Noninvasive Prenatal Paternity Testing: A Review on Genetic Markers
Abstract
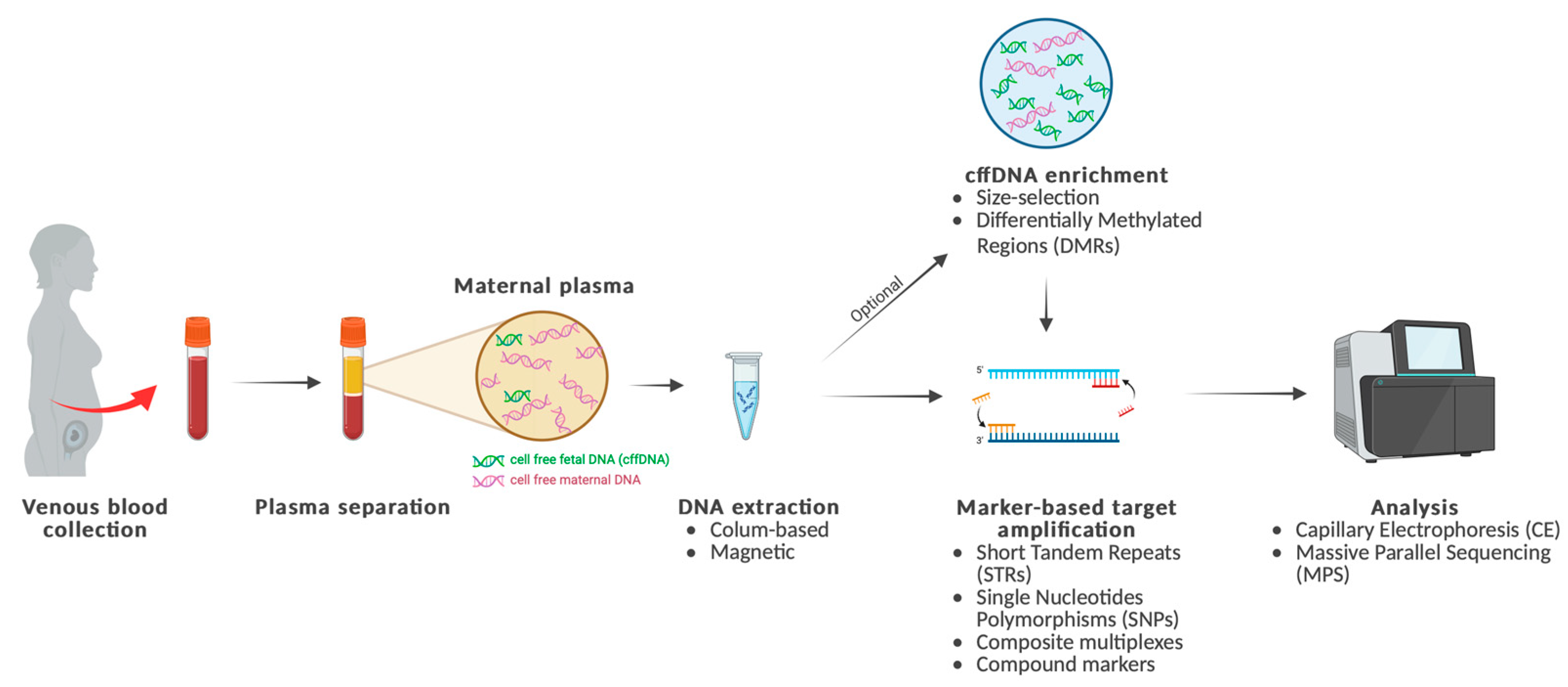
1. Introduction
2. Methodology
3. Application of Genetic Markers in NIPPT
3.1. Short Tandem Repeats (STRs)
| Type of Marker | Panel | Number of Markers Analyzed | Technology | Gestational Age (Weeks) | Total Number of Samples | Results Evaluation | Reference |
|---|---|---|---|---|---|---|---|
| Autosomal Short Tandem Repeats (a-STRs) | ampFLSTRTM SGM PlusTM Kit * | 11 | PCR-based method | 29–41 | 10 | N/A | Birch et al. [29] |
| AmpFlSTR TM Profiler Plus PCR Amplification Kit * | 16 | CE (310 Genetic Analyzer *) | 8–37 | 47 (1st Tri: 6) | N/A | Tang et al. [31] | |
| GlobalFiler™ PCR Amplification Kit */early access version of Ion AmpliSeq HID STR 25-plex * | 21/24 | CE (310 Genetic Analyzer *) and MPS (Ion Torrent PGM™ *) | 10; 16 | 2 (1st Tri: 1) | Posterior probability | Gysi et al. [33] | |
| Custom panel | 23 | MPS (NextSeq 500 system **) | 12–38 | 28 (1st Tri: 18) | Matching probability | Song et al. [34] | |
| Y-chromosome Short Tandem Repeats (Y-STRs) | Custom assay | 9 | CE (ABI Prism™ 377 DNA Sequencer) * | ND | 64 (1st Tri: 6) | N/A | Deng et al. [37] |
| ampFLSTR Yfiler™ */PowerPlex® Y23 *** and two custom panels | 67 (17/23/27) | CE (3500 Genetic Analyzer *) | 12–36 | 30 (1st Tri: 22) | CPI and posterior probability | Barra et al. [38] | |
| Custom panel | 12 | MPS (NextSeq 500 system **) | 21–37 | 24 (1st Tri: 0) | CPI and posterior probability | Song et al. [39] | |
| a-STRs and Y-STRs | ampFLSTRTM IdentifilerTM */ampFLSTR Yfiler™ * | 31 (15 STRs + 16 Y-STRs) | CE (310 Genetic Analyzer *) | 9–29 | 20 | ND | Wagner et al. [30] |
| Three custom panels | 34 (12/10/12) | MPS (Ion Torrent PGM *) | ≥9 | >180 | CPI | Whittle et al. [36] | |
| Single Nucleotide Polymorphisms (SNPs) | Custom panel | 92 | PCR-based method | ND | 154 | N/A | Tynan et al. [40] |
| Custom panel | 384 | PCR-based method | 8–14 | 30 | Probability of exclusion | Guo et al. [41] | |
| Microarray (HumanCytoSNP-12 array **) | 300,000 | Illumina Infinium technology ** | 6–21 | 21 (1st Tri: 11) | Accuracy (Diagnostic potency) | Ryan et al. [42] | |
| Three custom panels | 5000 to 8000 | MPS (Hiseq™ 2000 **) | 13–30 | 17 (1st Tri: 0) | CPI | Jiang et al. [43] | |
| Custom panel | 1479 | MPS (Hiseq™ 4000 **) | 11–18 | 7 (1st Tri: 3) | ND | Yang et al. [44] | |
| Custom panel | 720 | MPS (Ion Torrent PGM *) | 9–21 | 20 (1st Tri: 11) | ND | Yang et al. [45] | |
| Custom panel | 1795 | MPS (Hiseq™ 2000 **) | 9–21 | 34 (1st Tri: 11) | CPI | Qu et al. [46] | |
| Precision ID Identity Panel * | 124 | MPS (Ion S5™ System *) | 4–20 | 15 (1st Tri: 15) | Paternity Index | Christiansen et al. [47] | |
| Custom panel | 4151 | MPS (BGISEQ-500, MGI) | 6–35 | 358 (1st Tri: 155) | CPI | Chang et al. [48] | |
| Custom panel | 356 | MPS (MiniSeq™ **) | 7–20 | 15 (1st Tri: 10) | CPI and posterior probability | Tam et al. [49] | |
| Custom panel | 5226 | MPS (Hiseq™ 2000 **) | 9–18 | 15 (1st Tri: 11) | CPI | Xie et al. [50] | |
