Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme
Abstract
1. Introduction
2. Results
2.1. Bioinformatic Analysis of the Simultaneous Occurrence of Mutations in the APEX1 Gene and the hTERT Promoter
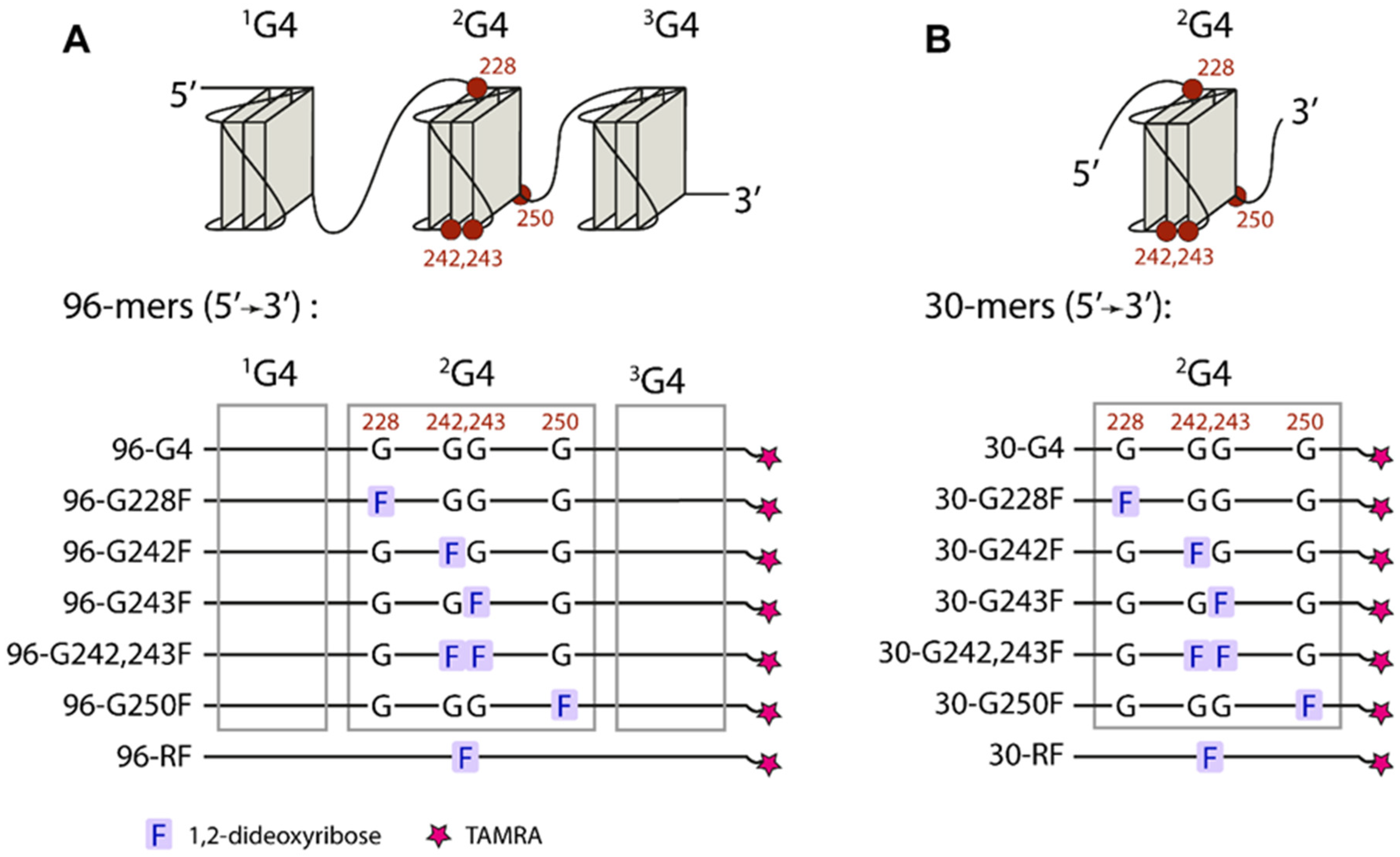
2.2. Design of DNA Models
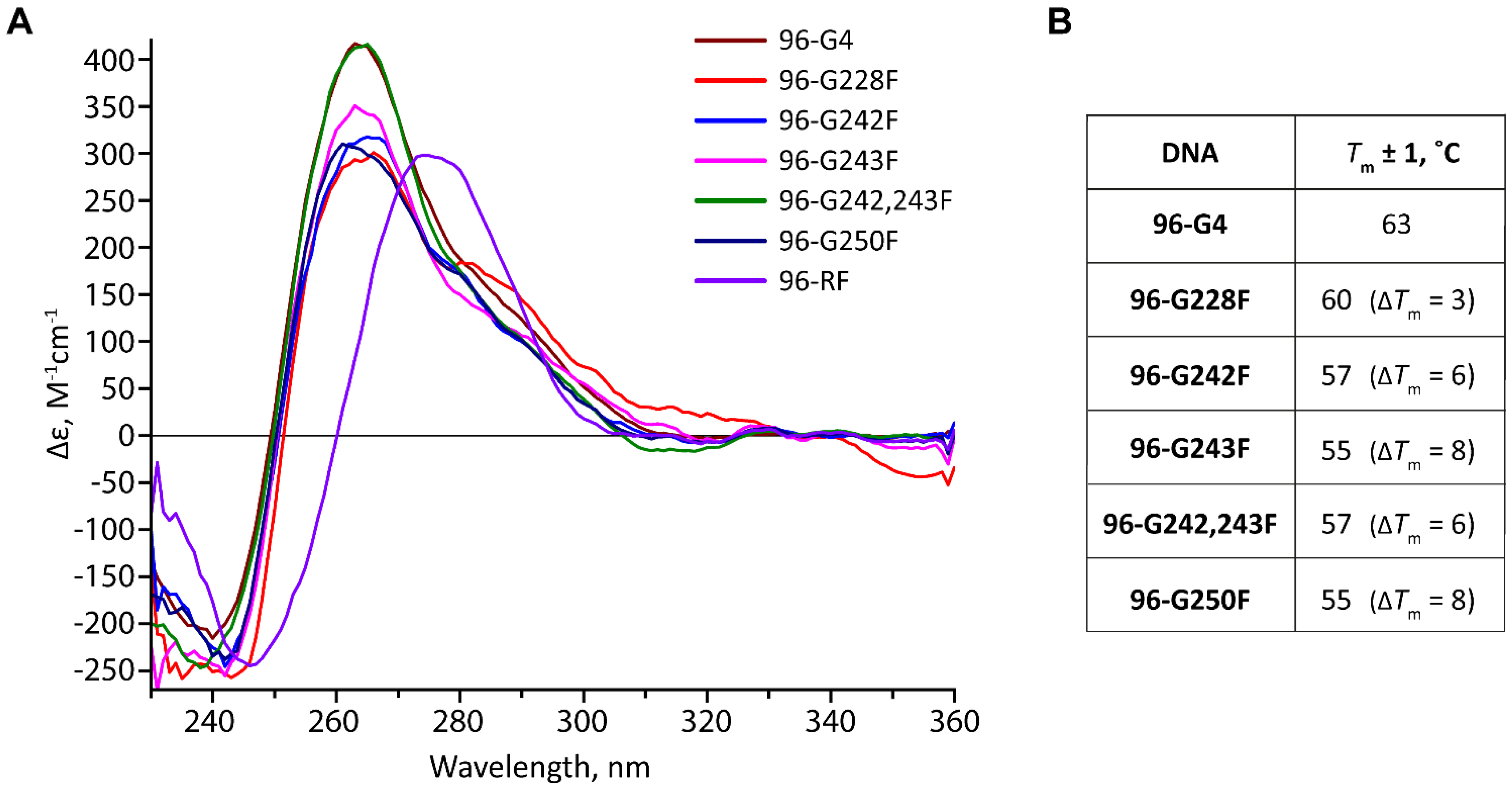
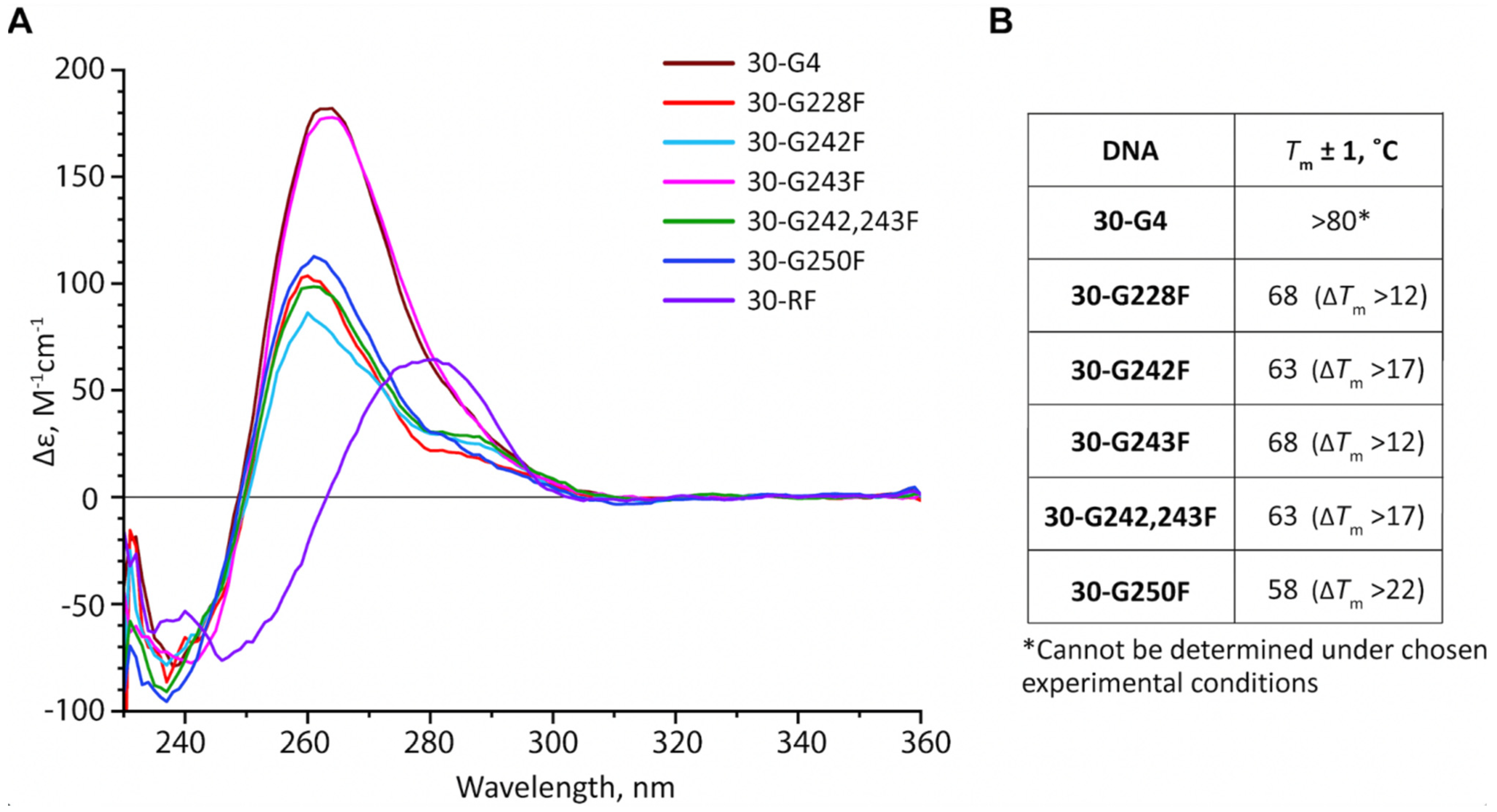
2.3. Position-Dependent Effects of AP Sites on the Topology and Thermal Stability of hTERT G4s
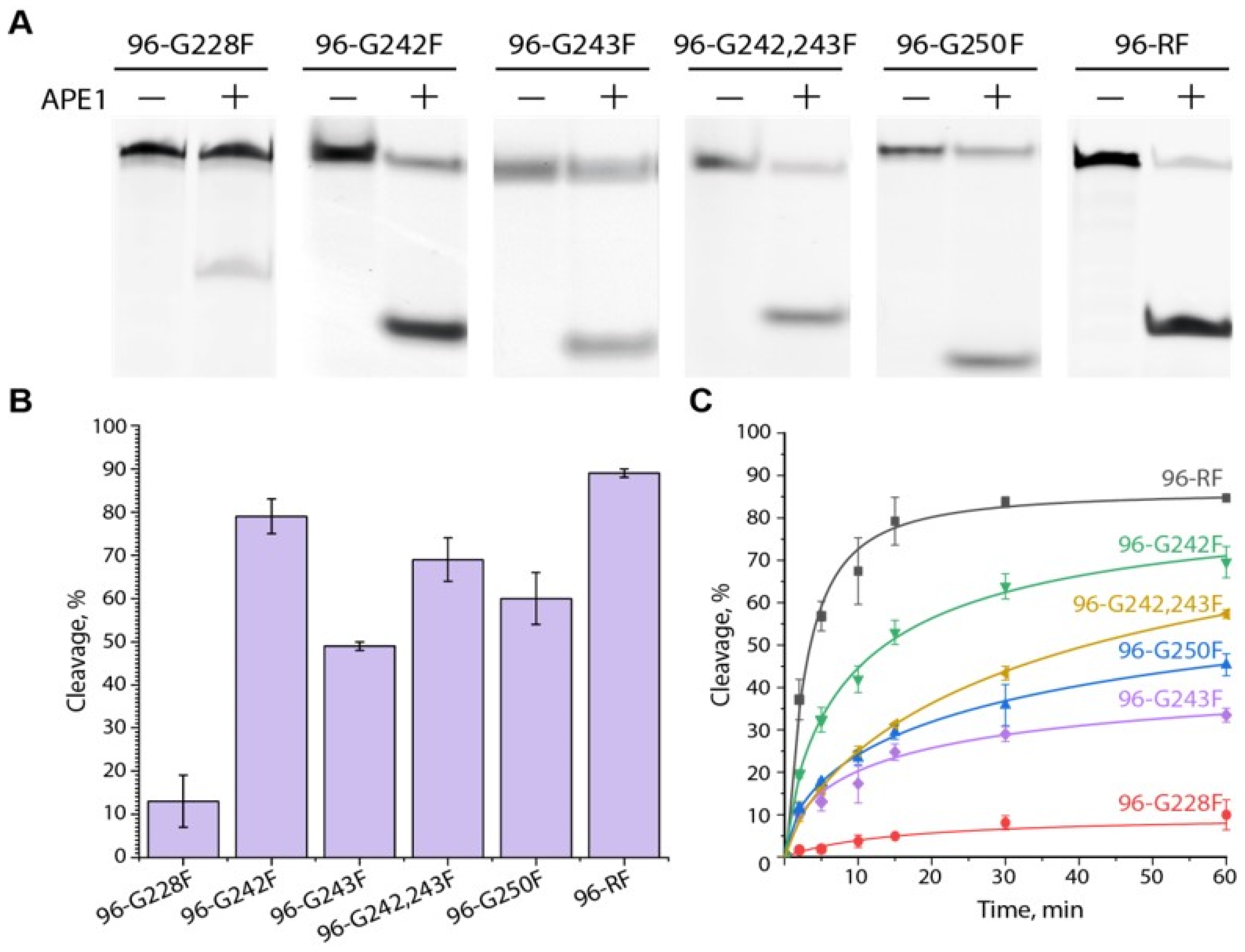
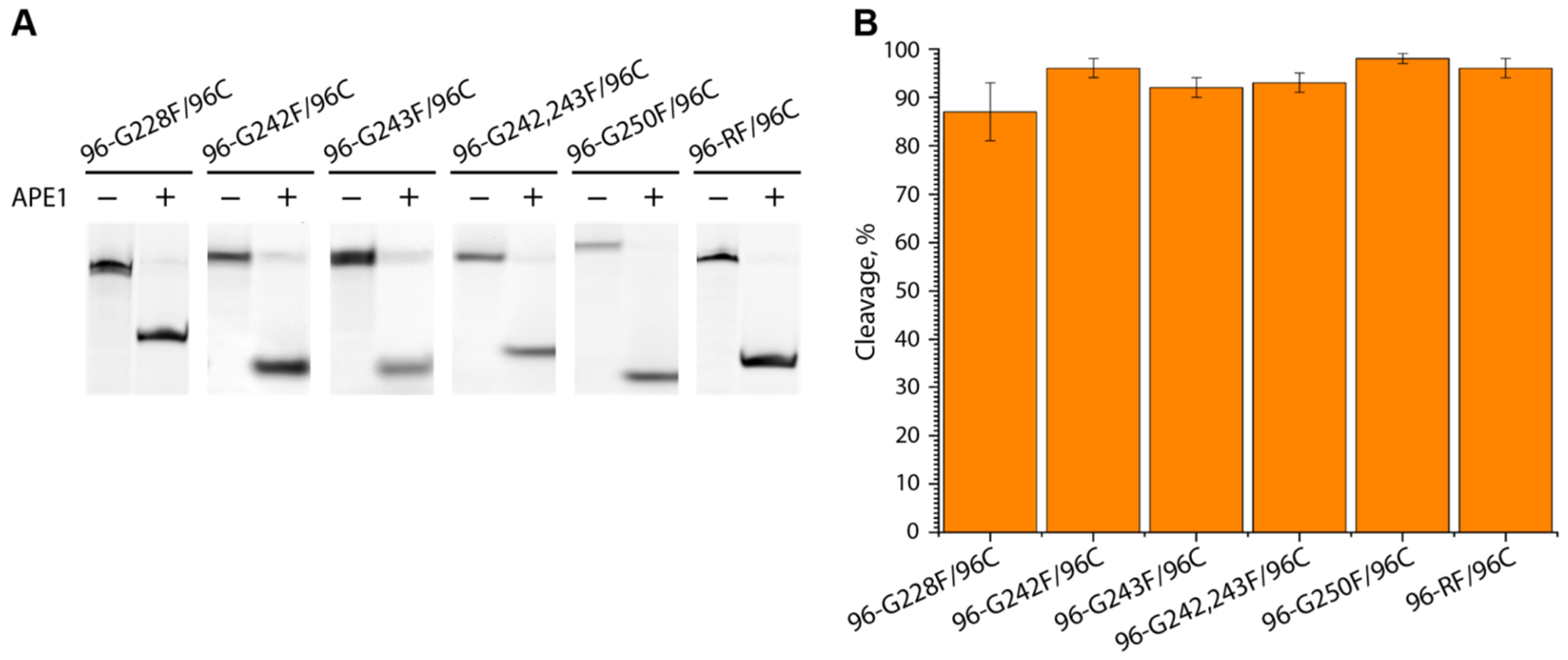
2.4. APE1-Mediated Cleavage of AP (F) Sites Located in the G4 Scaffold Formed by the G-Rich Strand of the hTERT Promoter, as Well as AP Sites Locate in the Respective DNA Duplexes
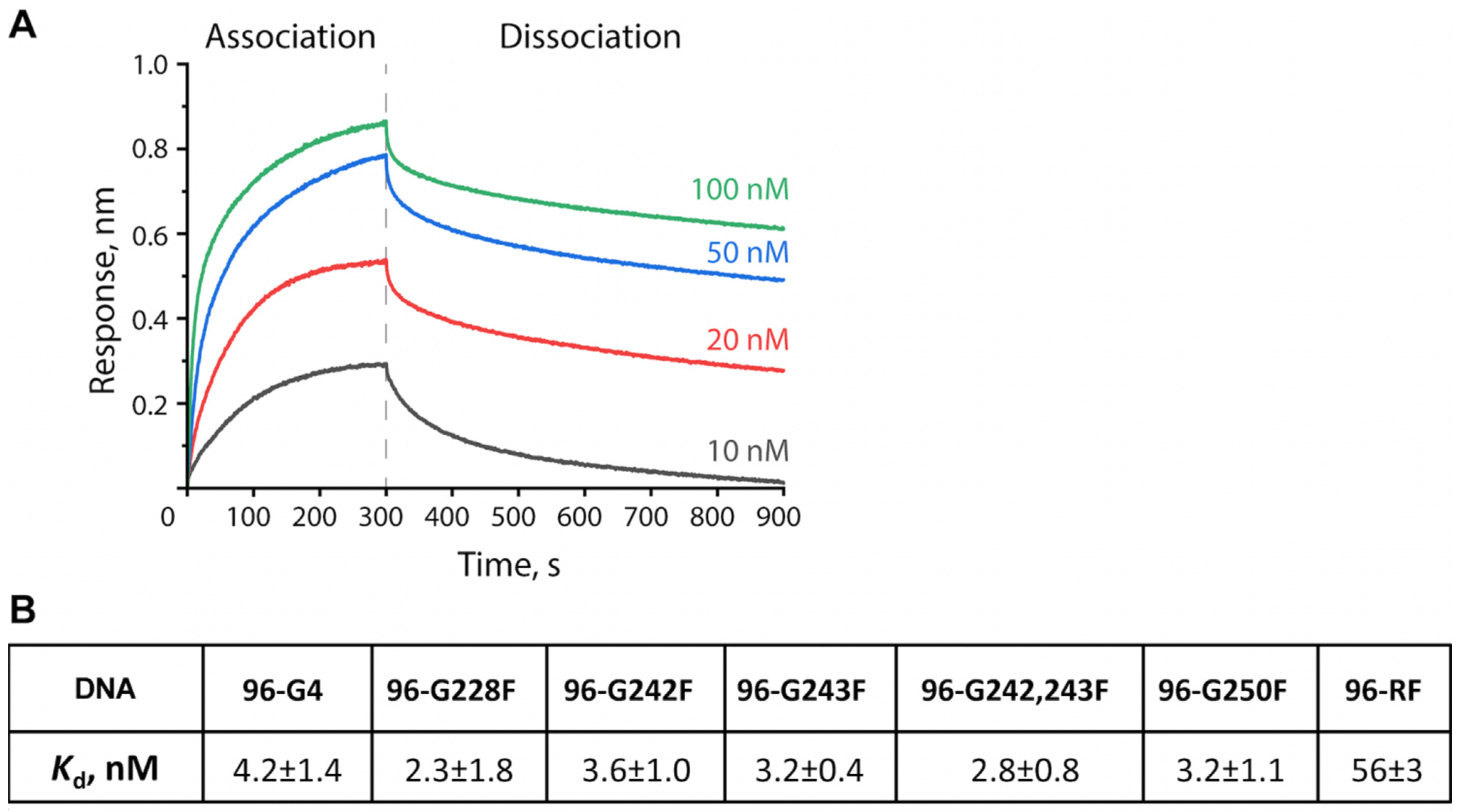
2.5. Efficiency of APE1 Binding to the 96-nt hTERT G4 and Its G>F Substitution–Bearing Analogs
2.6. Efficiency of APE1-Catalyzed Cleavage of the AP Sites in 30-nt hTERT 2G4 Constructs
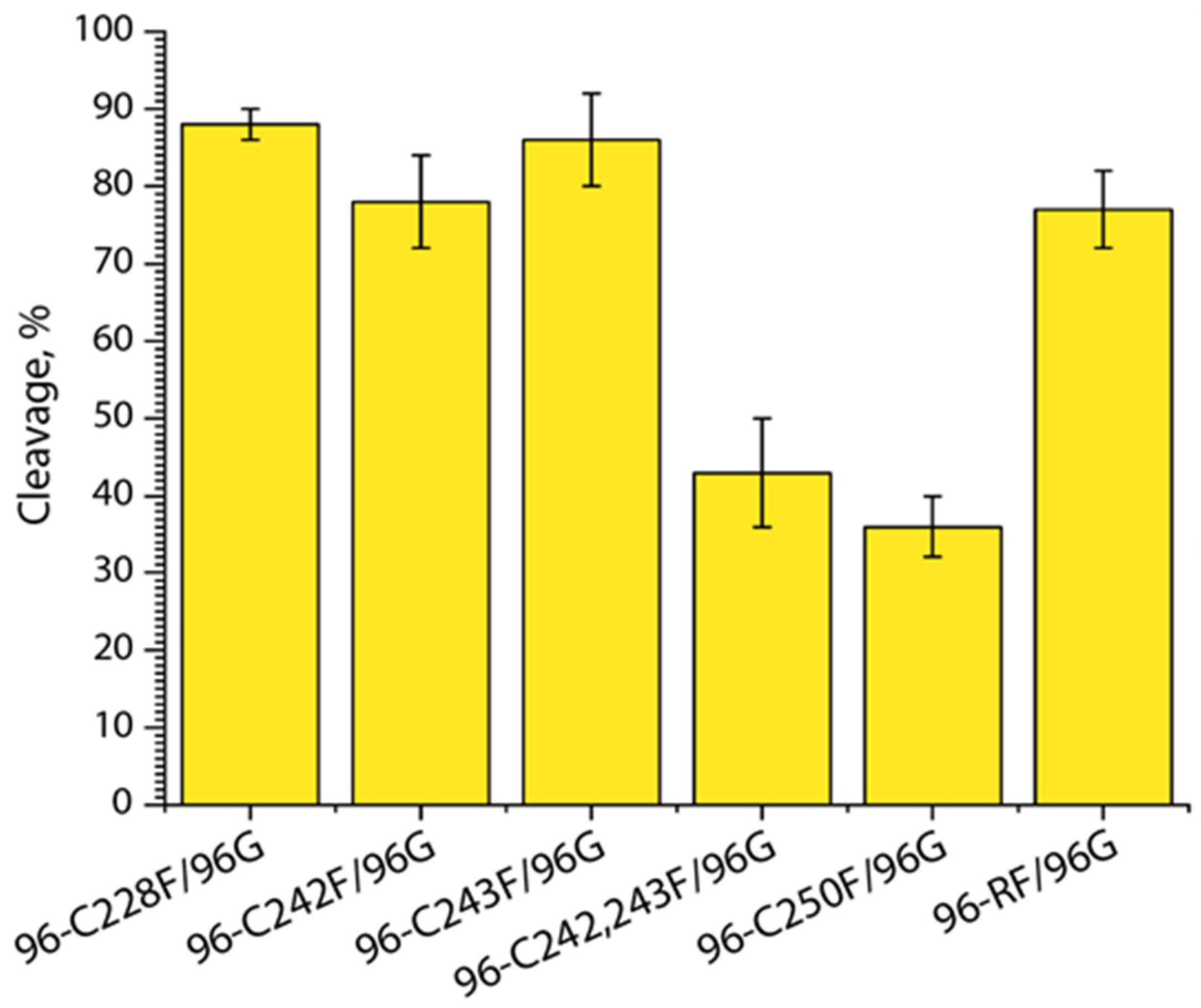
2.7. Reduced APE1-Catalyzed DNA Cleavage in the ds 96-bp hTERT Promoter Region Carrying AP (F) Sites in the C-Rich Strand
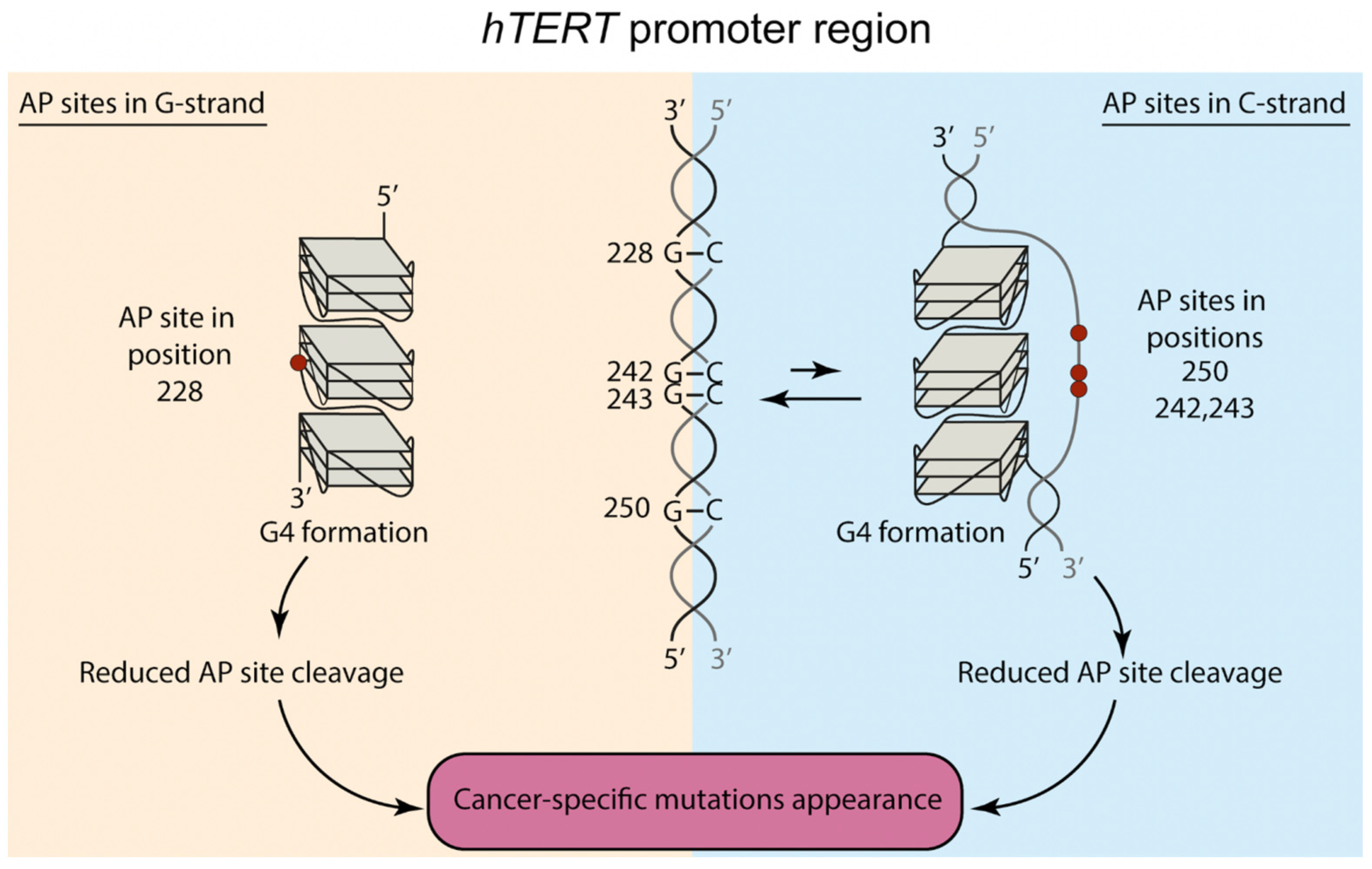
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis of Somatic Mutations in Isolates of Cancer Patients
4.2. DNA Sample Preparation
4.3. CD Measurements
4.4. APE1 Enzyme Isolation
4.5. DNA Cleavage by APE1
4.6. DNA-Binding Activity of APE1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thompson, P.S.; Cortez, D. New Insights into Abasic Site Repair and Tolerance. DNA Repair 2020, 90, 102866. [Google Scholar] [CrossRef]
- Krokan, H.E.; Bjoras, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Rate of Depurination of Native Deoxyribonucleic Acid. Biochemistry 1972, 11, 3610–3618. [Google Scholar] [CrossRef]
- Lindahl, T.; Karlstrom, O. Heat-Induced Depyrimidination of Deoxyribonucleic Acid in Neutral Solution. Biochemistry 1973, 12, 5151–5154. [Google Scholar] [CrossRef] [PubMed]
- Téoule, R. Radiation-Induced DNA Damage and Its Repair. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1987, 51, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Schaaper, R.M.; Kunkel, T.A.; Loeb, L.A. Infidelity of DNA Synthesis Associated with Bypass of Apurinic Sites. Proc. Natl. Acad. Sci. USA 1983, 80, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Shearman, C.W.; Loeb, L.A. Depurination Decreases Fidelity of DNA Synthesis in Vitro. Nature 1977, 270, 537–538. [Google Scholar] [CrossRef] [PubMed]
- Talpaert-Borlè, M. Formation, Detection and Repair of AP Sites. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1987, 181, 45–56. [Google Scholar] [CrossRef]
