Abstract
Plant protease inhibitors are a ubiquitous feature of plant species and exert a substantial influence on plant stress responses. However, the KTI (Kunitz trypsin inhibitor) family responding to abiotic stress has not been fully characterized in Populus yunnanensis. In this study, we conducted a genome-wide study of the KTI family and analyzed their gene structure, gene duplication, conserved motifs, cis-acting elements, and response to stress treatment. A total of 29 KTIs were identified in the P. yunnanensis genome. Based on phylogenetic analysis, the PyKTIs were divided into four groups (1,2, 3, and 4). Promoter sequence analysis showed that the PyKTIs contain many cis-acting elements related to light, plant growth, hormone, and stress responses, indicating that PyKTIs are widely involved in various biological regulatory processes. RNA sequencing and real-time quantitative polymerase chain reaction analysis showed that KTI genes were differentially expressed under the inverted cutting stress of P. yunnanensis. Transcriptome analysis of P. yunnanensis leaves revealed that PyKTI16, PyKTI18, and PyKTI19 were highly upregulated after inverted cutting. Through the GEO query of Populus transcriptome data, KTI genes played a positive defense role in MeJa, drought, time series, and pathogen stress. This study provided comprehensive information for the KTI family in P. yunnanensis, which should be helpful for the functional characterization of P. yunnanensis KTI genes in the future.
1. Introduction
Proteases play vital roles in many cellular processes, such as digestion, blood clotting, immune response, and cellular signaling. They are involved in protein turnover and regulation, ensuring proper functioning and homeostasis within living organisms. These activities of proteases are controlled by protease inhibitors [1]. Protease inhibitors are molecules that inhibit the action of proteases [2]. In the past couple of decades, the number of studies on plant proteases and their proteinaceous inhibitors has been interestingly increased. The Kunitz trypsin inhibitor (KTI) is a protease inhibitor that contains a unique Kunitz domain and specifically inhibits trypsin. According to the classification by Rawlings et al. (2004), KTI belongs to the I3 inhibitor family, primarily functioning in the regulation of extracellular and intracellular target protease activities [3,4]. It is involved in physiological processes such as plant cell growth, cell cycle control, apoptosis, protein degradation and transport, stress response, and developmental growth [5].
KTI, which was first isolated from soybeans by Kunitz in 1945 [6], belongs to the family of protease inhibitors (PIs) and exhibits the ability to inhibit serine proteases. It has been postulated that KTIs play a crucial role in conferring resistance against herbivores and pathogens in numerous plant species. Notably, a KTI protein derived from the roots of Pseudostellaria heterophylla exhibited remarkable inhibitory activity against Fusarium oxysporum [7]. Multiple Kunitz trypsin inhibitors have been identified in Solanum tuberosum tubers, and research has revealed their potential as antifungal and antimicrobial agents [8]. In Arabidopsis thaliana, the silencing of AtKTI1 resulted in reduced resistance to Pseudomonas syringae pv tomato DC3000, whereas the overexpression of AtKTI1 enhanced resistance against the pathogen [9]. Additionally, studies have demonstrated that NtKTI1 plays a crucial role in conferring resistance to various pathogens, including Rhizoctonia solani, Rhizopus nigricans, and Phytophthora parasitica var. nicotianae, in Nicotiana tabacum [10]. These findings highlight the importance of KTIs in plant defense mechanisms against diverse pathogens and emphasize their potential applications in crop protection strategies. The involvement of AtKTI4 and AtKTI5 in defense against spider mites was observed, with elevated levels of serine protease and cysteine protease inhibitor activity detected in leaf protein extracts upon overexpression of these two genes in Nicotiana benthamiana [11]. In Passiflora edulis fruit, a total of seven Kunitz trypsin inhibitors (KTIs) have been characterized, demonstrating their role in conferring resistance against Diatraea saccharalis [12]. This discovery sheds light on the potential of KTIs in contributing to the plant’s defense mechanisms against insect herbivory and highlights their significance in the context of plant–insect interactions.
Populus yunnanensis, a dioecious poplar species endemic to the southwestern region of China, exhibits a wide distribution in areas characterized by low latitudes and high elevations [13]. Due to its rapid growth and ability for easy-cutting propagation, P. yunnanensis holds a prominent position as a dominant tree species in both forestry production and environmental conservation efforts. Its suitability for various applications makes it a valuable resource in terms of both economic and ecological significance [14]. Upon conducting cutting experiments, we serendipitously observed that inverted cuttings of P. yunnanensis not only survived but also exhibited the ability to develop into fully formed trees [15]. This phenomenon was facilitated by the rooting process occurring at the original morphological apex and subsequent sprouting at the base, effectively sustaining the survival and growth of the cuttings. These findings provide valuable insights into the remarkable regenerative capabilities of P. yunnanensis and may have implications for propagation and cultivation practices. Subsequently, the inverted cuttings of P. yunnanensis exhibited delayed sprouting and a bending upward growth pattern. Eventually, lateral branches that emerged from the base bud of the stem exhibited strong growth, albeit slightly less vigorous than those of the upright branches [15,16]. The levels of many hormones in P. yunnanensis, including auxins (IAA), cytokinins (CTK), gibberellin (GAs), ethylene(ET), and brassinosteroids (BRs), underwent large changes in the inverted cuttings [16]. The growth response observed in plants subjected to inversion presents an intriguing area of research for investigating the mechanisms underlying this phenomenon and its potential applications in horticultural and forestry practices.
This research is the first to identify, analyze, and function the KTI gene family in P. yunnanensis at the whole-genome level. The expression of the KTI gene was analyzed in combination with the transcriptome of P. yunnanensis treated with inversion so as to provide basic research on the survival mechanism of inversion.
2. Results
2.1. Identification and Phylogenetic Tree of the PyunKTI Genes Family in Populus
In the genomes of A. thaliana, V. vinifera, Salix, and Populus, 301 sequences were confirmed as Kunitz genes, including 29 in Populus yunnanensis, 30 in Populus trichocarpa, 41 in Populus deltoides WV94, 35 in Populus deltoides, 16 in Populus tremula, 28 in Populus davidiana, 28 in Populus euphratica, 24 in Populus pruinosa, 16 in Salix purpurea, 24 in Salix purpurea, 18 in Salix suchowensis, 5 in V. vinifera, and 7 in A. thaliana (Tables S1 and S2). Phylogenetic trees were constructed using the 301 KTIs from 13 species, and four groups were divided (Figure 1). Our 301 KTIs were clustered into four groups as follows: 95 within group 1, 12 within group 2, 103 within group 3, and 91 within group 4 (Figure 2). The KTIs of Salix and Populus plants were clustered, indicating that the KTI sequences were conserved among Salicaceae.
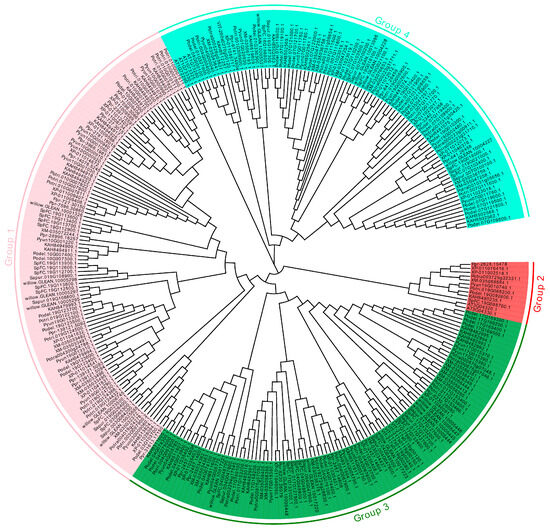
Figure 1.