| Custom panel | 861 | MPS (Ion S5™ System *) | ND (1st Tri) | 9 (1st Tri: 9) | CPI | Giannico et al. [51] | |
| Short Tandem Repeats (STRs) and SNPs | ForenSeqTM DNA Signature Prep **** | 152 (94 SNPs + 58 STRs) | MPS (MiSeq FGx ****) | 7–24 | 17 (1st Tri: 8) | CPI | Shen et al. [52] |
| Microhaplotypes (MHs) | Custom panel | 60 | MPS (Hiseq™ 2000 **) | 6–13 | 15 (1st Tri: 15) | CPI | Ou and Qu [53] |
| Custom panel | 60 | MPS (Hiseq™ 2000 **) | ≥10 | 19 | CPI | Bai et al. [54] | |
| Custom panel | 15 | CE (3130 Genetic Analyzer *) | >18 | 26 (1st Tri: 0) | ND | Zhang et al. [55] | |
| Deletion/Insertion Polymorphisms with STRs (DIP-STRs) | Custom panel | 28 | CE (3500xl Genetic Analyzer *) | 10–39 | 48 (1st Tri: 27) | ND | Moriot et al. [56] |
| Custom panel | 47 | CE (3500xl Genetic Analyzer *) | 7–13 | 87 (1st Tri: 87) | CPI and posterior probability | Damour et al. [57] | |
| DIP-STRs and SNP-STRs | Custom panel | 17 (6 DIP-STRs + 11 SNP-STRs) | CE (3130 and 3500 Genetic Analyzers *) | >18 | 21 (1st Tri: 0) | ND | Tan et al. [58] |
3.2. Single Nucleotide Polymorphism (SNPs)
3.3. Composite Multiplexes and Compound Markers
4. Discussion and Conclusions
5. Futures Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lo, Y.M.; Corbetta, N.; Chamberlain, P.F.; Rai, V.; Sargent, I.L.; Redman, C.W.; Wainscoat, J.S. Presence of fetal DNA in maternal plasma and serum. Lancet 1997, 350, 485–487. [Google Scholar] [CrossRef] [PubMed]
- Alberry, M.; Maddocks, D.; Jones, M.; Abdel Hadi, M.; Abdel-Fattah, S.; Avent, N.; Soothill, P.W. Free fetal DNA in maternal plasma in anembryonic pregnancies: Confirmation that the origin is the trophoblast. Prenat. Diagn. 2007, 27, 415–418. [Google Scholar] [CrossRef]
- Bianchi, D.W. Circulating fetal DNA: Its origin and diagnostic potential-a review. Placenta 2004, 25 (Suppl. A), S93–S101. [Google Scholar] [CrossRef]
- Wang, E.; Batey, A.; Struble, C.; Musci, T.; Song, K.; Oliphant, A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat. Diagn. 2013, 33, 662–666. [Google Scholar] [CrossRef]
- Barrett, A.N.; McDonnell, T.C.R.; Chan, K.C.A.; Chitty, L.S. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin. Chem. 2012, 58, 1026–1032. [Google Scholar] [CrossRef] [PubMed]
- Lun, F.M.; Chiu, R.W.; Chan, K.C.; Leung, T.Y.; Lau, T.K.; Lo, Y.M. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin. Chem. 2008, 54, 1664–1672. [Google Scholar] [CrossRef]
- Lo, Y.M.; Zhang, J.; Leung, T.N.; Lau, T.K.; Chang, A.M.; Hjelm, N.M. Rapid clearance of fetal DNA from maternal plasma. Am. J. Hum. Genet. 1999, 64, 218–224. [Google Scholar] [CrossRef] [PubMed]
- ACOG Practice Bulletin No. 88: Invasive prenatal testing for aneuploidy. Obstet. Gynecol. 2007, 110, 1459–1467. [CrossRef]
- Seeds, J.W. Diagnostic mid trimester amniocentesis: How safe? Am. J. Obstet. Gynecol. 2004, 191, 607–615. [Google Scholar] [CrossRef]
