altered methylation pattern in EXOC4 is associated with stroke outcome: An epigenome-wide association study. Clin. Epigenetics 2022, 14, 124. [Google Scholar] [CrossRef]
- Levy, D.; Larson, M.G.; Benjamin, E.J.; Newton-Cheh, C.; Wang, T.J.; Hwang, S.J.; Vasan, R.S.; Mitchell, G.F. Framingham Heart Study 100K Project: Genome-wide associations for blood pressure and arterial stiffness. BMC Med. Genet. 2007, 8 (Suppl. S1), S3. [Google Scholar] [CrossRef]
- Yet, I.; Menni, C.; Shin, S.Y.; Mangino, M.; Soranzo, N.; Adamski, J.; Suhre, K.; Spector, T.D.; Kastenmüller, G.; Bell, J.T. Genetic Influences on Metabolite Levels: A Comparison across Metabolomic Platforms. PLoS ONE 2016, 11, e0153672. [Google Scholar] [CrossRef]
- Kitamura, K.; Sato, K.; Sawabe, M.; Yoshida, M.; Hagiwara, N. P-Selectin Glycoprotein Ligand-1 (PSGL-1) Expressing CD4 T Cells Contribute Plaque Instability in Acute Coronary Syndrome. Circ. J. 2018, 82, 2128–2135. [Google Scholar] [CrossRef]
- Yeh, K.H.; Hsu, L.A.; Teng, M.S.; Wu, S.; Chou, H.H.; Ko, Y.L. Pleiotropic Effects of Common and Rare GCKR Exonic Mutations on Cardiometabolic Traits. Genes 2022, 13, 491. [Google Scholar] [CrossRef]
- Ercu, M.; Mücke, M.B.; Pallien, T.; Markó, L.; Sholokh, A.; Schächterle, C.; Aydin, A.; Kidd, A.; Walter, S.; Esmati, Y.; et al. Mutant Phosphodiesterase 3A Protects From Hypertension-Induced Cardiac Damage. Circulation 2022, 146, 1758–1778. [Google Scholar] [CrossRef]
- Traylor, M.; Amin Al Olama, A.; Lyytikäinen, L.P.; Marini, S.; Chung, J.; Malik, R.; Dichgans, M.; Kähönen, M.; Lehtimäki, T.; Anderson, C.D.; et al. Influence of Genetic Variation in PDE3A on Endothelial Function and Stroke. Hypertension 2020, 75, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lee, J.; Vincent, L.G.; Wang, Q.; Gu, M.; Lan, F.; Churko, J.M.; Sallam, K.I.; Matsa, E.; Sharma, A.; et al. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised β-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell 2015, 17, 89–100. [Google Scholar] [CrossRef]