Phylogenetic relationship of KTI gene family. The MEGA 11 with the neighbor-joining method was used to conduct the phylogenetic tree. Different background colors represented different groups of the KTI gene family.
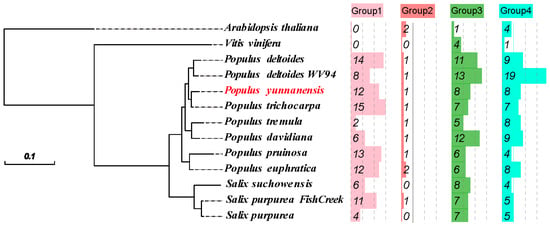
Figure 2.
Phylogeny during the evolution of angiosperms. Different colors represent the proportion of each species in the four groups.
2.2. Characterization of the PyunKTIs Gene Family in P. yunnanensis
The Kunitz genes in P. yunnanensis were named PyunKTI1~29 (Table 1). Bioinformatics analysis showed that the amino acid lengths of PyunKTI1~29 were 183–259. Most of the KTI proteins (75.86%) were predicted to be acidic, and basic protein (24.14%) was distributed in group 3 and group 4. The instability index (II) and GRAVY of the 29 PyunKTIs proteins were greater than 20 and less than 0.5, respectively. In addition, the prediction of subcellular localization showed that all PyunKTIs proteins were located in the vacuole.

Table 1.
Identification and characterization of the 29 PyunKTIs proteins.
The signal peptides prediction of the 29 PyunKTI proteins almost belongs to the “standard” secretory signal peptides transported by the sec translocon and cleaved by Signal Peptidase I (SP I) and were differentiated into four different groups: Group 1 has SP positions at the 1 bp–26 bp/27 bp/28 bp amino acid site. Group 2 does not have SP positions. Group 3 has SP positions at 1–24/25, and, specifically, PyunKTI4 has positions at 1–19. Group 4 has all SP positions at 1–21 (Table S3).
The hydrophobicity analysis of 29 KTI genes using Prot-Scale was used to further confirm more precise regions, combined with the prediction results of TMHMM for the transmembrane region (Table S4). In general, the transmembrane region of the protein is hydrophobic, while the part outside the membrane is hydrophilic [17]. The results showed that the predicted results of TMHMM and Prot-Scale were consistent, and the results of groups 1, 3, and 4 were less different. The amino acids from 1 to 30 had obvious hydrophobic characteristics and were in the transmembrane region, which was predicted to be the signal peptide sequence. In group 2, the amino acid sequence of PyunKTI26 was hydrophilic from 1 to 40 and hydrophobic at sequence sites 40 to 60, possibly a transmembrane helix region, but not a signal peptide. These findings suggest that the characteristic features of PyunKTI proteins may include instability, hydrophilicity, and nuclear localization.
PDB files of V. vinifera (5YH4) and Populus (Q5ZFE7) homologous proteins with the best alignment results were downloaded from the PDB database. ESPript3.0 was used to refine the protein alignment file and add a secondary structure. The secondary structures of the 29 PyunKTI proteins were composed of an α-helix (6.51–20.79%), extended strand (28.64–37.07%), beta turn (4.43–8.91%), and random coil (37.84–53.59%) (Table S5 and Figure S1). In protein alignment files, the α-helix was mainly in the front end of the protein alignment sequence, the grape PDB file did not contain an α-helix, and the beta turn was predicted to be 4 in the Populus PDB file, but 12 in V. vinifera (Figure 3).
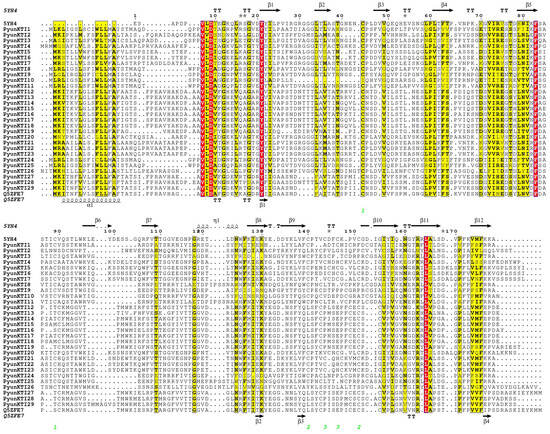
Figure 3.
Sequence alignment of functional sites of PDB and PyunKTI proteins of V. vinifera (5YH4) and Populus (Q5ZFE7) homologous proteins. A red background indicates complete amino acid sequence identity at the position; a yellow background indicates higher similarity.
Protein tertiary structure prediction of the 29 PyunKTI proteins with Swiss-model homologous modeling. Except for PyunKTI2, the other PyunKTI prediction results were modeled by miraculin-like protein from V. vinifera (5yh4.1). The global model quality estimate ranged from 0.44 to 0.78, the identity ranged from 30.91% to 72.73%, and the accuracy of the model reached more than 80%. The prediction results could be used to find functional sites and predict functional relationships (Table S6).
2.3. Gene Structures and Conserved Motifs of the PyunKTI Genes Family
The exon–intron distribution of the 29 PyunKTI genes was analyzed (Figure 4). All Kunitz genes do not contain introns, except for PyunKTI1, PyunKTI4, and PyunKTI5, which contain one intron. The analysis of sequence diversity outside of the KTI domain indicates a high degree of conservation within the KTI gene family following genome duplication. In total, 15 distinct conserved motifs were identified among the 29 PyunKTI proteins (Figure 4 and Table S7). The number of motifs varied across the PyunKTI proteins, ranging from 6 to 10. However, when considering the phylogenetic tree, it was observed that PyunKTI proteins within the same branch displayed a high similarity in motif distribution. Notably, the group 2 proteins exhibited the fewest motifs (6 motifs), while the group 3 proteins displayed the highest number of motifs (9 or 10 motifs). Conserved motifs 4 and 5 were present in all 29 PyunKTI proteins, while motifs 2, 6, and 8 were consistently found across the PyunKTI proteins. Additionally, certain motifs showed group-specificity: motifs 1, 9, and 12 were unique to nearly all group 1 PyunKTIs, motif 15 was exclusive to the group 3 PyunKTIs, and motif 13 was specific to all group 4 PyunKTIs, with these six motifs being absent in other groups. Based on these results, motifs 2, 4, 5, 6, and 8 were relatively conserved in the KTI gene family during evolution.
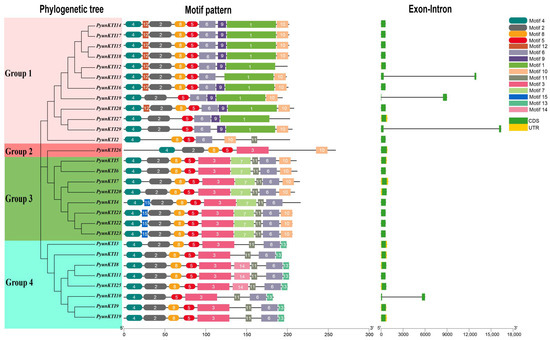
Figure 4.
Phylogenetic tree, conserved motif, and gene structure of the PyunKTI proteins. Different colors on the phylogenetic tree represent different groups of PyunKTI genes family. In the motif pattern, the motif numbers 1–15 are displayed in different colored boxes. In exon–intron analysis, the black lines represent introns, the green boxes represent the coding sequences, and the yellow boxes represent the non-coding sequences.
2.4. Chromosomal Location and Duplication of the PyunKTI Gene Family
The distribution of 29 PyunKTIs on the 19 P. yunnanensis chromosomes was uneven (Figure 5). There was no correlation between the chromosome length and the number of PyunKTI genes. For example, P. yunnanensis chromosome 10 was not the longest chromosome but contained eight PyunKTIs (PyunKTI10-PyunKTI18). However, the longest P. yunnanensis chromosome 1, contained only one PyunKTI (PyunKTI1). Detailed information on the chromosomal distribution is shown in Table 1.