- Sczepanski, J.T.; Jacobs, A.C.; Greenberg, M.M. Self-Promoted DNA Interstrand Cross-Link Formation by an Abasic Site. J. Am. Chem. Soc. 2008, 130, 9646–9647. [Google Scholar] [CrossRef]
- Price, N.E.; Johnson, K.M.; Wang, J.; Fekry, M.I.; Wang, Y.; Gates, K.S. Interstrand DNA–DNA Cross-Link Formation Between Adenine Residues and Abasic Sites in Duplex DNA. J. Am. Chem. Soc. 2014, 136, 3483–3490. [Google Scholar] [CrossRef] [PubMed]
- Khodyreva, S.N.; Prasad, R.; Ilina, E.S.; Sukhanova, M.V.; Kutuzov, M.M.; Liu, Y.; Hou, E.W.; Wilson, S.H.; Lavrik, O.I. Apurinic/Apyrimidinic (AP) Site Recognition by the 5′-dRP/AP Lyase in Poly(ADP-Ribose) Polymerase-1 (PARP-1). Proc. Natl. Acad. Sci. USA 2010, 107, 22090–22095. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Pramanik, S.; Harris, H.L.; Tarpley, M.; Sarkar, A.; Spagnol, G.; Sorgen, P.L.; Chowdhury, D.; Band, V.; Klinkebiel, D.; et al. Endogenous Oxidized DNA Bases and APE1 Regulate the Formation of G-Quadruplex Structures in the Genome. Proc. Natl. Acad. Sci. USA 2020, 117, 11409–11420. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and Decay of the Primary Structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Demple, B.; Herman, T.; Chen, D.S. Cloning and Expression of APE, the cDNA Encoding the Major Human Apurinic Endonuclease: Definition of a Family of DNA Repair Enzymes. Proc. Natl. Acad. Sci. USA 1991, 88, 11450–11454. [Google Scholar] [CrossRef]
- Fleming, A.M.; Howpay Manage, S.A.; Burrows, C.J. Binding of AP Endonuclease-1 to G-Quadruplex DNA Depends on the N-Terminal Domain, Mg²⁺, and Ionic Strength. ACS Bio. Med. Chem. Au 2021, 1, 44–56. [Google Scholar] [CrossRef]
- Tell, G.; Quadrifoglio, F.; Tiribelli, C.; Kelley, M.R. The Many Functions of APE1/Ref-1: Not Only a DNA Repair Enzyme. Antioxid. Redox Signal. 2009, 11, 601–619. [Google Scholar] [CrossRef] [PubMed]
- Bhakat, K.K.; Mantha, A.K.; Mitra, S. Transcriptional Regulatory Functions of Mammalian AP-Endonuclease (APE1/Ref-1), an Essential Multifunctional Protein. Antioxid. Redox Signal. 2009, 11, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Hoitsma, N.M.; Whitaker, A.M.; Beckwitt, E.C.; Jang, S.; Agarwal, P.K.; Van Houten, B.; Freudenthal, B.D. AP-Endonuclease 1 Sculpts DNA through an Anchoring Tyrosine Residue on the DNA Intercalating Loop. Nucleic Acids Res. 2020, 48, 7345–7355. [Google Scholar] [CrossRef] [PubMed]