- Ercu, M.; Walter, S.; Klussmann, E. Mutations in Phosphodiesterase 3A (PDE3A) Cause Hypertension Without Cardiac Damage. Hypertension 2023, 80, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Min, J.Y.; Min, K.B. Interaction between the RGS6 gene and psychosocial stress on obesity-related traits. Endocr. J. 2017, 64, 357–362. [Google Scholar] [CrossRef]
- Sengar, A.S.; Kumar, M.; Rai, C.; Chakraborti, S.; Kumar, D.; Kumar, P.; Mukherjee, S.; Mondal, K.; Stewart, A.; Maity, B. RGS6 drives cardiomyocyte death following nucleolar stress by suppressing Nucleolin/miRNA-21. J. Transl. Med. 2024, 22, 204. [Google Scholar] [CrossRef] [PubMed]
- Annevelink, C.E.; Sapp, P.A.; Petersen, K.S.; Shearer, G.C.; Kris-Etherton, P.M. Diet-derived and diet-related endogenously produced palmitic acid: Effects on metabolic regulation and cardiovascular disease risk. J. Clin. Lipidol. 2023, 17, 577–586. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, M.; Jung, S.; Lee, S.H.; Lee, J.H. Altered plasma lysophosphatidylcholines and amides in non-obese and non-diabetic subjects with borderline-to-moderate hypertriglyceridemia: A case-control study. PLoS ONE 2015, 10, e0123306. [Google Scholar] [CrossRef]
- Piccinin, E.; Cariello, M.; De Santis, S.; Ducheix, S.; Sabbà, C.; Ntambi, J.M.; Moschetta, A. Role of Oleic Acid in the Gut-Liver Axis: From Diet to the Regulation of Its Synthesis via Stearoyl-CoA Desaturase 1 (SCD1). Nutrients 2019, 11, 2283. [Google Scholar] [CrossRef]
- Lopez, S.; Bermudez, B.; Montserrat-de la Paz, S.; Jaramillo, S.; Abia, R.; Muriana, F.J. Virgin Olive Oil and Hypertension. Curr. Vasc. Pharmacol. 2016, 14, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, M.I.; Gonzalo-Gobernado, R.; Montaner, J. Neuroprotective diets for stroke. Neurochem. Int. 2017, 107, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.J.; de las Fuentes, L.; Schwander, K.L.; Simino, J.; Rao, D.C. Gene-smoking interactions identify several novel blood pressure loci in the Framingham Heart Study. Am. J. Hypertens. 2015, 28, 343–354. [Google Scholar] [CrossRef]
- Fowler, E.D.; Zissimopoulos, S. Molecular, Subcellular, and Arrhythmogenic Mechanisms in Genetic RyR2 Disease. Biomolecules 2022, 12, 1030. [Google Scholar] [CrossRef]