- Brambati, B.; Simoni, G.; Travi, M.; Danesino, C.; Tului, L.; Privitera, O.; Stioui, S.; Tedeschi, S.; Russo, S.; Primignani, P. Genetic diagnosis by chorionic villus sampling before 8 gestational weeks: Efficiency, reliability, and risks on 317 completed pregnancies. Prenat. Diagn. 1992, 12, 789–799. [Google Scholar] [CrossRef]
- Firth, H.V.; Boyd, P.A.; Chamberlain, P.F.; MacKenzie, I.Z.; Morriss-Kay, G.M.; Huson, S.M. Analysis of limb reduction defects in babies exposed to chorionic villus sampling. Lancet 1994, 343, 1069–1071. [Google Scholar] [CrossRef] [PubMed]
- Lissmann, R.; Lokot, M.; Marston, C. Understanding the lived experience of pregnancy and birth for survivors of rape and sexual assault. BMC Pregnancy Childbirth 2023, 23, 796. [Google Scholar] [CrossRef]
- Lathrop, A. Pregnancy resulting from rape. J. Obstet. Gynecol. Neonatal Nurs. 1998, 27, 25–31. [Google Scholar] [CrossRef]
- Krueger, M.M. Pregnancy as a result of rape. J. Sex Educ. Ther. 1988, 14, 23–27. [Google Scholar] [CrossRef]
- D’Angelo, D.V.; Liu, Y.; Basile, K.C.; Smith, S.G.; Chen, J.; Friar, N.W.; Stevens, M. Rape and sexual coercion related pregnancy in the United States. Am. J. Prev. Med. 2024, 66, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Holmes, M.M.; Resnick, H.S.; Kilpatrick, D.G.; Best, C.L. Rape-related pregnancy: Estimates and descriptive characteristics from a national sample of women. Am. J. Obstet. Gynecol. 1996, 175, 320–325. [Google Scholar] [CrossRef]
- Shulman, L.P.; Muram, D.; Speck, P.M. Counseling sexual assault victims who become pregnant after the assault: Benefits and limitations of first-trimester paternity determination. J. Interpers. Violence 1992, 7, 205–210. [Google Scholar] [CrossRef]
- Blake, M.d.T.; Drezett, J.; Machi, G.S.; Pereira, V.X.; Raimundo, R.D.; Oliveira, F.R.; Sarubbi Junior, V.; Adami, F.; de Abreu, L.C. Factors associated to late-term abortion after rape: Literature review. Reprodução Clim. 2014, 29, 60–65. [Google Scholar] [CrossRef]
- Rouhani, S.A.; Scott, J.; Burkhardt, G.; Onyango, M.A.; Haider, S.; Greiner, A.; Albutt, K.; VanRooyen, M.; Bartels, S.A. A quantitative assessment of termination of sexual violence-related pregnancies in eastern Democratic Republic of Congo. Confl. Health 2016, 10, 9. [Google Scholar] [CrossRef]
- Zhang, S.; Han, S.; Zhang, M.; Wang, Y. Non-invasive prenatal paternity testing using cell-free fetal DNA from maternal plasma: DNA isolation and genetic marker studies. Leg. Med. 2018, 32, 98–103. [Google Scholar] [CrossRef]
- Repiská, G.; Sedláčková, T.; Szemes, T.; Celec, P.; Minárik, G. Selection of the optimal manual method of cell free fetal DNA isolation from maternal plasma. Clin. Chem. Lab. Med. (CCLM) 2013, 51, 1185–1189. [Google Scholar] [CrossRef]
- Hatami, A.; Saadatmand, M.; Garshasbi, M. Cell-free fetal DNA (cffDNA) extraction from whole blood by using a fully automatic centrifugal microfluidic device based on displacement of magnetic silica beads. Talanta 2024, 267, 125245. [Google Scholar] [CrossRef] [PubMed]