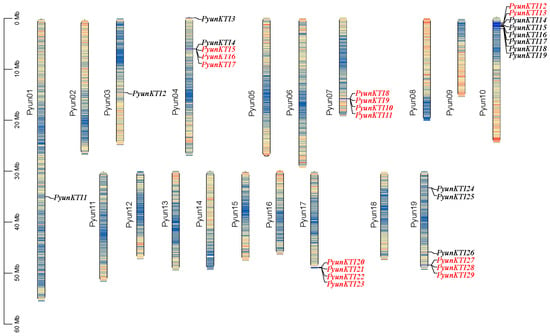
Figure 5.
Chromosomal distribution of P. yunnanensis PyunKTI genes. Tandem replication genes are shown in red.
As previously noted in the literature, gene duplication frequently accompanies the expansion of gene families [18]. Segmental or whole-genome duplication (WGD) and tandem duplication have been identified as the primary modes of duplication [19]. To gain insight into the mechanisms underlying the expansion of the KTIs gene family, we conducted analyses on the chromosomal information from P. yunnanensis, P. trichocarpa, A. thaliana, and V. vinifera to examine segmental or WGD duplication, as well as dispersed, proximal, and tandem events. Regarding PyunKTIs, 5 segmental or WGD (17.24%), 7 proximal (24.14%), 3 dispersed (10.34%), and 14 tandem duplications (48.28%) genes were detected. In 30 KTI genes of P. trichocarpa, 16 tandem (55.17%), 6 segmental or WGD (20.69%), 3 dispersed (10.34%), and 4 proximal duplication (13.79%) genes were detected. In seven Kunitz genes of A. thaliana, two dispersed (28.57%), one proximal (14.29%), and four tandem duplication (57.14%) genes were detected. In five Kunitz genes of V. vinifera, three tandem (60.00%), one dispersed (20.00%), as well as one proximal (20.00%) duplication gene were detected (Figure S2 and Table S8).
2.5. Tandem Repeat and Synteny Gene Pairs
A total of 16 tandem repeat gene pairs were analyzed among the 29 PyunKTIs gene members in P. yunnanensis. The Ka/Ks ratios of the KTI tandem repeat gene pairs ranged from 0.35 to 1.18, with 81.82% of the pairs having Ka/Ks ratios of <1, but the PyunKTI12/PyunKTI13 and PyunKTI28/PyunKTI29 gene pairs had Ka/Ks ratios of >1 (Table S9).
The synteny of the KTI gene family in P. yunnanensis, P. trichocarpa, A. thaliana, and V. vinifera was analyzed. A total of three segmental duplication gene pairs in 29 PyunKTIs were found, such as PyunKTI1/PyunKTI24, PyunKTI4/PyunKTI21, and PyunKTI5/PyunKTI20, respectively (Figure 6). There were 21 Synteny KTI gene pairs between P. yunnanensis and P. trichocarpa, 0 pairs between P. yunnanensis and V. vinifera, and 0 pairs between P. yunnanensis and A. thaliana (Figure 7). The analysis of the Ka/Ks ratio in the segmental duplication gene pairs of KTI revealed a range from 0.15 to 2.79. Notably, 95.65% of the gene pairs exhibited a Ka/Ks ratio below 1, indicating a prevalent occurrence of purifying selective pressure within the KTI gene family during evolution. However, it is worth mentioning that the PyunKTI9/Potri.007G111600.1 gene pair displayed a Ka/Ks ratio greater than 1 (Table S9), suggesting the possibility of positive selection acting on this particular gene pair.
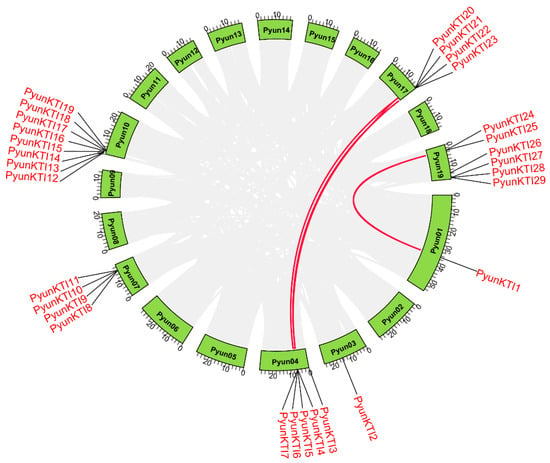
Figure 6.
Synteny analysis of the PyunKTI genes in P. yunnanensis.
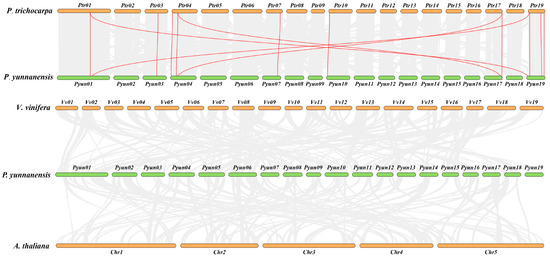
Figure 7.
Synteny analysis of the PyunKTI genes in P. yunnanensis, P. trichocarpa, A. thaliana, and V. vinifera. All collinear genes were labeled gray, while the collinear KTI gene pairs were labeled in red.
2.6. Promoter Analysis of the PyunKTIs Genes
The 2000-bp promoters of the 29 PyunKTI genes were found to contain four types of cis-acting regulatory elements (CREs) associated with light responsiveness, plant growth and development, phytohormone responsiveness, and stress responsiveness. (Figure 8 and Figure S3 and Table S10). In terms of plant growth and development, it was observed that the promoters of 6 out of the 29 PyunKTI genes (20.69%) contained an RY element, which is known for its involvement in seed-specific regulation. Promoters of 16 (55.17%) PyunKTI genes may have been involved in secondary xylem development. The MYC binding site involved in drought-inducibility existed in the promoters of 89.66% of PyunKTI genes. In addition, the promoters of 17 (58.62%), 22 (75.86%), 20 (68.97%), 16 (55.17%), 13 (44.83%), 5 (17.24%), 12 (41.38%), and 5 (17.24%) PyunKTI genes contained ethylene responsiveness (ERE), abscisic acid responsiveness (ABRE), MeJA responsiveness (CGTCA-motif and TGACG-motif), auxin responsiveness (TGA-element), gibberellin responsiveness (TATC-box), salicylic acid responsiveness (TCA-element), and a dehydration responsive element (DRE core), respectively. Furthermore, stress-responsive anoxic-induction elements (ARE), stress-responsive elements (STRE), Fungal elicitor-responsive elements (W box), low-temperature-responsive elements (LTR), MYB binding sites involved in drought-inducibility (MBSs), and defense and stress response elements (TC-rich repeats) were found in the promoters of 25 (86.21%), 14 (48.28%), 8 (27.59%), 8 (27.59%), 7 (24.14%), and 6 (20.69%) PyunKTI genes, respectively. Then, three CREs of the stress response regarding the wound-responsive element (WUN-motif, WRE3, box S) were found in the promoters of 10 (34.48%), 7 (24.14%), and 6 (20.69%) PyunKTI genes, respectively. The promoters of all 29 PyunKTI genes were found to contain light response elements. These findings suggest that the KTI genes may play a broad role in responding to diverse stressors and in regulating plant growth and development.
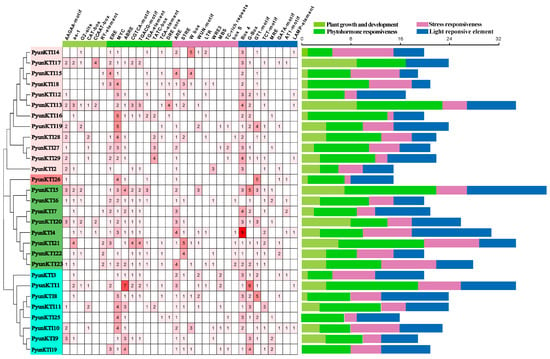
Figure 8.