- Sanchez-Alonso, J.L.; Loucks, A.; Schobesberger, S.; van Cromvoirt, A.M.; Poulet, C.; Chowdhury, R.A.; Trayanova, N.; Gorelik, J. Nanoscale regulation of L-type calcium channels differentiates between ischemic and dilated cardiomyopathies. EBioMedicine 2020, 57, 102845. [Google Scholar] [CrossRef]
- Knezevic, I.I.; Predescu, S.A.; Neamu, R.F.; Gorovoy, M.S.; Knezevic, N.M.; Easington, C.; Malik, A.B.; Predescu, D.N. Tiam1 and Rac1 are required for platelet-activating factor-induced endothelial junctional disassembly and increase in vascular permeability. J. Biol. Chem. 2009, 284, 5381–5394. [Google Scholar] [CrossRef] [PubMed]
- Adeyemo, A.; Gerry, N.; Chen, G.; Herbert, A.; Doumatey, A.; Huang, H.; Zhou, J.; Lashley, K.; Chen, Y.; Christman, M.; et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009, 5, e1000564. [Google Scholar] [CrossRef]
- Piechocki, M.; Przewłocki, T.; Pieniążek, P.; Trystuła, M.; Podolec, J.; Kabłak-Ziembicka, A. A Non-Coronary, Peripheral Arterial Atherosclerotic Disease (Carotid, Renal, Lower Limb) in Elderly Patients-A Review: Part I-Epidemiology, Risk Factors, and Atherosclerosis-Related Diversities in Elderly Patients. J. Clin. Med. 2024, 13, 1471. [Google Scholar] [CrossRef]
- Piechocki, M.; Przewłocki, T.; Pieniążek, P.; Trystuła, M.; Podolec, J.; Kabłak-Ziembicka, A. A Non-Coronary, Peripheral Arterial Atherosclerotic Disease (Carotid, Renal, Lower Limb) in Elderly Patients-A Review PART II-Pharmacological Approach for Management of Elderly Patients with Peripheral Atherosclerotic Lesions Outside Coronary Territory. J. Clin. Med. 2024, 13, 1508. [Google Scholar] [CrossRef]
- Lin, Z.; Li, J.; Liu, F.; Cao, J.; Chen, S.; Chen, J.; Huang, K.; Wang, Y.; Li, H.; Wang, Y.; et al. Metabolomics signature of blood pressure salt sensitivity and its link to cardiovascular disease: A dietary salt-intervention trial. Sci. China Life Sci. 2024, 67, 1666–1675. [Google Scholar] [CrossRef]
