The Influence of a Genetic Variant in CCDC78 on LMNA-Associated Skeletal Muscle Disease
Abstract
1. Introduction
2. Results
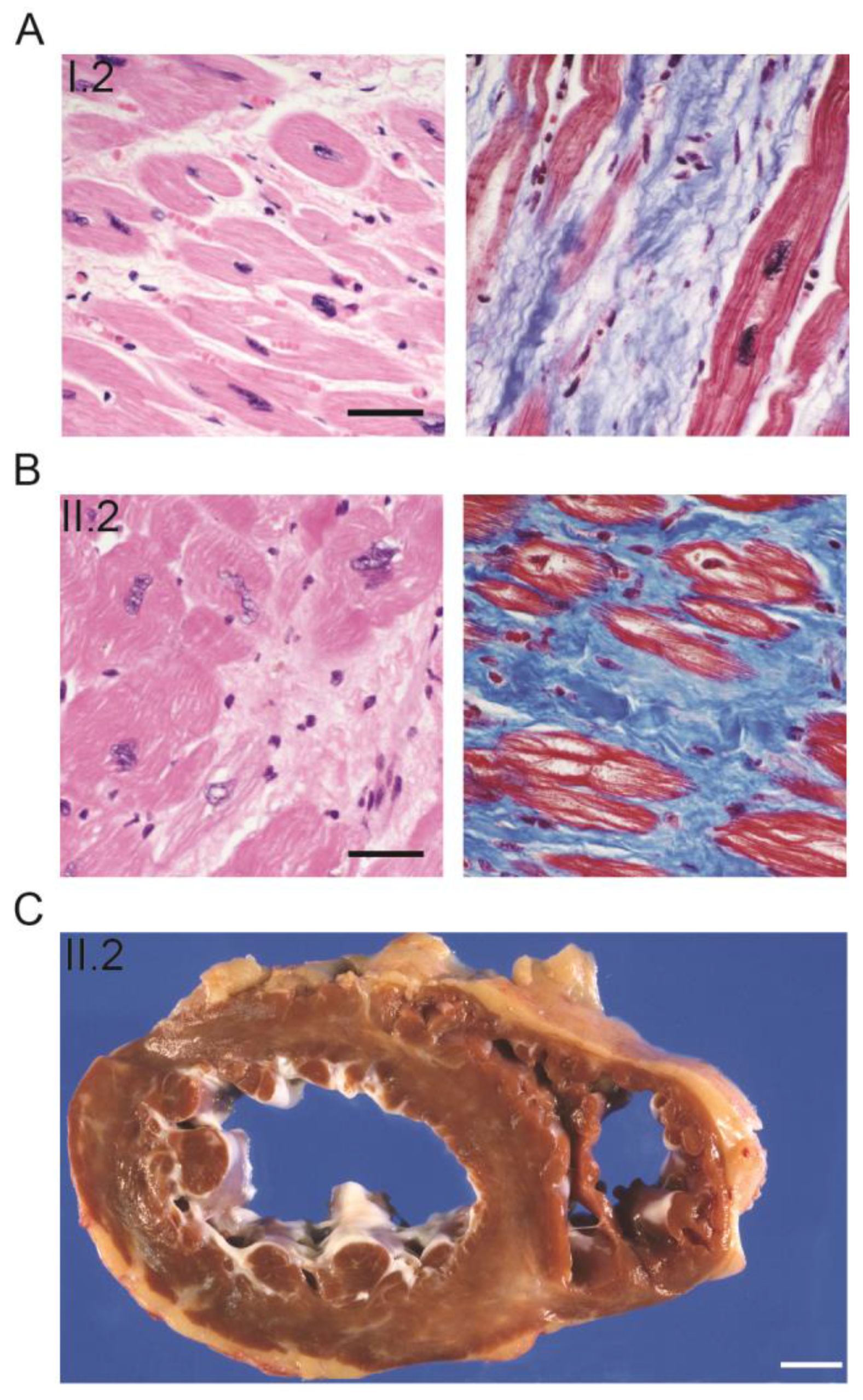