- Mol, C.D.; Izumi, T.; Mitra, S.; Tainer, J.A. DNA-Bound Structures and Mutants Reveal Abasic DNA Binding by APE1 DNA Repair and Coordination. Nature 2000, 403, 451–456. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Cuneo, M.J.; Dyrkheeva, N.S.; Wilson, S.H. Capturing Snapshots of APE1 Processing DNA Damage. Nat. Struct. Mol. Biol. 2015, 22, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Permata, T.B.M.; Hagiwara, Y.; Sato, H.; Yasuhara, T.; Oike, T.; Gondhowiardjo, S.; Held, K.D.; Nakano, T.; Shibata, A. Base Excision Repair Regulates PD-L1 Expression in Cancer Cells. Oncogene 2019, 38, 4452–4466. [Google Scholar] [CrossRef] [PubMed]
- Hadi, M.Z. Functional Characterization of Ape1 Variants Identified in the Human Population. Nucleic Acids Res. 2000, 28, 3871–3879. [Google Scholar] [CrossRef]
- Kladova, O.A.; Alekseeva, I.V.; Saparbaev, M.; Fedorova, O.S.; Kuznetsov, N.A. Modulation of the Apurinic/Apyrimidinic Endonuclease Activity of Human APE1 and of Its Natural Polymorphic Variants by Base Excision Repair Proteins. Int. J. Mol. Sci. 2020, 21, 7147. [Google Scholar] [CrossRef]
- McNeill, D.R.; Whitaker, A.M.; Stark, W.J.; Illuzzi, J.L.; McKinnon, P.J.; Freudenthal, B.D.; Wilson, D.M. Functions of the Major Abasic Endonuclease (APE1) in Cell Viability and Genotoxin Resistance. Mutagenesis 2020, 35, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Caston, R.A.; Gampala, S.; Armstrong, L.; Messmann, R.A.; Fishel, M.L.; Kelley, M.R. The Multifunctional APE1 DNA Repair–Redox Signaling Protein as a Drug Target in Human Disease. Drug Discov. Today 2021, 26, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Malfatti, M.C.; Bellina, A.; Antoniali, G.; Tell, G. Revisiting Two Decades of Research Focused on Targeting APE1 for Cancer Therapy: The Pros and Cons. Cells 2023, 12, 1895. [Google Scholar] [CrossRef] [PubMed]
- Berquist, B.R.; McNeill, D.R.; Wilson, D.M. Characterization of Abasic Endonuclease Activity of Human Ape1 on Alternative Substrates, as Well as Effects of ATP and Sequence Context on AP Site Incision. J. Mol. Biol. 2008, 379, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Marenstein, D.R.; Wilson III, D.M.; Teebor, G.W. Human AP Endonuclease (APE1) Demonstrates Endonucleolytic Activity against AP Sites in Single-Stranded DNA. DNA Repair 2004, 3, 527–533. [Google Scholar] [CrossRef]
- Fan, J.; Matsumoto, Y.; Wilson, D.M. Nucleotide Sequence and DNA Secondary Structure, as Well as Replication Protein A, Modulate the Single-Stranded Abasic Endonuclease Activity of APE1. J. Biol. Chem. 2006, 281, 3889–3898. [Google Scholar] [CrossRef]