- Xie, Y.; Qi, H.; Peng, W.; Li, B.; Wen, F.; Zhang, F.; Zhang, L. SNPs in lncRNA KCNQ1OT1 Modulate Its Expression and Confer Susceptibility to Salt Sensitivity of Blood Pressure in a Chinese Han Population. Nutrients 2022, 14, 3990. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, F.; Zhang, B.; Qi, H.; Xie, Y.; Peng, W.; Li, B.; Wen, F.; Li, P.; Sun, Y.; et al. Associations of Metabolites Related Salt Sensitivity of Blood Pressure and Essential Hypertension in Chinese Population: The EpiSS Study. Nutrients 2025, 17, 1289. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.L.; Lyu, Y.; Jiang, Y.; Xiang, J.; Ji, S.; Kang, S.; Lyu, X.; He, C.; Li, P.; et al. mGWAS identification of six novel single nucleotide polymorphism loci with strong correlation to gastric cancer. Cancer Metab. 2021, 9, 34. [Google Scholar] [CrossRef]
- Tian, Z.; Liu, Y.; Usa, K.; Mladinov, D.; Fang, Y.; Ding, X.; Greene, A.S.; Cowley, A.W., Jr.; Liang, M. Novel role of fumarate metabolism in dahl-salt sensitive hypertension. Hypertension 2009, 54, 255–260. [Google Scholar] [CrossRef]
- Xue, H.; Geurts, A.M.; Usa, K.; Wang, F.; Lin, Y.; Phillips, J.; Henderson, L.; Baker, M.A.; Tian, Z.; Liang, M. Fumarase Overexpression Abolishes Hypertension Attributable to endothelial NO synthase Haploinsufficiency in Dahl Salt-Sensitive Rats. Hypertension 2019, 74, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Gökcen, B.B.; Şanlier, N. Coffee consumption and disease correlations. Crit. Rev. Food Sci. Nutr. 2019, 59, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.E.; Flury, A.; Marmet, C.; Poquet, L.; Rimoldi, S.F.; Sartori, C.; Rexhaj, E.; Brenner, R.; Allemann, Y.; Zimmermann, D.; et al. Mediation of coffee-induced improvements in human vascular function by chlorogenic acids and its metabolites: Two randomized, controlled, crossover intervention trials. Clin. Nutr. 2017, 36, 1520–1529. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Song, H.; Pan, X.; Xue, H.; Wan, Y.; Wang, T.; Tian, Z.; Hou, E.; Lanza, I.R.; Liu, P.; et al. Urinary Metabolites Associated with Blood Pressure on a Low- or High-Sodium Diet. Theranostics 2018, 8, 1468–1480. [Google Scholar] [CrossRef]
- Qi, H.; Liu, B.; Guo, C.; Liu, Z.; Cao, H.; Liu, K.; Sun, W.; Zhang, L. Effects of environmental and genetic risk factors for salt sensitivity on blood pressure in northern China: The systemic epidemiology of salt sensitivity (EpiSS) cohort study. BMJ Open 2018, 8, e023042. [Google Scholar] [CrossRef]