- Legler, T.J.; Liu, Z.; Heermann, K.-H.; Hempel, M.; Gutensohn, K.; Kiesewetter, H.; Pruss, A. Specific magnetic bead-based capture of free fetal DNA from maternal plasma. Transfus. Apher. Sci. 2009, 40, 153–157. [Google Scholar] [CrossRef]
- Liang, B.; Li, H.; He, Q.; Li, H.; Kong, L.; Xuan, L.; Xia, Y.; Shen, J.; Mao, Y.; Li, Y.; et al. Enrichment of the fetal fraction in non-invasive prenatal screening reduces maternal background interference. Sci. Rep. 2018, 8, 17675. [Google Scholar] [CrossRef] [PubMed]
- Dhallan, R.; Au, W.C.; Mattagajasingh, S.; Emche, S.; Bayliss, P.; Damewood, M.; Cronin, M.; Chou, V.; Mohr, M. Methods to increase the percentage of free fetal DNA recovered from the maternal circulation. J. Am. Med. Assoc. 2004, 291, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Du, Z.; Song, Y.; Gao, S.; Yu, S.; Zhu, H.; Ren, M.; Zhang, G. Size-selective separation and overall-amplification of cell-free fetal DNA fragments using PCR-based enrichment. Sci. Rep. 2017, 7, 40936. [Google Scholar] [CrossRef]
- Chan, K.C.; Zhang, J.; Hui, A.B.; Wong, N.; Lau, T.K.; Leung, T.N.; Lo, K.W.; Huang, D.W.; Lo, Y.M. Size distributions of maternal and fetal DNA in maternal plasma. Clin. Chem. 2004, 50, 88–92. [Google Scholar] [CrossRef]
- Ou, X.; Gao, J.; Chen, Y.; Gao, Y.; Qu, D.; Sun, H. Detecting hypermethylated fetal 1 sequences in maternal plasma: Implications for noninvasive paternity testing in pregnancy. Transfusion 2013, 53, 1856–1858. [Google Scholar] [CrossRef]
- Birch, L.; English, C.A.; O’Donoghue, K.; Barigye, O.; Fisk, N.M.; Keer, J.T. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clin. Chem. 2005, 51, 312–320. [Google Scholar] [CrossRef]
- Wagner, J.; Dzijan, S.; Marjanovic, D.; Lauc, G. Non-invasive prenatal paternity testing from maternal blood. Int. J. Leg. Med. 2009, 123, 75–79. [Google Scholar] [CrossRef]
- Tang, D.L.; Li, Y.; Zhou, X.; Li, X.; Zheng, F. Multiplex fluorescent PCR for noninvasive prenatal detection of fetal-derived paternally inherited diseases using circulatory fetal DNA in maternal plasma. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009, 144, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M.; Shen, Y.; McCord, B.R. The development of reduced size STR amplicons as tools for analysis of degraded DNA. J. Forensic Sci. 2003, 48, 1054–1064. [Google Scholar] [CrossRef]
- Gysi, M.; Arora, N.; Sulzer, A.; Voegeli, P.; Kratzer, A. Non-invasive prenatal paternity testing with STRs: A pilot study. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e291–e292. [Google Scholar] [CrossRef]
- Song, W.; Xiao, N.; Zhou, S.; Yu, W.; Wang, N.; Shao, L.; Liang, X. Non-invasive prenatal paternity testing using mini-STR-based next-generation sequencing: A pilot study. J. Lab. Med. 2022, 46, 337–344. [Google Scholar] [CrossRef]
- Dumache, R.; Stoicanescu, D.; Muresan, C.; Ciocan, V.; Enache, A. Circulating Cell-Free Plasma DNA in Noninvasive Prenatal Paternity Testing with Short Tandem Repeats (STRs). Clin. Lab. 2020, 66, 2521–2525. [Google Scholar] [CrossRef]