- Broxson, C.; Hayner, J.N.; Beckett, J.; Bloom, L.B.; Tornaletti, S. Human AP Endonuclease Inefficiently Removes Abasic Sites within G4 Structures Compared to Duplex DNA. Nucleic Acids Res. 2014, 42, 7708–7719. [Google Scholar] [CrossRef]
- Howpay Manage, S.A.; Zhu, J.; Fleming, A.M.; Burrows, C.J. Promoters vs. Telomeres: AP-Endonuclease 1 Interactions with Abasic Sites in G-Quadruplex Folds Depend on Topology. RSC Chem. Biol. 2023, 4, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Dolinnaya, N.G.; Ogloblina, A.M.; Yakubovskaya, M.G. Structure, Properties, and Biological Relevance of the DNA and RNA G-Quadruplexes: Overview 50 Years after Their Discovery. Biochem. Mosc. 2016, 81, 1602–1649. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-Throughput Sequencing of DNA G-Quadruplex Structures in the Human Genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-Molecule Visualization of DNA G-Quadruplex Formation in Live Cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Hänsel-Hertsch, R.; Di Antonio, M.; Balasubramanian, S. DNA G-Quadruplexes in the Human Genome: Detection, Functions and Therapeutic Potential. Nat. Rev. Mol. Cell Biol. 2017, 18, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA Secondary Structures: Stability and Function of G-Quadruplex Structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef]
- Bielskutė, S.; Plavec, J.; Podbevšek, P. Oxidative Lesions Modulate G-Quadruplex Stability and Structure in the Human BCL2 Promoter. Nucleic Acids Res. 2021, 49, 2346–2356. [Google Scholar] [CrossRef]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of G-Quadruplexes on the Regulation of Genome Integrity, DNA Damage and Repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martínez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-Quadruplex Structures Mold the DNA Methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957. [Google Scholar] [CrossRef]
- Zyner, K.G.; Simeone, A.; Flynn, S.M.; Doyle, C.; Marsico, G.; Adhikari, S.; Portella, G.; Tannahill, D.; Balasubramanian, S. G-Quadruplex DNA Structures in Human Stem Cells and Differentiation. Nat. Commun. 2022, 13, 142. [Google Scholar] [CrossRef]
- Linke, R.; Limmer, M.; Juranek, S.; Heine, A.; Paeschke, K. The Relevance of G-Quadruplexes for DNA Repair. Int. J. Mol. Sci. 2021, 22, 12599. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA Glycosylases Remove Oxidative Damages from Quadruplex DNA and Exhibit Preferences for Lesions in the Telomeric Sequence Context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA Damage Is Epigenetic by Regulating Gene Transcription via Base Excision Repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhu, J.; Howpay Manage, S.A.; Burrows, C.J. Human NEIL3 Gene Expression Regulated by Epigenetic-Like Oxidative DNA Modification. J. Am. Chem. Soc. 2019, 141, 11036–11049. [Google Scholar] [CrossRef]