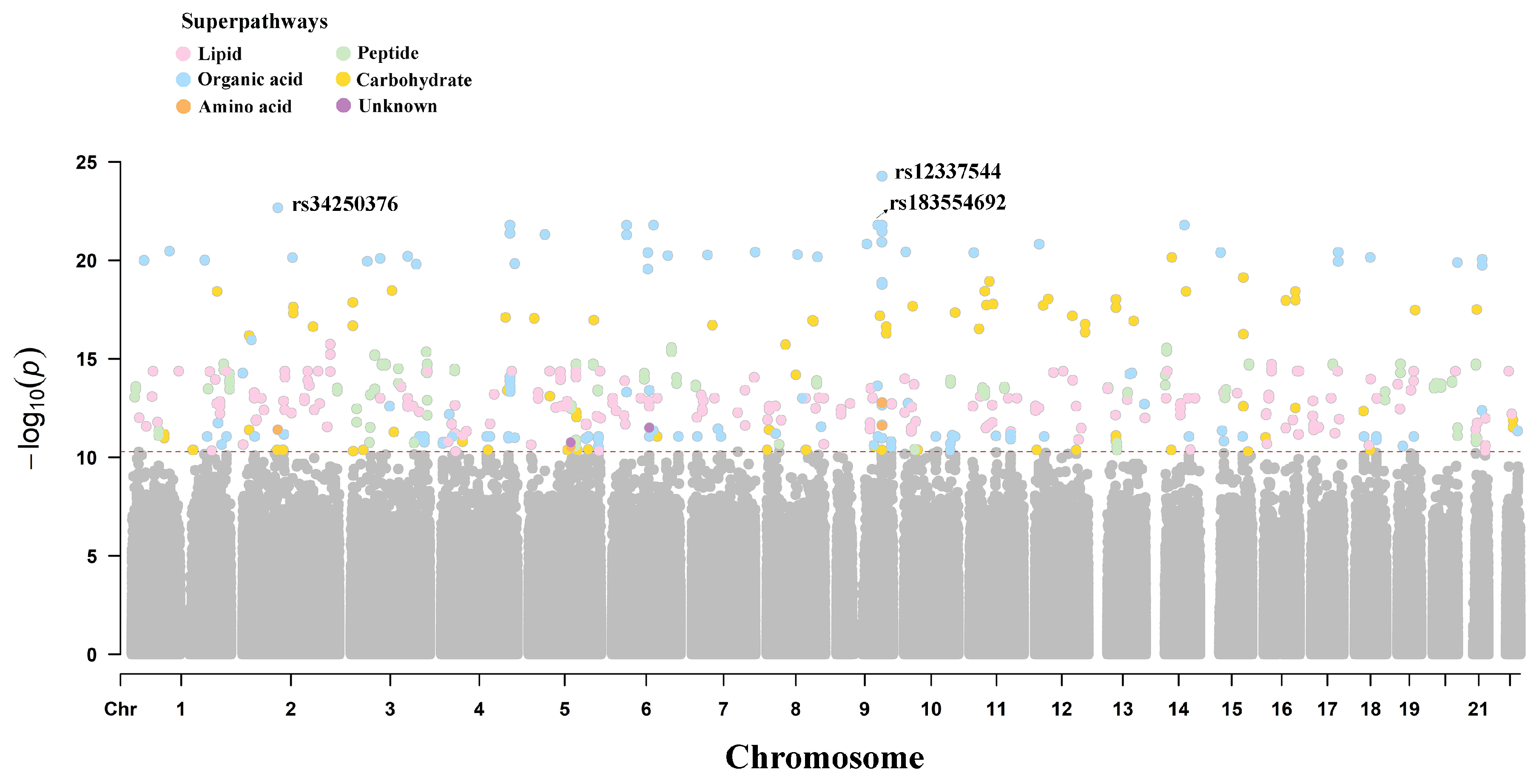

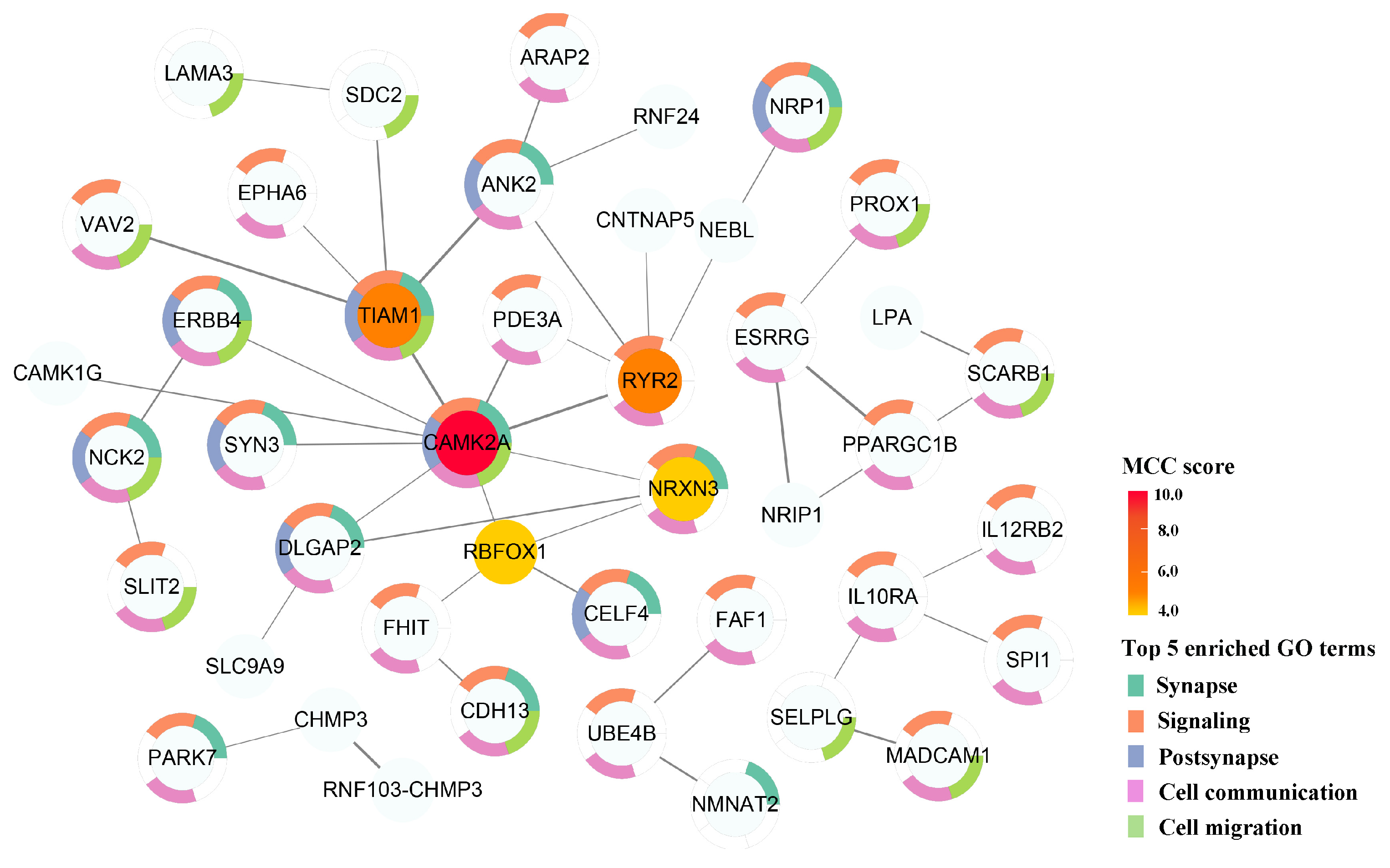

| Variables | Total (n = 54) | SS (n = 30) | SR (n = 24) | p |
|---|---|---|---|---|
| Age (year) | 57.57 ± 5.43 | 57.73 ± 5.77 | 57.38 ± 5.11 | 0.812 a |
| Gender (Male, n, %) | 17 (31.5) | 8 (33.3) | 9 (30.0) | 1.000 c |
| BMI (kg/m2) | 26.62 ± 3.52 | 26.39 ± 3.26 | 26.92 ± 3.86 | 0.586 a |
| FBG (mmol/L) | 5.45 ± 0.63 | 5.44 ± 0.61 | 5.47 ± 0.66 | 0.924 b |
| TC (mmol/L) | 5.48 ± 0.94 | 5.37 ± 1.03 | 5.61 ± 0.80 | 0.360 a |
| TGs (mmol/L) | 1.83 ± 1.03 | 1.86 ± 1.09 | 1.79 ± 0.96 | 0.721 b |
| LDL-C (mmol/L) | 2.32 ± 0.91 | 2.24 ± 0.88 | 2.42 ± 0.95 | 0.461 a |
| HDL-C (mmol/L) | 1.95 ± 1.17 | 1.93 ± 1.21 | 1.97 ± 1.14 | 0.614 b |
| SBP (mmHg) | 122.01 ± 20.45 | 114.19 ± 21.10 | 131.78 ± 14.94 | 0.001 a |
| DBP (mmHg) | 78.11 ± 12.02 | 71.72 ± 10.72 | 86.10 ± 8.25 | <0.001 a |
| MAP (mmHg) | 92.74 ± 13.39 | 85.87 ± 12.52 | 101.33 ± 8.78 | <0.001 a |
| ΔMAP1 (mmHg) | −2.62 ± 15.14 | 10.16 ± 3.31 | −18.59 ± 5.92 | <0.001 b |
| ΔMAP2 (mmHg) | −2.07 ± 10.78 | −10.48 ± 5.44 | 8.44 ± 4.81 | <0.001 a |
| Smoking (n, %) | 7 (13.0) | 5 (16.7) | 2 (8.3) | 0.443 d |
| Drinking (n, %) | 26 (48.1) | 16 (53.3) | 10 (41.7) | 0.563 c |
| Hypertension (n, %) | 28 (51.9) | 15 (50.0) | 13 (54.2) | 0.976 c |
| Family history of hypertension (n, %) | 29 (53.7) | 18 (60.0) | 11 (45.8) | 0.446 c |
| GIMs | Numbers of mQTLs | SNP * | CHR | Position | Minor Allele | Other Allele | MAF | p |
|---|---|---|---|---|---|---|---|---|
| Arabinonic acid | 67 | rs3792688 | 14 | 36788896 | G | C | 0.010 | 7.13 × 10−21 |
| Feruloylquinic acid | 46 | rs11076702 | 16 | 88774569 | A | C | 0.019 | 4.20 × 10−15 |
| Phe-lle | 43 | rs11758014 | 6 | 148830586 | C | G | 0.010 | 2.73 × 10−16 |
| Phe-Phe | 42 | rs111865319 | 14 | 23507554 | T | G | 0.010 | 4.08 × 10−16 |
| LysoPC (0:0/22:5n-3) | 34 | rs17773637 | 2 | 103168041 | A | T | 0.010 | 1.40 × 10−13 |
| Androsterone sulfate | 29 | rs193142735 | 7 | 28077870 | T | A | 0.010 | 1.34 × 10−13 |
| Fumaric acid | 29 | rs117266991 | 6 | 93172013 | G | A | 0.028 | 3.96 × 10−14 |