- Whittle, M.R.; Francischini, C.W.; Sumita, D.R. Routine implementation of noninvasive prenatal paternity testing with STRs. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e233–e234. [Google Scholar] [CrossRef]
- Deng, Z.; Wu, G.; Li, Q.; Zhang, X.; Liang, Y.; Li, D.; Gao, S.; Lan, Y. Noninvasive genotyping of 9 Y-chromosome specific STR loci using circulatory fetal DNA in maternal plasma by multiplex PCR. Prenat. Diagn. 2006, 26, 362–368. [Google Scholar] [CrossRef]
- Barra, G.B.; Santa Rita, T.H.; Chianca, C.F.; Velasco, L.F.; de Sousa, C.F.; Nery, L.F.; Costa, S.S. Fetal male lineage determination by analysis of Y-chromosome STR haplotype in maternal plasma. Forensic Sci. Int. Genet. 2015, 15, 105–110. [Google Scholar] [CrossRef]
- Song, W.; Xiao, N.; Zhou, S.; Yu, W.; Wang, N.; Shao, L.; Duan, Y.; Chen, M.; Pan, L.; Xia, Y.; et al. Non-invasive prenatal paternity testing by analysis of Y-chromosome mini-STR haplotype using next-generation sequencing. PLoS ONE 2022, 17, e0266332. [Google Scholar] [CrossRef]
- Tynan, J.A.; Mahboubi, P.; Cagasan, L.L.; van den Boom, D.; Ehrich, M.; Oeth, P. Restriction enzyme-mediated enhanced detection of circulating cell-free fetal DNA in maternal plasma. J. Mol. Diagn. 2011, 13, 382–389. [Google Scholar] [CrossRef]
- Guo, X.; Bayliss, P.; Damewood, M.; Varney, J.; Ma, E.; Vallecillo, B.; Dhallan, R. A noninvasive test to determine paternity in pregnancy. N. Engl. J. Med. 2012, 366, 1743–1745. [Google Scholar] [CrossRef]
- Ryan, A.; Baner, J.; Demko, Z.; Hill, M.; Sigurjonsson, S.; Baird, M.L.; Rabinowitz, M. Informatics-based, highly accurate, noninvasive prenatal paternity testing. Genet. Med. 2013, 15, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xie, Y.; Li, X.; Ge, H.; Deng, Y.; Mu, H.; Feng, X.; Yin, L.; Du, Z.; Chen, F.; et al. Noninvasive prenatal paternity testing (NIPAT) through maternal plasma DNA sequencing: A pilot study. PLoS ONE 2016, 11, e0159385. [Google Scholar] [CrossRef]
- Yang, D.; Liang, H.; Gao, Y.; Lin, S.; He, Z.; Gao, J.; Sun, H.; Li, Q.; Ma, X.; Ou, X. Noninvasive fetal genotyping of paternally inherited alleles using targeted massively parallel sequencing in parentage testing cases. Transfusion 2017, 57, 1505–1514. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Liang, H.; Lin, S.; Li, Q.; Ma, X.; Gao, J.; Sun, H.; Chen, Q.; Wu, J.; Ou, X. An SNP panel for the analysis of paternally inherited alleles in maternal plasma using ion Torrent PGM. Int. J. Leg. Med. 2018, 132, 343–352. [Google Scholar] [CrossRef]
- Qu, N.; Xie, Y.; Li, H.; Liang, H.; Lin, S.; Huang, E.; Gao, J.; Chen, F.; Shi, Y.; Ou, X. Noninvasive prenatal paternity testing using targeted massively parallel sequencing. Transfusion 2018, 58, 1792–1799. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, S.L.; Jakobsen, B.; Børsting, C.; Udengaard, H.; Buchard, A.; Kampmann, M.-L.; Grøndahl, M.L.; Morling, N. Non-invasive prenatal paternity testing using a standard forensic genetic massively parallel sequencing assay for amplification of human identification SNPs. Int. J. Leg. Med. 2019, 133, 1361–1368. [Google Scholar] [CrossRef]