- Zhou, J.; Fleming, A.M.; Averill, A.M.; Burrows, C.J.; Wallace, S.S. The NEIL Glycosylases Remove Oxidized Guanine Lesions from Telomeric and Promoter Quadruplex DNA Structures. Nucleic Acids Res. 2015, 43, 4039–4054. [Google Scholar] [CrossRef]
- Burra, S.; Marasco, D.; Malfatti, M.C.; Antoniali, G.; Virgilio, A.; Esposito, V.; Demple, B.; Galeone, A.; Tell, G. Human AP-Endonuclease (Ape1) Activity on Telomeric G4 Structures Is Modulated by Acetylatable Lysine Residues in the N-Terminal Sequence. DNA Repair 2019, 73, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, S.; Chen, Y.; Song, H.; Khutsishvili, I.; Marky, L.A.; Ray, S.; Natarajan, A.; Singh, P.K.; Bhakat, K.K. The Human AP-Endonuclease 1 (APE1) Is a DNA G-Quadruplex Structure Binding Protein and Regulates KRAS Expression in Pancreatic Ductal Adenocarcinoma Cells. Nucleic Acids Res. 2022, 50, 3394–3412. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of Telomeres and Telomerase in Cancer, and Advances in Telomerase-Targeted Therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef]
- Borah, S.; Xi, L.; Zaug, A.J.; Powell, N.M.; Dancik, G.M.; Cohen, S.B.; Costello, J.C.; Theodorescu, D.; Cech, T.R. TERT Promoter Mutations and Telomerase Reactivation in Urothelial Cancer. Science 2015, 347, 1006–1010. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Killela, P.J.; Reitman, Z.J.; Jiao, Y.; Bettegowda, C.; Agrawal, N.; Diaz, L.A.; Friedman, A.H.; Friedman, H.; Gallia, G.L.; Giovanella, B.C.; et al. TERT Promoter Mutations Occur Frequently in Gliomas and a Subset of Tumors Derived from Cells with Low Rates of Self-Renewal. Proc. Natl. Acad. Sci. USA 2013, 110, 6021–6026. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, T.; Ge, N.; Liu, L.; Yuan, X.; Liu, J.; Kong, F.; Wang, C.; Ren, H.; Yan, K.; et al. TERT Promoter Mutations Are Associated with Distant Metastases in Upper Tract Urothelial Carcinomas and Serve as Urinary Biomarkers Detected by a Sensitive castPCR. Oncotarget 2014, 5, 12428–12439. [Google Scholar] [CrossRef] [PubMed]
- Rachakonda, S.; Hoheisel, J.D.; Kumar, R. Occurrence, Functionality and Abundance of the TERT Promoter Mutations. Int. J. Cancer 2021, 149, 1852–1862. [Google Scholar] [CrossRef]