| TGs (12:0/12:0/12:0) | 24 | rs11076702 | 16 | 88774569 | A | C | 0.019 | 4.20 × 10−15 |
| Glycoursodeoxycholate 3-glucuronide | 23 | rs111941908 | 2 | 29660776 | G | A | 0.019 | 9.95 × 10−14 |
| Oleic acid | 19 | rs148784825 | 1 | 196024278 | C | T | 0.019 | 4.41 × 10−15 |
| LysoPC (0:0/14:0) | 2 | rs7523503 | 1 | 200899804 | A | G | 0.151 | 4.60 × 10−11 |
| 16α-hydroxy DHEA 3-sulfate isomer | 2 | rs3782520 | 12 | 109027340 | T | C | 0.074 | 1.25 × 10−11 |
| L-Cystine | 2 | rs17812386 | 9 | 113942888 | A | G | 0.029 | 1.65 × 10−13 |
| X-MZ140RT42 | 2 | rs117266991 | 6 | 93172013 | G | A | 0.028 | 3.16 × 10−12 |
| Glycoursocholic acid | 1 | rs117458538 | 16 | 86932146 | G | A | 0.056 | 6.90 × 10−12 |
| DG (18:0/18:0) | 1 | rs146258959 | 18 | 34962385 | C | T | 0.028 | 2.40 × 10−11 |
| Palmitic amide | 1 | rs118008665 | 17 | 59640888 | T | C | 0.028 | 5.92 × 10−12 |
| N-Acetyl-l-aspartic acid | 1 | rs145741649 | 5 | 105503858 | A | G | 0.020 | 2.89 × 10−11 |
| GIMs | Model 1 a | Model 2 b | ||||
|---|---|---|---|---|---|---|
| β | ORs (95%CI) | p | β | ORs (95%CI) | p | |
| Arabinonic acid | −0.087 | 0.916 (0.866, 0.970) | 0.004 | −0.085 | 0.919 (0.871, 0.970) | 0.003 |
| Feruloylquinic acid | −0.052 | 0.949 (0.915, 0.984) | 0.007 | −0.054 | 0.947 (0.915, 0.981) | 0.003 |
| Phe-lle | −0.097 | 0.908 (0.881, 0.936) | <0.001 | −0.093 | 0.911 (0.887, 0.935) | <0.001 |
| Phe-Phe | −0.088 | 0.915 (0.892, 0.939) | <0.001 | −0.087 | 0.916 (0.894, 0.939) | <0.001 |
| LysoPC (0:0/22:5n-3) | 0.092 | 1.097 (1.046, 1.150) | <0.001 | 0.093 | 1.098 (1.047, 1.151) | <0.001 |
| Androsterone sulfate | −0.065 | 0.937 (0.909, 0.967) | <0.001 | −0.066 | 0.937 (0.908, 0.966) | <0.001 |
| Fumaric acid | −0.563 | 0.570 (0.389, 0.834) | 0.006 | −0.564 | 0.569 (0.388, 0.834) | 0.006 |
| TGs (12:0/12:0/12:0) | −0.182 | 0.833 (0.794, 0.875) | <0.001 | −0.180 | 0.835 (0.796, 0.877) | <0.001 |
| Glycoursodeoxycholate 3-glucuronide | −0.053 | 0.948 (0.928, 0.969) | <0.001 | −0.056 | 0.946 (0.926, 0.966) | <0.001 |
| Oleic acid | −0.044 | 0.957 (0.934, 0.981) | 0.001 | −0.045 | 0.956 (0.934, 0.979) | <0.001 |