- Chang, L.; Yu, H.; Miao, X.; Zhang, J.; Li, S. Development and comprehensive evaluation of a noninvasive prenatal paternity testing method through a scaled trial. Forensic Sci. Int. Genet. 2019, 43, 102158. [Google Scholar] [CrossRef]
- Tam, J.C.W.; Chan, Y.M.; Tsang, S.Y.; Yau, C.I.; Yeung, S.Y.; Au, K.K.; Chow, C.K. Noninvasive prenatal paternity testing by means of SNP-based targeted sequencing. Prenat. Diagn. 2020, 40, 497–506. [Google Scholar] [CrossRef]
- Xie, Y.; Qu, N.; Lin, S.; Jiang, H.; Zhang, Y.; Zhang, X.; Liang, H.; Chen, F.; Ou, X. Noninvasive prenatal paternity testing by maternal plasma DNA sequencing in twin pregnancies. Electrophoresis 2020, 41, 1095–1102. [Google Scholar] [CrossRef]
- Giannico, R.; Forlani, L.; Andrioletti, V.; Cotroneo, E.; Termine, A.; Fabrizio, C.; Cascella, R.; Salvaderi, L.; Linarello, P.; Varrone, D.; et al. NIPAT as non-invasive prenatal paternity testing using a panel of 861 SNVs. Genes 2023, 14, 312. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Li, R.; Li, H.; Gao, Y.; Chen, H.; Qu, N.; Peng, D.; Wu, R.; Sun, H. Noninvasive prenatal paternity testing with a combination of well-established SNP and STR markers using massively parallel sequencing. Genes 2021, 12, 454. [Google Scholar] [CrossRef]
- Ou, X.; Qu, N. Noninvasive prenatal paternity testing by target sequencing microhaps. Forensic Sci. Int. Genet. 2020, 48, 102338. [Google Scholar] [CrossRef]
- Bai, Z.; Zhao, H.; Lin, S.; Huang, L.; He, Z.; Wang, H.; Ou, X. Evaluation of a microhaplotype-based noninvasive prenatal test in twin gestations: Determination of paternity, zygosity, and fetal fraction. Genes 2020, 12, 26. [Google Scholar] [CrossRef]
- Zhang, R.; Tan, Y.; Wang, L.; Jian, H.; Zhu, J.; Xiao, Y.; Tan, M.; Xue, J.; Yang, F.; Liang, W. Set of 15 SNP-SNP Markers for Detection of Unbalanced Degraded DNA Mixtures and Noninvasive Prenatal Paternity Testing. Front Genet 2021, 12, 800598. [Google Scholar] [CrossRef] [PubMed]
- Moriot, A.; Hall, D. Analysis of fetal DNA in maternal plasma with markers designed for forensic DNA mixture resolution. Genet. Med. 2019, 21, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Damour, G.; Baumer, K.; Legardeur, H.; Hall, D. Early noninvasive prenatal paternity testing by targeted fetal DNA analysis. Sci. Rep. 2023, 13, 12139. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, L.; Bai, P.; Li, Z.; Zhang, R.; Yang, F.; Wang, L.; Liang, W. Detection of cell-free fetal DNA in maternal plasma using two types of compound markers. Electrophoresis 2021, 42, 1158–1167. [Google Scholar] [CrossRef]
- Gao, S.; Li, B.; Mao, L.; Wang, W.; Zou, D.; Zheng, J.; Zhou, M.; Yu, S.; Zheng, F.; Yin, Y.; et al. A theoretical base for non-invasive prenatal paternity testing. Forensic Sci. Int. 2023, 346, 111649. [Google Scholar] [CrossRef]