- Pavlova, A.V.; Savitskaya, V.Y.; Dolinnaya, N.G.; Monakhova, M.V.; Litvinova, A.V.; Kubareva, E.A.; Zvereva, M.I. G-Quadruplex Formed by the Promoter Region of the hTERT Gene: Structure-Driven Effects on DNA Mismatch Repair Functions. Biomedicines 2022, 10, 1871. [Google Scholar] [CrossRef]
- Wang, K.; Maayah, M.; Sweasy, J.B.; Alnajjar, K.S. The Role of Cysteines in the Structure and Function of OGG1. J. Biol. Chem. 2021, 296, 100093. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fleming, A.M.; Burrows, C.J. Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-Dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017, 139, 2569–2572. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These “Spare Tires” Have an Evolved Function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef]
- Nishio, M.; Kaori, T.; Kazunori, I. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030. [Google Scholar] [CrossRef] [PubMed]
- Sidorenko, V.S.; Nevinsky, G.A.; Zharkov, D.O. Mechanism of Interaction between Human 8-Oxoguanine-DNA Glycosylase and AP Endonuclease. DNA Repair 2007, 6, 317–328. [Google Scholar] [CrossRef]
- Dick, D.A. The Distribution of Sodium, Potassium and Chloride in the Nucleus and Cytoplasm of Bufo bufo Oocytes Measured by Electron Microprobe Analysis. J. Physiol. 1978, 284, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Bischof, H.; Rehberg, M.; Stryeck, S.; Artinger, K.; Eroglu, E.; Waldeck-Weiermair, M.; Gottschalk, B.; Rost, R.; Deak, A.T.; Niedrist, T.; et al. Novel Genetically Encoded Fluorescent Probes Enable Real-Time Detection of Potassium in Vitro and in Vivo. Nat. Commun. 2017, 8, 1422. [Google Scholar] [CrossRef]
- Li, W.; Miyoshi, D.; Nakano, S.; Sugimoto, N. Structural Competition Involving G-Quadruplex DNA and Its Complement. Biochemistry 2003, 42, 11736–11744. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Sahoo, B.; Varun, K.A.S.; Maiti, S.; Maiti, S. Effect of Loop Length Variation on Quadruplex-Watson Crick Duplex Competition. Nucleic Acids Res. 2008, 36, 4433–4442. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Nair, D.R.; Maiti, S. Effect of Flanking Bases on Quadruplex Stability and Watson–Crick Duplex Competition. FEBS J. 2009, 276, 3628–3640. [Google Scholar] [CrossRef]
- Kreig, A.; Calvert, J.; Sanoica, J.; Cullum, E.; Tipanna, R.; Myong, S. G-Quadruplex Formation in Double-Strand DNA Probed by NMM and CV Fluorescence. Nucleic Acids Res. 2015, 43, 7961–7970. [Google Scholar] [CrossRef]