| LysoPC (0:0/14:0) | −0.353 | 0.702 (0.553, 0.891) | 0.005 | −0.353 | 0.702 (0.553, 0.891) | 0.005 |
| 16α-hydroxy DHEA 3-sulfate isomer | −0.037 | 0.964 (0.881, 1.055) | 0.428 | −0.027 | 0.973 (0.887, 1.068) | 0.567 |
| L-Cystine | −0.207 | 0.813 (0.711, 0.930) | 0.004 | −0.198 | 0.820 (0.726, 0.927) | 0.002 |
| X-MZ140RT42 | −0.528 | 0.590 (0.428, 0.814) | 0.002 | −0.533 | 0.587 (0.423, 0.815) | 0.002 |
| Glycoursocholic acid | −0.050 | 0.951 (0.895, 1.011) | 0.117 | −0.050 | 0.951 (0.895, 1.011) | 0.117 |
| DG (18:0/18:0) | 0.068 | 1.071 (0.798, 1.437) | 0.652 | 0.068 | 1.071 (0.798, 1.437) | 0.652 |
| Palmitic amide | 0.140 | 1.150 (0.673, 1.965) | 0.611 | 0.140 | 1.150 (0.673, 1.965) | 0.611 |
| N-Acetyl-l-aspartic acid | −0.486 | 0.615 (0.466, 0.811) | 0.001 | −0.486 | 0.615 (0.466, 0.811) | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Zhang, B.; Wen, F.; Qi, H.; Zhang, F.; Xie, Y.; Peng, W.; Li, B.; Qu, A.; Yao, X.; et al. Novel Metabolites Genetically Linked to Salt Sensitivity of Blood Pressure: Evidence from mGWAS in Chinese Population. Int. J. Mol. Sci. 2025, 26, 4538. https://doi.org/10.3390/ijms26104538
Yang X, Zhang B, Wen F, Qi H, Zhang F, Xie Y, Peng W, Li B, Qu A, Yao X, et al. Novel Metabolites Genetically Linked to Salt Sensitivity of Blood Pressure: Evidence from mGWAS in Chinese Population. International Journal of Molecular Sciences. 2025; 26(10):4538. https://doi.org/10.3390/ijms26104538
Chicago/Turabian StyleYang, Xiaojun, Bowen Zhang, Fuyuan Wen, Han Qi, Fengxu Zhang, Yunyi Xie, Wenjuan Peng, Boya Li, Aibin Qu, Xinyue Yao, and et al. 2025. "Novel Metabolites Genetically Linked to Salt Sensitivity of Blood Pressure: Evidence from mGWAS in Chinese Population" International Journal of Molecular Sciences 26, no. 10: 4538. https://doi.org/10.3390/ijms26104538
APA StyleYang, X., Zhang, B., Wen, F., Qi, H., Zhang, F., Xie, Y., Peng, W., Li, B., Qu, A., Yao, X., & Zhang, L. (2025). Novel Metabolites Genetically Linked to Salt Sensitivity of Blood Pressure: Evidence from mGWAS in Chinese Population. International Journal of Molecular Sciences, 26(10), 4538. https://doi.org/10.3390/ijms26104538