- Sanchez, J.J.; Phillips, C.; Borsting, C.; Balogh, K.; Bogus, M.; Fondevila, M.; Harrison, C.D.; Musgrave-Brown, E.; Salas, A.; Syndercombe-Court, D.; et al. A multiplex assay with 52 single nucleotide polymorphisms for human identification. Electrophoresis 2006, 27, 1713–1724. [Google Scholar] [CrossRef]
- Børsting, C.; Sanchez, J.J.; Hansen, H.E.; Hansen, A.J.; Bruun, H.Q.; Morling, N. Performance of the SNPforID 52 SNP-plex assay in paternity testing. Forensic Sci. Int. Genet. 2008, 2, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Novroski, N.; Cihlar, J. Evolution of single-nucleotide polymorphism use in forensic genetics. Wiley Interdiscip. Rev. Forensic Sci. 2022, 4, e1459. [Google Scholar] [CrossRef]
- Drabek, J.; Cereda, G. Interpreting noninvasive prenatal paternity tests. Genet. Med. 2014, 16, 793–794. [Google Scholar] [CrossRef] [PubMed]
- Ryan, A.; Demko, Z.; Sigurjonsson, S.; Rabinowitz, M. Response to Drabek and Cereda. Genet. Med. 2014, 16, 794. [Google Scholar] [CrossRef] [PubMed]
- Goya, R.; Sun, M.G.F.; Morin, R.D.; Leung, G.; Ha, G.; Wiegand, K.C.; Senz, J.; Crisan, A.; Marra, M.A.; Hirst, M.; et al. SNVMix: Predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics 2010, 26, 730–736. [Google Scholar] [CrossRef]
- Dørum, G.; Kaur, N.; Gysi, M. Pedigree-based relationship inference from complex DNA mixtures. Int. J. Leg. Med. 2017, 131, 629–641. [Google Scholar] [CrossRef]
- Hernandis, E.; Dørum, G.; Egeland, T. relMix: An open source software for DNA mixtures with related contributors. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 221–223. [Google Scholar] [CrossRef]
- Kaur, N.; Bouzga, M.M.; Dørum, G.; Egeland, T. Relationship inference based on DNA mixtures. Int. J. Leg. Med. 2016, 130, 323–329. [Google Scholar] [CrossRef]
- Oldoni, F.; Kidd, K.K.; Podini, D. Microhaplotypes in forensic genetics. Forensic Sci. Int. Genet. 2019, 38, 54–69. [Google Scholar] [CrossRef]
- Wang, J.Y.T.; Whittle, M.R.; Puga, R.D.; Yambartsev, A.; Fujita, A.; Nakaya, H.I. Noninvasive prenatal paternity determination using microhaplotypes: A pilot study. BMC Med. Genom. 2020, 13, 157. [Google Scholar] [CrossRef]
- Ou, X.; Bai, Z. A case of heteropaternal superfecundation identified by microhap sequencing of maternal plasma cell-free DNA: A case of HS identified by microhap sequencing. Forensic Sci. Int. Genet. 2021, 51, 102458. [Google Scholar] [CrossRef] [PubMed]
- Damour, G.; Mauffrey, F.; Hall, D. Identification and characterization of novel DIP-STRs from whole-genome sequencing data. Forensic Sci. Int. Genet. 2023, 64, 102849. [Google Scholar] [CrossRef] [PubMed]
- Oldoni, F.; Castella, V.; Hall, D. A novel set of DIP-STR markers for improved analysis of challenging DNA mixtures. Forensic Sci. Int. Genet. 2015, 19, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.; Castella, V. DIP–STR: A new marker for resolving unbalanced DNA mixtures. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e1–e2. [Google Scholar] [CrossRef]
- ülküer, ü.; KurtuluŞ-ülküer, M.; Elma, C.; Kesici, T.; MenevŞe, S. Short tandem repeat (STR) polymorphisms in Turkish population. J. Genet. 2004, 83, 197–199. [Google Scholar] [CrossRef]