2.1. Clinical Features of a Multigenerational Family with Cardiac and Skeletal Muscle Defects
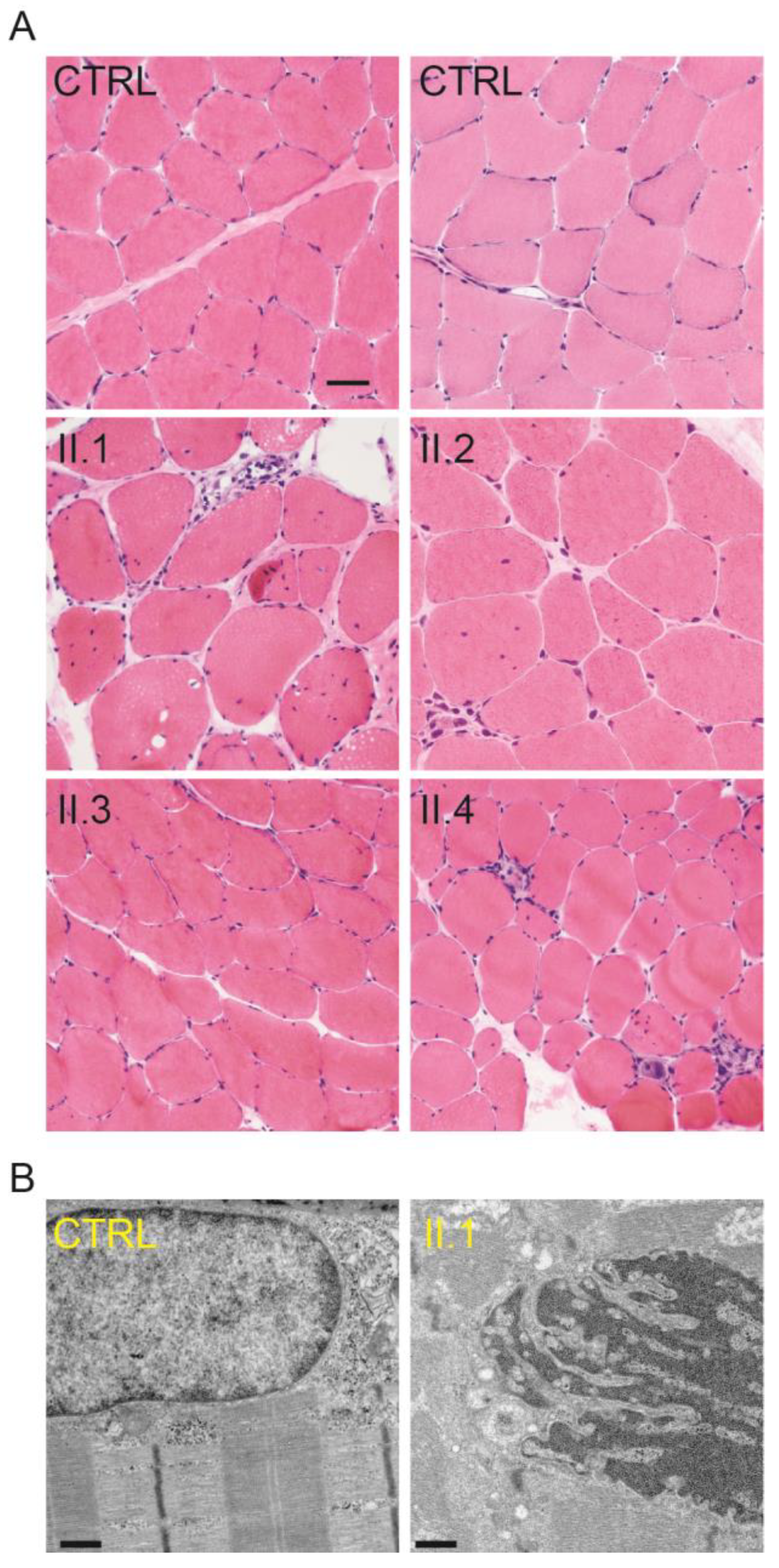