Cis-regulatory element (CRE) analysis of PyunKTI genes family. The number of CREs in the promoter region of the PyunKTI genes. The number of each CRE was shown in the heatmap box, and white represents that there was no corresponding CRE. Different colors on the bar chart represent different types of CREs in the PyunKTI genes family.
2.7. Expression Profile of PyunKTI Genes from P. yunnanensis with Upright and Inverted Cuttings
This article sequenced and analyzed the raw transcriptome reads to assess the differences between the primary and fast-growing periods of upright and inverted cuttings. An RNA-seq analysis of the two processed cuttings (Figure 9) revealed that the expression levels in upright and inverted cuttings were markedly different at the same growth period. The top two principal components (PCs) obtained in the principal components analysis (PCA) could explain 90.8% of the total variation (Figure 9A). Notably, principal component 1 (PC 1) accounted for a substantial 80.8% of the observed variation, effectively distinguishing the different growth periods. Furthermore, principal component 2 (PC 2), explaining 10% of the variation, successfully differentiated the leaf samples obtained from the upright and inverted cuttings. These findings demonstrate significant differences in the expression patterns of PyunKTI genes across various growth periods while also revealing subtle variations associated with the direction of the cuttings.
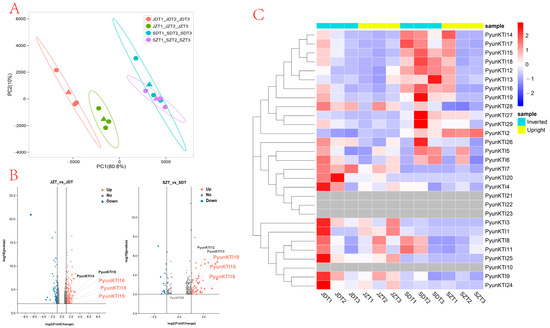
Figure 9.
Expression profile of PyunKTI genes from P. yunnanensis with upright and inverted cuttings. (A) PCA analysis of the PyunKTI genes in P. yunnanensis. (B) Volcano plot analysis of the PyunKTI genes in P. yunnanensis. (C) Heatmap analysis showing the expression patterns and coexpressed relationships of each KTI gene.
As depicted in Figure 9B,C, a considerable number of PyunKTI genes exhibited higher expression levels in the inverted cuttings compared to the upright cuttings during the same growth period. Taking group 1 as an example, PyunKTI16, PyunKTI18, and PyunKTI19 were found to be upregulated in both upright and inverted cuttings, as evidenced by the gene expression data obtained from the leaves. In the volcano plot, in the comparison of the JZT vs. JDT group and SZT vs. JDT group in July, log2 (FoldChange) > 1 was used as the criteria to screen for differential genes. PyunKTI16, PyunKTI18, and PyunKTI19 were all significantly upregulated.
2.8. RT-qPCR Analysis of the PyunKTI Genes Expression Level
The expression levels of the PyunKTI16, PyunKTI18, and PyunKTI19 genes under upright and inversion insertion treatments are shown in Figure 10. The results showed that, in the case of upright insertion, the expression of genes increased slightly with the change in developmental stage, from the early growth period to the rapid growth period. However, at the same developmental stage, the gene expression in the inversion insertion treatment increased sharply. Consistent results were obtained in the analysis of the transcriptome data, which showed that PyunKTI16, PyunKTI18, and PyunKTI19 had significant responses to inverted stress.
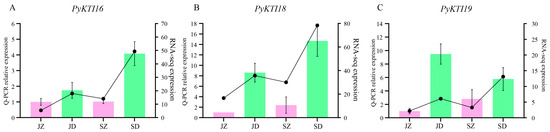
Figure 10.
Inverted treatment increases the expression of KTI genes. (A–C) show the qPCR relative expression and transcriptome results of PyunKTI16, PyunKTI18, and PyunKTI19, respectively. JZ, JD, SZ, and SD denote July upright insertion, July inversion insertion, September upright insertion, and September inversion insertion, respectively. Pink represents the upright insertion results, green represents the inversion insertion results, and the broken line represents the transcriptome results.
2.9. Transcriptome Analysis in GEO Database of PyunKTI Genes
The phylogenetic tree on the KTI gene family of P. yunnanensis and P. trichocarpa was grouped based on homologous multiple sequence comparisons (Figure 11A). Different levels of expression in major tissues (shoot tip, leaf, and root) of the KTIs in P. trichocarpa were analyzed by RNA-seq (GSE81077). The Kunitz genes of P. trichocarpa were divided into four groups (group 1/2/3/4) in the phylogenetic tree of P. yunnanensis and P. trichocarpa (Figure 12).
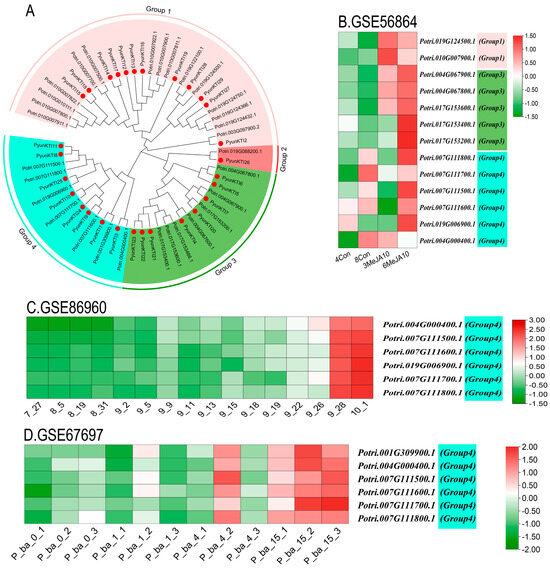
Figure 11.
KTI gene expression in the GEO database in the related transcriptome with P. trichocarpa as the reference gene. (A) NJ Phylogenetic tree of P. yunnanensis and P. trichocarpa. (B) GSE56864: Effect of Methyl Jasmonate on the poplar root transcriptome. 4Con and 8Con are the control roots; 3MeJA10 and 6MeJA10 are the Methyl Jasmonate treated roots. (C) GSE86960: A time series of autumn senescence leaves from P. tremula. Samples were collected from the growing season in July to the aging season in October. (D) GSE67697: Transcriptome analysis of poplar during leaf spot infection with Sphaerulina spp. Samples were collected at 0, 1, 3, 4, and 15 days after infection with the pathogen.
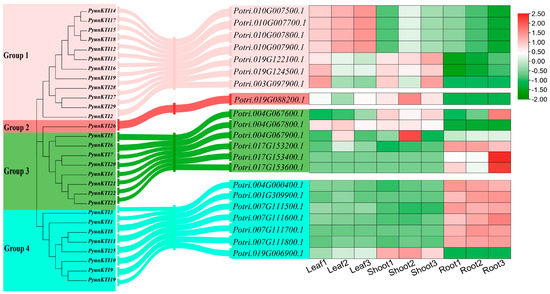
Figure 12.
The grouping relationship between the KTI gene of P. yunnanensis and the KTI gene of P. trichocarpa and the expression pattern of the KTI gene in various tissue parts of P. trichocarpa.