- Zheng, K.; Chen, Z.; Hao, Y.; Tan, Z. Molecular Crowding Creates an Essential Environment for the Formation of Stable G-Quadruplexes in Long Double-Stranded DNA. Nucleic Acids Res. 2010, 38, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovská, I.; Renciuk, D.; Vorlíčková, M. Circular Dichroism and Conformational Polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Liu, Y.; Prasad, R.; Beard, W.A.; Kedar, P.S.; Hou, E.W.; Shock, D.D.; Wilson, S.H. Coordination of Steps in Single-Nucleotide Base Excision Repair Mediated by Apurinic/Apyrimidinic Endonuclease 1 and DNA Polymerase β. J. Biol. Chem. 2007, 282, 13532–13541. [Google Scholar] [CrossRef]
- Bazlekowa-Karaban, M.; Prorok, P.; Baconnais, S.; Taipakova, S.; Akishev, Z.; Zembrzuska, D.; Popov, A.V.; Endutkin, A.V.; Groisman, R.; Ishchenko, A.A.; et al. Mechanism of Stimulation of DNA Binding of the Transcription Factors by Human Apurinic/Apyrimidinic Endonuclease 1, APE1. DNA Repair 2019, 82, 102698. [Google Scholar] [CrossRef]
- Miroshnikova, A.D.; Kuznetsova, A.A.; Vorobjev, Y.N.; Kuznetsov, N.A.; Fedorova, O.S. Effects of Mono- and Divalent Metal Ions on DNA Binding and Catalysis of Human Apurinic/Apyrimidinic Endonuclease 1. Mol. BioSystems 2016, 12, 1527–1539. [Google Scholar] [CrossRef]
- Pastor, A.; Singh, A.K.; Fisher, M.T.; Chaudhuri, T.K. Protein Folding on Biosensor Tips: Folding of Maltodextrin Glucosidase Monitored by Its Interactions with GroEL. FEBS J. 2016, 283, 3103–3114. [Google Scholar] [CrossRef] [PubMed]
- Beernink, P.T.; Segelke, B.W.; Hadi, M.Z.; Erzberger, J.P.; Wilson, D.M.; Rupp, B. Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism. J. Mol. Biol. 2001, 307, 1023–1034. [Google Scholar] [CrossRef]
- Sowers, M.L.; Conrad, J.W.; Chang-Gu, B.; Cherryhomes, E.; Hackfeld, L.C.; Sowers, L.C. DNA Base Excision Repair Intermediates Influence Duplex–Quadruplex Equilibrium. Molecules 2023, 28, 970. [Google Scholar] [CrossRef] [PubMed]
- Popov, A.V.; Grin, I.R.; Dvornikova, A.P.; Matkarimov, B.T.; Groisman, R.; Saparbaev, M.; Zharkov, D.O. Reading Targeted DNA Damage in the Active Demethylation Pathway: Role of Accessory Domains of Eukaryotic AP Endonucleases and Thymine-DNA Glycosylases. J. Mol. Biol. 2020, 432, 1747–1768. [Google Scholar] [CrossRef] [PubMed]
- Redstone, S.C.J.; Fleming, A.M.; Burrows, C.J. Oxidative Modification of the Potential G-Quadruplex Sequence in the PCNA Gene Promoter Can Turn on Transcription. Chem. Res. Toxicol. 2019, 32, 437–446. [Google Scholar] [CrossRef]