- Chandra, D.; Mishra, V.C.; Raina, A.; Raina, V. Mutation rate evaluation at 21 autosomal STR loci: Paternity testing experience. Leg. Med. 2022, 58, 102080. [Google Scholar] [CrossRef]
- Brinkmann, B.; Pfeiffer, H.; Schürenkamp, M.; Hohoff, C. The evidential value of STRs. Int. J. Leg. Med. 2001, 114, 173–177. [Google Scholar] [CrossRef]
- Thomson, J.A.; Pilotti, V.; Stevens, P.; Ayres, K.L.; Debenham, P.G. Validation of short tandem repeat analysis for the investigation of cases of disputed paternity. Forensic Sci. Int. 1999, 100, 1–16. [Google Scholar] [CrossRef]
- Dierig, L.; Kunz, S.N.; Wiegand, P. Comparison of massively parallel sequencing to capillary electrophoresis for short tandem repeat genotyping of trace DNA. Electrophoresis 2024, 45, 451–462. [Google Scholar] [CrossRef]
- Ballard, D.; Winkler-Galicki, J.; Wesoly, J. Massive parallel sequencing in forensics: Advantages, issues, technicalities, and prospects. Int. J. Leg. Med. 2020, 134, 1291–1303. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Yang, H.; Yu, H.; Fan, J.; Wang, J.; Liu, Z.; Liu, J.; Ren, J.; Zhang, G. Developmental validation of a DIP-InDel/DIP-STR system for the detection of unbalanced DNA mixtures. Electrophoresis 2025, 46, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, J.; Wang, J.; Chen, D.; Liu, Z.; Shi, J.; Li, Z.; Li, W.; Zhang, G.; Du, B. A set of 14 DIP-SNP markers to detect unbalanced DNA mixtures. Biochem. Biophys. Res. Commun. 2018, 497, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Morling, N.; Allen, R.W.; Carracedo, A.; Geada, H.; Guidet, F.; Hallenberg, C.; Martin, W.; Mayr, W.R.; Olaisen, B.; Pascali, V.L.; et al. Paternity Testing Commission of the International Society of Forensic Genetics: Recommendations on genetic investigations in paternity cases. Forensic Sci. Int. 2002, 129, 148–157. [Google Scholar] [CrossRef]
- Gjertson, D.W.; Brenner, C.H.; Baur, M.P.; Carracedo, A.; Guidet, F.; Luque, J.A.; Lessig, R.; Mayr, W.R.; Pascali, V.L.; Prinz, M.; et al. ISFG: Recommendations on biostatistics in paternity testing. Forensic Sci. Int. Genet. 2007, 1, 223–231. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrara, L.; Hall, D. Noninvasive Prenatal Paternity Testing: A Review on Genetic Markers. Int. J. Mol. Sci. 2025, 26, 4518. https://doi.org/10.3390/ijms26104518
Carrara L, Hall D. Noninvasive Prenatal Paternity Testing: A Review on Genetic Markers. International Journal of Molecular Sciences. 2025; 26(10):4518. https://doi.org/10.3390/ijms26104518
Chicago/Turabian StyleCarrara, Laura, and Diana Hall. 2025. "Noninvasive Prenatal Paternity Testing: A Review on Genetic Markers" International Journal of Molecular Sciences 26, no. 10: 4518. https://doi.org/10.3390/ijms26104518
APA StyleCarrara, L., & Hall, D. (2025). Noninvasive Prenatal Paternity Testing: A Review on Genetic Markers. International Journal of Molecular Sciences, 26(10), 4518. https://doi.org/10.3390/ijms26104518