The functional expression of KTI genes in P. yunnanensis under different stress treatments was analyzed to screen and classify the KTI genes in the transcriptome in P. trichocarpa. Groups 1/2/3/4 may play different functions. In the GSE56864 data (Figure 11B), the PyunKTI genes were significantly upregulated after the Methyl Jasmonate treatment of sample roots, which were mainly concentrated in groups 3 and 4. In the GSE86960 dataset (Figure 11C), a gene expression analysis was conducted to examine the changes in P. tremula during autumn senescence. Members of the KTI gene family were gradually upregulated with time, with the highest gene expression at the end of leaf senescence, and these genes were concentrated only in group 4, from late summer and late into autumn. Using the GSE67697 data (Figure 11D), we conducted a transcriptome analysis of poplar during leaf spot infection with Sphaerulina spp. After inoculation with Sphaerulina spp., members of the KTI gene family were progressively upregulated with increasing time, reaching the highest gene expression at 15 days, and these genes were concentrated only in group 4. In the GSE97463 data (Figure 13A), differential gene expression analysis of drought-responsive sense and antisense genes in Populus was noted. In the short-term drought treatment, members of the KTI gene family were significantly upregulated in the apex, concentrated only in group 4. Regarding the long-term drought treatment, members of the KTI gene family were significantly upregulated in the roots, mainly concentrated in group 4, with some members upregulated in group 3 and group 1. Regarding the GSE79401 data (Figure 13B), plants on day 5 were under a mild drought state, and on day 7, they were in a severe drought state. Members of the KTI gene family were already significantly upregulated under mild drought, mainly in group 1 and group 4. Under prolonged drought treatment, more members of the KTI gene family were upregulated and more highly expressed, mainly in groups 1, 3, and 4.
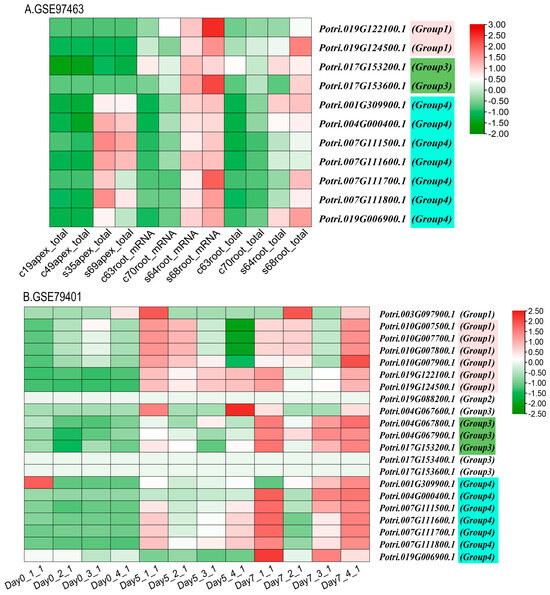
Figure 13.
KTI gene expression in the transcriptome of drought-treated P. trichocarpa as a reference gene in the GEO database. (A) GSE97463: Differential gene expression analysis of drought-responsive sense and antisense genes in Populus. Sample name starting with c represents the control group, and sample name starting with s represents the drought treatment group. (B) GSE79401: RNA-seq of drought-treated P. trichocarpa. Samples were collected at 0, 5, and 7 days for the drought treatment.
3. Discussion
3.1. Conserved Fundamental Features and Diverse Expression Patterns of KTIs
The traditional classes of plant protein protease inhibitors encompass various types, such as Kunitz, Bowman–Birk, potato I and II, squash, barley, cystatins, and miscellaneous. Since the discovery of soybean KTI in 1945, numerous other inhibitors have been identified and classified as KTIs due to their characteristic structural features. These features include a molecular weight ranging from approximately 18 to 24 kDa, two disulfide bonds, a single reactive site, multiple β-trefoil folds, and most importantly, the presence of the trypsin inhibitor domain [20,21]. In this study, we found that PyunKTI2 proteins of P. yunnanensis exhibited high sequence similarities to other known KTIs [11,22,23] in conserved regions, which were recognized as structural features of KTIs.
The number of KTI family members and the level of activity exhibited significantly. The number of KTI family members in the Populus species is more than in other species and the KTI members found in A. thaliana and V. vinifera were fewer than 25% in P. yunnanensis. Most of the KTI genes in the 16 species existed as tandem repeats, and the genes underwent purifying selection within P. yunnanensis and between P. yunnanensis and P. trichocarpa. Gene family expansion is often associated with gene duplication, which occurs through two main patterns: WGD and tandem duplication [18]. In this study, the expansion of the P. yunnanensis, P. trichocarpa, A. thaliana, and V. vinifera KTI gene families occurred mainly through tandem repeat events. The chromosomal localization results showed that 29 KTIs were unevenly distributed on seven chromosomes (Chr 1, Chr 3, Chr 4, Chr 7, Chr 10, Chr 17, and Chr 19). KTI1, KTI2, and KTI3 were present on chromosomes individually, while all the other genes were present in clusters of genes, which reflects the selection of KTIs during gene amplification.
The statistical results of ZHANG et al. [24] showed that most angiosperms contain KTI members, and the number of KTI members of monocotyledon and dicotyledon plants is significantly different. The number of KTI members of monocotyledon plants is generally small, only 1 to 3, while the number of KTI members of dicotyledon plants is more; for example, Medicago sativa has the largest number of KTI members (51), followed by Glycine max (50). In this paper, we analyzed the KTI gene members of 13 species of Populus and Salix, which belong to the most abundant type of dicotyledonous plants. It is speculated that KTI plays a more important role in the growth and development of dicotyledonous plants, especially Salicaceae.
An analysis of the conserved structural domains revealed that the number and type of conserved motifs varied among the PyKTIs. However, gene members within the same group shared similar conserved motif types, suggesting the presence of analogous structures and functions within the same group [25]. All PyKTIs contained motifs 4, 2, and 5, which may represent the key elements conserved during evolution. Motif 7 is exclusive to group 3, while motifs 1, 9, and 12 are found only in group 1, and motifs 13 and 14 are also unique to group 1. The similar motif composition within the same group suggests that the proteins are structurally conserved and likely share similar functions.
The overall secondary structure of the Kunitz inhibitor family consists of 43% β structures, 55% loops, and 2% helices, and more than 50% of the structural part is made up of loops [20]. This could be the reason behind the dynamic nature of these proteins. The average secondary structure of the 29 PyunKTI proteins was composed of 11.17% α-helix, 32.63% extended strand, 6.83% beta turn, and 49.37% random coil. The secondary structure of the poplar KTI is different from other types of KTI genes. The α-helix structure is composed of a segment of MK***FLL*A (M: Methionine; K: Lysine; F: Phenylalanine; L: Leucine; A: Alanine) at the front end of the gene, and the α-helix protein is the main part of the transmembrane protein. The alpha-helical structure of a protein present in the cellular lipid bilayer, as a transmembrane protein, usually has both hydrophobic and hydrophilic side chains. The hydrophobic side chains interact with the lipids present in the membrane, while the hydrophilic side chains remain exposed to the outer surface.
As a subset of the serine protease inhibitors, Kunitz participates in the regulation of many stress-resistant processes, such as plant growth and development, the synthesis of secondary metabolites, and plant hormone signaling pathways and their interactions [20]. In this study, the characteristics of the KTI family in P. yunnanensis and the regulatory role of KTI in response to the inverted cuttings’ stress were analyzed, providing gene resources for P. yunnanensis resistance breeding and advancing the functional study of the P. yunnanensis KTI genes.