- Davletgildeeva, A.T.; Kuznetsova, A.A.; Fedorova, O.S.; Kuznetsov, N.A. Activity of Human Apurinic/Apyrimidinic Endonuclease APE1 Toward Damaged DNA and Native RNA With Non-Canonical Structures. Front. Cell Dev. Biol. 2020, 8, 590848. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Herrmann, C.J.; Simonovic, M.; Szklarczyk, D.; von Mering, C. Version 4.0 of PaxDb: Protein Abundance Data, Integrated across Model Organisms, Tissues, and Cell-Lines. Proteomics 2015, 15, 3163–3168. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Tran, R.; Omaga, C.A.; Howpay Manage, S.A.; Burrows, C.J.; Conboy, J.C. Second Harmonic Generation Interrogation of the Endonuclease APE1 Binding Interaction with G-Quadruplex DNA. Anal. Chem. 2022, 94, 15027–15032. [Google Scholar] [CrossRef]
- Conrad, J.W.; Sowers, M.L.; Yap, D.Y.; Cherryhomes, E.; Pettitt, B.M.; Khanipov, K.; Sowers, L.C. Transition Mutations in the hTERT Promoter Are Unrelated to Potential I-Motif Formation in the C-Rich Strand. Biomolecules 2023, 13, 1308. [Google Scholar] [CrossRef] [PubMed]

| Position of hTERT Promoter Mutation | Patients with Simultaneous Mutations in the hTERT and APEX1 |
|---|---|
| G228 | 1 out of 122 (0.81%) |
| G242 | 1 out of 122 (0.81%) |
| G243 | 1 out of 122 (0.81%) |
| G250 | 2 out of 122 (1.63%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Savitskaya, V.Y.; Novoselov, K.A.; Dolinnaya, N.G.; Monakhova, M.V.; Snyga, V.G.; Diatlova, E.A.; Peskovatskova, E.S.; Golyshev, V.M.; Kitaeva, M.I.; Eroshenko, D.A.; et al. Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme. Int. J. Mol. Sci. 2025, 26, 337. https://doi.org/10.3390/ijms26010337
Savitskaya VY, Novoselov KA, Dolinnaya NG, Monakhova MV, Snyga VG, Diatlova EA, Peskovatskova ES, Golyshev VM, Kitaeva MI, Eroshenko DA, et al. Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme. International Journal of Molecular Sciences. 2025; 26(1):337. https://doi.org/10.3390/ijms26010337
Chicago/Turabian StyleSavitskaya, Viktoriia Yu., Kirill A. Novoselov, Nina G. Dolinnaya, Mayya V. Monakhova, Viktoriia G. Snyga, Evgeniia A. Diatlova, Elizaveta S. Peskovatskova, Victor M. Golyshev, Mariia I. Kitaeva, Daria A. Eroshenko, and et al. 2025. "Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme" International Journal of Molecular Sciences 26, no. 1: 337. https://doi.org/10.3390/ijms26010337
APA StyleSavitskaya, V. Y., Novoselov, K. A., Dolinnaya, N. G., Monakhova, M. V., Snyga, V. G., Diatlova, E. A., Peskovatskova, E. S., Golyshev, V. M., Kitaeva, M. I., Eroshenko, D. A., Zvereva, M. I., Zharkov, D. O., & Kubareva, E. A. (2025). Position-Dependent Effects of AP Sites Within an hTERT Promoter G-Quadruplex Scaffold on Quadruplex Stability and Repair Activity of the APE1 Enzyme. International Journal of Molecular Sciences, 26(1), 337. https://doi.org/10.3390/ijms26010337