3.2. Involved in Plant Growth and Development
Because plant KTIs can regulate the activity of serine protease, avoid excessive protein degradation, and promote cell homeostasis, it has received more and more attention in recent years, and many reports indicate that it is closely related to plant growth and development. It has been found that serine protease plays an important role in plant stress, senescence, programmed cell death, and sexual reproduction [26]. Phaseolus vulgaris subtilisin-like protease 2 (PvSLP2) was much more active in aged leaves than in mature and young leaves, which was related to the degree of leaf senescence [27]. Islam et al. found that the decreased expression of Trifolium repens Kunitz proteinase inhibitor 2 (Tr-KTI2) affected a series of developmental traits, including the stem length, branch number, and petiole length, as well as increased morphological diversity and decreased developmental stability, indicating that KTI plays an important role in genetic variation and the developmental stability of plants [28]. Ethylene and mechanical injury can activate the transcriptional activity of HECATE 1 (HEC1), a member of the A. thaliana bHLH transcription factor family, and promote Kunitz protease inhibitor 1 (Kunitz-PI1) accumulation of apical curls and cotyledon apices [29,30]. In this study, transcriptome and qRT-PCR at primary and rapid growth showed that PyunKTI1, PyunKTI2, and PyunKTI4 were upregulated during the growth process of P. yunnanensis.
3.3. Involvement in Biological and Environmental Stresses
KTI can also enhance plants’ tolerance to various stresses, including drought, salinity, and low temperatures [28,31]. We hypothesized that some PyKTI genes might be regulated by stress because their promoters contain cis-acting elements related to stress responses. For example, we found that the promoters of PyKTI3 and PyKTI1 contain two MBSs, and the promoters of PyKTI11, PyKTI25, and PyKTI10 contain four MYCs. All five PyKTIs belong to group 4 in the evolutionary tree, and MBSs and MYCs are cis-acting elements related to drought stress (Figure 8). Potri.001G309900.1, Potri.004G000400.1, Potri.007G111500.1, Potri.007G111600.1, Potri.007G111700.1, Potri.007G111800.1, and Potri.019G006900.1 in P. trichocarpa also belonged to group 4 in the evolutionary tree, and these seven genes showed upregulated expression in both the apex and roots of P. trichocarpa after the drought treatment and continued to be upregulated as the drought treatment continued (Figure 11A and Figure 13).
Numerous studies have shown that conditions like high temperatures, water stress, and the application of Methyl Jasmonate significantly elevate the mRNA and protein levels of KTI in various plants, including Brassica oleracea, Glycine max, Solanum tuberosum, and Passiflora edulis. These findings indicate that KTI plays a crucial role in the response to abiotic stress [12,32,33,34,35]. PtKTI12 is important for wobble uridine modification and stress tolerance under water deficit conditions [36]. We analyzed the expression of Cassia obtusifolia CoTI under different stress conditions and found that its expression was significantly affected by salt stress, drought, and abscisic acid [37]. Islam et al. used RNA interference technology to reduce the mRNA content of the KTI gene in Trifolium repens, which resulted in the destruction of homeostasis and the release of a large number of intracellular emergency molecules (H2O2) and activated the flavonoid metabolic pathway [28]. Sustained expression of the trypsin protease inhibitor in transgenic tobacco confers greater resistance to salt stress, acid-base stress, and drought stress [31]. In this paper, the inversion of P. yunnanensis is a type of stress compared with plants, and its survival mechanism has not been clearly studied. However, transcriptome and qRT-PCR studies of upright cuttings and inverted cuttings showed that the upregulation response of KTI family members was obvious in inverted cuttings. Accordingly, we hypothesized that the KTI genes exert the same stress tolerance function in P. yunnanensis.
4. Materials and Methods
4.1. Identification and Characterization of the PyunKTI Gene Family
Arabidopsis thaliana and Vitis vinifera were selected as model species. The A. thaliana, V. vinifera, Salix, and Populus genomes were downloaded from Phytozome, NCBI, and PlantGe-nIE. A hidden Markov model (HMM) was used to identify the Kunitz candidates, and the HMM profiles of the Kunitz (PF00017) domains were downloaded from the Pfam protein database (https://www.ebi.ac.uk/interpro/, accessed on 15 June 2024) [38]. The putative Kunitz proteins, identified by HMMER v3.0 software [39], were submitted to the CDD database and BLAST program (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 15 June 2024) to check for complete Kunitz domains [40]. The family members of P. yunnanensis were confirmed and sorted according to their chromosomal position for downstream analysis. The physicochemical properties of the putative Kunitzs, including the number of amino acids (No. of aa), molecular weight (MW), isoelectric point (pI), instability index, and grand average of hydropathicity (GRAVY), were analyzed using the online ExPASy-ProtParam tool (http://web.expasy.org/protParam/, accessed on 15 June 2024). The subcellular localization, secondary structure, and signal peptides of PyunKTI proteins were predicted using Plant-mPloc, Prabi, and SignalP 6.0, respectively. Homologous proteins in the PDB database were downloaded using the PDB file, using the ESPript3.0 beautify protein ratio on file, and adding a secondary structure.
4.2. Analyses of Phylogenetics, Gene Structure, Motif Composition, Chromosomal Distribution, Gene Duplication, and Synteny
To investigate the evolutionary relationships among the 301 candidate Kunitz proteins from 13 plant species, multiple sequence alignments were conducted using ClustaW with default parameters [41]. Subsequently, a phylogenetic tree was constructed using MEGA 11 software, employing the neighbor-joining (NJ) method with pair-wise deletion, Poisson correction, and 1000 bootstrap replicates for statistical support [42,43]. The resulting phylogenetic tree was visualized using the EvolView v3 software [44]. Furthermore, comprehensive analyses were performed to explore additional aspects of the Kunitz proteins, including the gene structure, motif composition, chromosomal distribution, gene duplication, and synteny.
The genomic DNA sequences and coding sequences (CDS) of the 29 PyunKTI genes were utilized to investigate the gene structure in P. yunnanensis, a process that involves analyzing the arrangement of various subunits within the gene. To identify and characterize the conserved motifs in the Kunitz proteins, we employed the online Multiple Expectation Maximization for Motif Elicitation tool (MEME). The MEME tool uses statistical algorithms and machine learning techniques to identify patterns or motifs within protein sequences. In this study, we set the parameters to search for 15 motifs with the optimum motif width of 8–80 residues, allowing for any number of repetitions [45]. Tattoos were used to visualize the gene structure and motif composition of PyunKTI proteins [46]. The chromosomal positions of the PyunKTIs were mapped using MapChart v5.6.0 software, and the Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the duplication pattern and synteny of the PyunKTIs [47]. Finally, to assess the selective pressure acting on the PyunKTIs, we calculated the ratio of nonsynonymous to synonymous substitutions (Ka/Ks) using KaKs_Calculator2.0 [48].
4.3. Identification of Cis-Regulatory Elements (CREs) in PyunKTIs Gene Promoters
To identify cis-regulatory elements (CREs) in the promoter regions of the 29 PyunKTI genes, the 2000-bp sequences upstream from the start codon were analyzed using the PlantCARE tool [49]. The obtained results were further visualized using EvolView v3 [44].
4.4. Transcriptome Analysis About PyunKTI Genes
In the Gene Expression Omnibus (GEO) database, transcriptome items were screened for “poplar” keywords and various stress treatments. These GEO data are all based on P. trichocarpa as the reference genome, and P. trichocarpa and P. yunnanensis were constructed as a phylogenetic tree and classified according to the previous groups. The expression datasets of the Kunitz gene family in different treatments were constructed. GSE81077 provides data for the RNA-seq of major tissues and xylem cell types of P. trichocarpa [50]. GSE56864 focuses on the effect of Methyl Jasmonate on the poplar root transcriptome [51]. GSE86960 analyzes the expression of a gene in P. tremula in senescent leaves from different years [52]. GSE67697 investigates the transcriptome analysis of poplar during leaf spot infection with Sphaerulina [53]. GSE97463 explores the differential gene expression analysis of drought-responsive sense and antisense genes in Populus [54]. GSE79401 provides RNA-seq data for drought-treated P. trichocarpa [55]. Xylem tissue was collected from the plants (1) without drought treatment (control, day 0), (2) under mild drought stress (stage 3, day 5), (3) under severe drought stress (stage 4, day 7), and was followed by RNA-seq analysis.
4.5. The Expression Profiles of PyunKTI Genes Were Analyzed Using the Available Transcriptome Datasets
The cuttings of 3 clones of P. yunnanensis were cultivated in the greenhouse for one year (Southwest Forestry University, Kunming, China. 102.76 E, 25.06 N). In February 2022, we obtained about 15 cm cuttings and preserved these in the soil. The cuttings with different tissues were used in the experiments performed in July 2023 (the early growth period). We obtained morphologically uniformly sized roots, as well as stems, lateral buds, branches, young leaves (first to third leaves counted from the stem tip), and stem tips. All plant samples collected were brought back in liquid nitrogen and then stored at −80 °C until their next use.
One clone was treated as a biological replicate, and three replicates were established. All samples were sent to BioMark Technologies (Beijing, China) for RNA-seq. The genome of P. yunnanensis was used as a reference annotation library for data analysis (BioProject: PRJNA886471). Fragments per kilobase of transcript per million mapped (FPKM) were used as an index to measure the expression level of the transcripts or genes. Based on the transcriptome datasets, the expression level of the Kunitz genes in differential expression levels of genes under different growth stages of P. yunnanensis between positive and inverse insertions was converted with log2 (FPKM).
4.6. RT-qPCR Analysis of the PyunKTI Genes
Gene-specific primers for PyuKTI16, PyuKTI18, and PyuKTI19 were obtained from the study using Primer-BLAST [56], and the primer details can be found in Table S11, which provide information regarding the primers used for amplification. To investigate the dynamic expression patterns of the three KTI genes, reverse-transcription quantitative PCR (RT-qPCR) was performed. For normalization purposes, we chose the endogenous control gene HIS as the internal standard. The Ct values were amplified and quantified using the Rotor-Gene qPCR system, utilizing the Fast Super EvaGreen qPCR Master Mix (US Everbright Inc., Suzhou, China). Two-step amplification was carried out as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 10 s, and 60 °C for 30 s. This experiment was repeated for three biological replicates, each involving three technical repeats. To determine the relative expression levels, the 2−ΔΔCt method was employed [57].
5. Conclusions
We identified 29 PyunKTI genes from P. yunnanensis, which can be classified into four groups. Tandem repeat events mainly achieve the amplification of PyunKTI genes. Biologically, PyunKTI plays a wide range of roles in coping with various stresses and regulating plant growth and development. The results of this study will contribute to a better understanding of the functional and molecular mechanisms of the PyunKTI genes in stress resistance and their potential use in the genetic evolution of P.yunnanensis.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms26010188/s1.
Author Contributions
Conceptualization, H.G.; Data curation, D.Z. and S.M.; Formal analysis, S.M.; Funding acquisition, D.Z.; Investigation, X.Z. and S.Z.; Methodology, H.G., S.M., and L.Z.; Project administration, D.Z. and D.L.; Software, X.Z. and R.X.; Supervision, D.Z. and D.L.; Validation, H.G. and C.W.; Visualization, H.G. and S.M.; Writing—original draft, H.G. and S.M.; Writing—review and editing, H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Youth Talents Special Project of Yunnan Province, “Xingdian Talents Support Program” (XDYC-QNRC-2022-0232 and YNWR-QNBJ-2020194).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genomic data and RNA-seq data for P. yunnanensis are available from the corresponding author. The data supporting the results of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bateman, K.S.; Michael, N.G.J. Plant Protein Proteinase Inhibitors: Structure and Mechanism of Inhibition. Curr. Protein Pept. Sci. 2011, 12, 341–347. [Google Scholar] [CrossRef]
- Turk, B. Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 2006, 5, 785–799. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Tolle, D.P.; Barrett, A.J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004, 378, 705–716. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J.; Alex, B. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012, 40, D343–D350. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2013, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed]
- Kunitz, M. Crystallization of a Trypsin Inhibitor from Soybean. Science 1945, 101, 668–669. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Ng, T.B. Concurrent isolation of a Kunitz-type trypsin inhibitor with antifungal activity and a novel lectin from Pseudostellaria heterophylla roots. Biochem. Biophys. Res. Commun. 2006, 342, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Bártová, V.; Bárta, J.; Jarošová, M. Antifungal and antimicrobial proteins and peptides of potato (Solanum tuberosum L.) tubers and their applications. Appl. Microbiol. Biotechnol. 2019, 103, 5533–5547. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. Kunitz Trypsin Inhibitor: An Antagonist of Cell Death Triggered by Phytopathogens and Fumonisin B1 in Arabidopsis. Mol. Plant 2008, 1, 482–495. [Google Scholar] [CrossRef]
- Huang, H.; Qi, S.D.; Qi, F.; Wu, C.A.; Zheng, C.C. NtKTI1, a Kunitz trypsin inhibitor with antifungal activity from Nicotiana tabacum, plays an important role in tobacco’s defense response. FEBS J. 2010, 277, 4076–4088. [Google Scholar] [CrossRef]
- Ana, A.; Lucia, T.M.; Pablo, G.M.; Manuel, M.; Isabel, D.; Santamaria, M.E. Arabidopsis Kunitz Trypsin Inhibitors in Defense Against Spider Mites. Front. Plant Sci. 2018, 9, 986. [Google Scholar]
- Botelho-Júnior, S.; Machado, O.L.T.; Fernandes, K.V.S.; Lemos, F.J.A.; Perdizio, V.A.; Oliveira, A.N.E.A.; Monteiro, L.R.; Filho, M.L.; Jacinto, T. Defense response in non-genomic model species: Methyl jasmonate exposure reveals the passion fruit leaves’ ability to assemble a cocktail of functionally diversified Kunitz-type trypsin inhibitors and recruit two of them against papain. Planta 2014, 240, 345–356. [Google Scholar] [CrossRef]
- Jiang, H.; Korpelainen, H.; Li, C. Populus yunnanensis males adopt more efficient protective strategies than females to cope with excess zinc and acid rain. Chemosphere 2013, 91, 1213–1220. [Google Scholar] [CrossRef]
- Li, X.; Yang, Y.; Sun, X.; Lin, H.; Chen, J.; Ren, J.; Hu, X.; Yang, Y. Comparative Physiological and Proteomic Analyses of Poplar (Populus yunnanensis) Plantlets Exposed to High Temperature and Drought. PLoS ONE 2014, 9, e107605. [Google Scholar] [CrossRef] [PubMed]
- Zhou, A.-P.; Zong, D.; Gan, P.-H.; Zou, X.-L.; Fei, X.; Zhong, Y.-Y.; He, C.-Z. Physiological analysis and transcriptome profiling of inverted cuttings of Populus yunnanensis reveal that cell wall metabolism plays a crucial role in responding to inversion. Genes 2018, 9, 572. [Google Scholar] [CrossRef]
- Zhou, A.P.; Gan, P.H.; Zong, D.; Fei, X.; Zhong, Y.Y.; Li, S.Q.; Yu, J.-D.; He, C.Z. Bark tissue transcriptome analyses of inverted Populus yunnanensis cuttings reveal the crucial role of plant hormones in response to inversion. PeerJ 2019, 7, e7740. [Google Scholar] [CrossRef]
- Mbaye, M.N.; Hou, Q.; Basu, S.; Teheux, F.; Rooman, M. A Comprehensive Computational Study of Amino Acid Interactions in Membrane Proteins. Sci. Rep. 2019, 9, 12043. [Google Scholar] [CrossRef]
- Wang, P.; Moore, B.M.; Panchy, N.L.; Meng, F.; Lehti-Shiu, M.D.; Shiu, S.H. Factors Influencing Gene Family Size Variation Among Related Species in a Plant Family, Solanaceae. Genome Biol. Evol. 2018, 10, 2596–2613. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Depamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2010, 50, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Bendre, A.D.; Ramasamy, S.; Suresh, C.G. Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int. J. Biol. Macromol. 2018, 113, S0141813018303222. [Google Scholar] [CrossRef] [PubMed]
- Oliva, M.; Silva, M.; Sallai, R.C.; Brito, M.V.; Sampaio, M.U. A novel subclassification for Kunitz proteinase inhibitors from leguminous seeds. Biochimie 2010, 92, 1667–1673. [Google Scholar] [CrossRef]
- Kim, S.H.; Hara, S.; Hase, S.; Ikenaka, T.; Toda, H.; Kitamura, K.; Kaizuma, N. Comparative study on amino acid sequences of Kunitz-type soybean trypsin inhibitors, Tia, Tib, and Tic. J. Biochem. 1985, 98, 435–448. [Google Scholar] [CrossRef]
- Kobayashi, H. A Kunitz-type Protease Inhibitor, Bikunin, Inhibits Ovarian Cancer Cell Invasion by Blocking the Calcium-dependent Transforming Growth Factor-β1 Signaling Cascade. J. Biol. Chem. 2003, 278, 7790–7799. [Google Scholar] [CrossRef]
- Zhang, X.; Liao, D.-Y.; Gou, X.-Z.; Zhou, J.-Y.; Liao, H. Structure and function of plant Kunitz-type protease inhibitors. Plant Physiol. J. 2018, 54, 1391–1400. [Google Scholar]
- Stegmaier, P.; Kel, A.E.; Wingender, E. Systematic DNA-binding domain classification of transcription factors. Genome Inform. Int. Conf. Genome Inform. 2004, 15, 276–286. [Google Scholar]
- Luciński, R.; Misztal, L.; Samardakiewicz, S.A.; Jackowski, G. Involvement of Deg5 protease in wounding-related disposal of PsbF apoprotein. Plant Physiol. Biochem. 2011, 49, 311–320. [Google Scholar] [CrossRef]
- Budi, M.K.; Saboti, J.; Megli, V.; Kos, J.; Kidri, M. Characterization of two novel subtilases from common bean (Phaseolus vulgaris L.) and their responses to drought. Plant Physiol. Biochem. 2013, 62, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Leung, S.; Burgess, E.P.; Laing, W.A.; Richardson, K.A.; Hofmann, R.W.; Dijkwel, P.P.; McManus, M.T. Knock-down of transcript abundance of a family of Kunitz proteinase inhibitor genes in white clover (Trifolium repens) reveals a redundancy and diversity of gene function. New Phytol. 2015, 208, 1188–1201. [Google Scholar] [CrossRef]
- Boex-Fontvieille, E.; Rustgi, S.; Von Wettstein, D.; Pollmann, S.; Reinbothe, S.; Reinbothe, C. Corrigendum: An Ethylene-Protected Achilles’ Heel of Etiolated Seedlings for Arthropod Deterrence. Front. Plant Sci. 2018, 9, 1741. [Google Scholar] [CrossRef]
- Edouard, B.F.; Sachin, R.; Steffen, R.; Christiane, R. A Kunitz-type protease inhibitor regulates programmed cell death during flower development in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 6119–6135. [Google Scholar]
- Srinivasan, T.; Kumar, K.R.R.; Kirti, P.B. Constitutive Expression of a Trypsin Protease Inhibitor Confers Multiple Stress Tolerance in Transgenic Tobacco. Plant Cell Physiol. 2009, 50, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Afsana, I.; Susanna, L.; Aluh, N.; Dijkwel, P.P.; Mcmanus, M.T. Kunitz Proteinase Inhibitors Limit Water Stress Responses in White Clover (Trifolium repens L.) Plants. Front. Plant Sci. 2017, 8, 1683. [Google Scholar]
- Komatsu, S.; Yamamoto, R.; Nanjo, Y.; Mikami, Y.; Yunokawa, H.; Sakata, K. A comprehensive analysis of the soybean genes and proteins expressed under flooding stress using transcriptome and proteome techniques. J. Proteome Res. 2009, 8, 4766–4778. [Google Scholar] [CrossRef]
- Kang, S.G.; Choi, J.H.; Suh, S.G. A Leaf-specific 27 kDa Protein of Potato Kunitz-Type Proteinase Inhibitor Is Induced in Response to Abscisic Acid, Ethylene, Methyl Jasmonate, and Water Deficit. Mol. Cells 2002, 13, 144–147. [Google Scholar] [CrossRef]
- Sheshukova, E.V.; Komarova, T.V.; Ershova, N.M.; Shindyapina, A.V.; Dorokhov, Y.L. An Alternative Nested Reading Frame May Participate in the Stress-Dependent Expression of a Plant Gene. Front. Plant Sci. 2017, 8, 2137. [Google Scholar] [CrossRef]
- Wang, H.; Xu, C.; Zhang, Y.; Yan, X.; Zheng, B. PtKTI12 genes influence wobble uridine modifications and drought stress tolerance in hybrid poplar. Tree Physiol. 2020, 40, 1778–1791. [Google Scholar] [CrossRef]
- Liu, Z.; Zhu, Q.; Li, Y.; Yu, J.; Wang, W.; Tan, R.; Zhou, J.; Liao, H. Isolation and in silico characterization of a shikimate kinase from Cassia obtusifolia. Acta Physiol. Plant. 2015, 37, 85. [Google Scholar] [CrossRef]
- Sara, E.G.; Jaina, M.; Alex, B.; Eddy, S.R.; Aurélien, L.; Potter, S.C.; Matloob, Q.; Richardson, L.J.; Salazar, G.A.; Alfredo, S. The Pfam protein families database in 2019. Nuclc Acids Res. 2018, 47, D427–D432. [Google Scholar]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, 29–37. [Google Scholar] [CrossRef]
- Aron, M.B.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Farideh, C.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple Sequence Alignment Using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2003, 1, 2–3. [Google Scholar] [CrossRef]
- Koichiro, T.; Glen, S.; Sudhir, K. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nuclc Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Mikael, B.; Buske, F.A.; Martin, F.; Grant, C.E.; Luca, C.; Jingyuan, R.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 9. [Google Scholar] [CrossRef]
- Yupeng, W.; Haibao, T.; Debarry, J.D.; Xu, T.; Jingping, L.; Xiyin, W.; Tae-Ho, L.; Huizhe, J.; Barry, M.; Hui, G. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef]
- Plett, J.M.; Daguerre, Y.; Wittulsky, S.; Vayssières, A.; Martin, F. Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc. Natl. Acad. Sci. USA 2014, 111, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Edlund, E.; Novak, O.; Karady, M.; Ljung, K.; Jansson, S. Contrasting patterns of cytokinins between years in senescing aspen leaves. Plant Cell Environ. 2017, 40, 622. [Google Scholar] [CrossRef] [PubMed]
- Foster, A.J.; Gervais, P.; Philippe, T.; Armand, S.; Martina, S. Transcriptome Analysis of Poplar during Leaf Spot Infection with Sphaerulina spp. PLoS ONE 2015, 10, e0138162. [Google Scholar] [CrossRef]
- Yinan, Y.; Su, C. Widespread antisense transcription of Populus genome under drought. Mol. Genet. Genom. 2018, 293, 1017–1033. [Google Scholar]
- Li, S.; Lin, Y.C.J.; Wang, P.; Zhang, B.; Li, M.; Chen, S.; Shi, R.; Tunlaya-Anukit, S.; Liu, X.; Wang, Z.; et al. The AREB1 Transcription Factor Influences Histone Acetylation to Regulate Drought Responses and Tolerance in Populus trichocarpa. Plant Cell 2019, 31, 663–686. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).