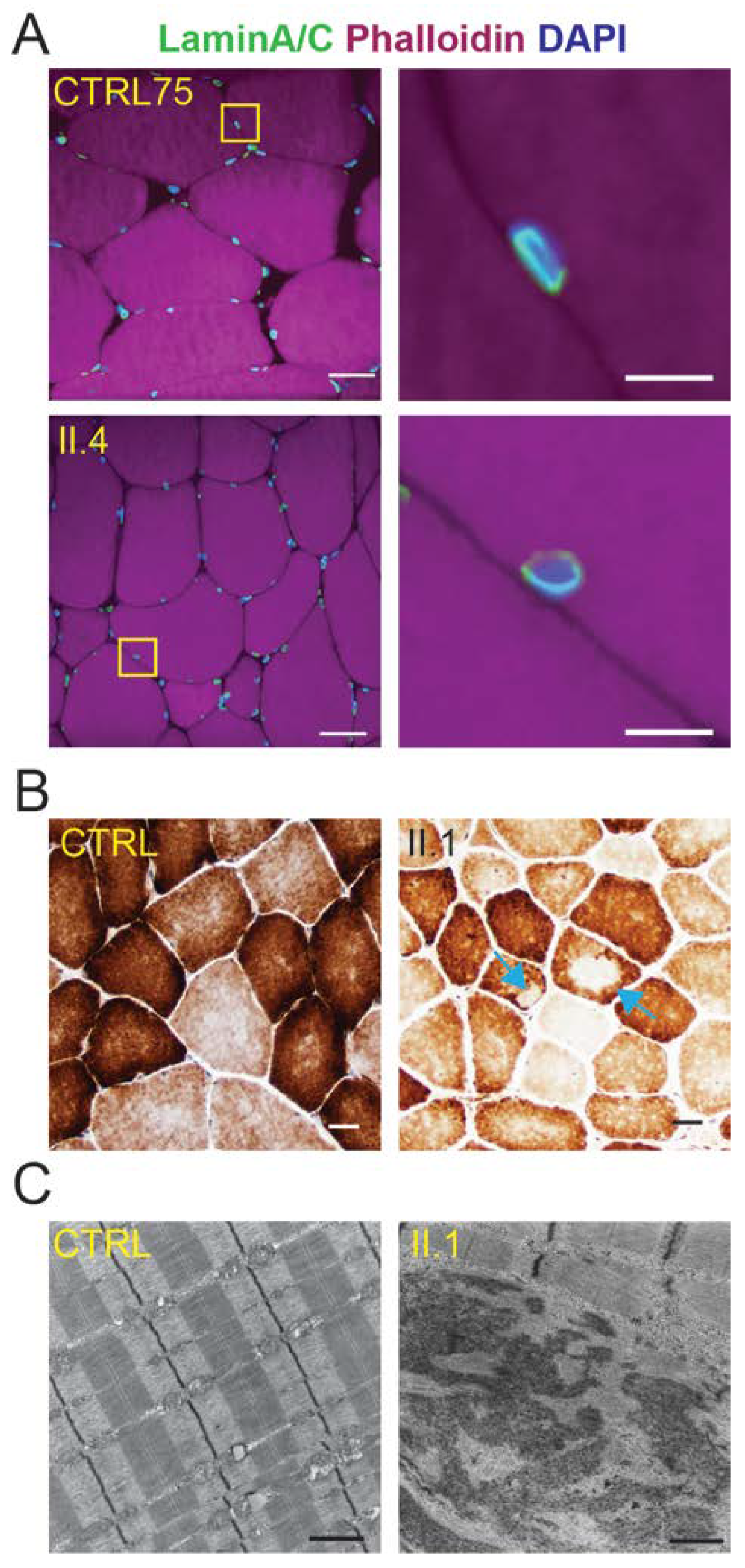
2.2. Skeletal Muscle Pathology of the Four Generation II Siblings Contains Dystrophic Features
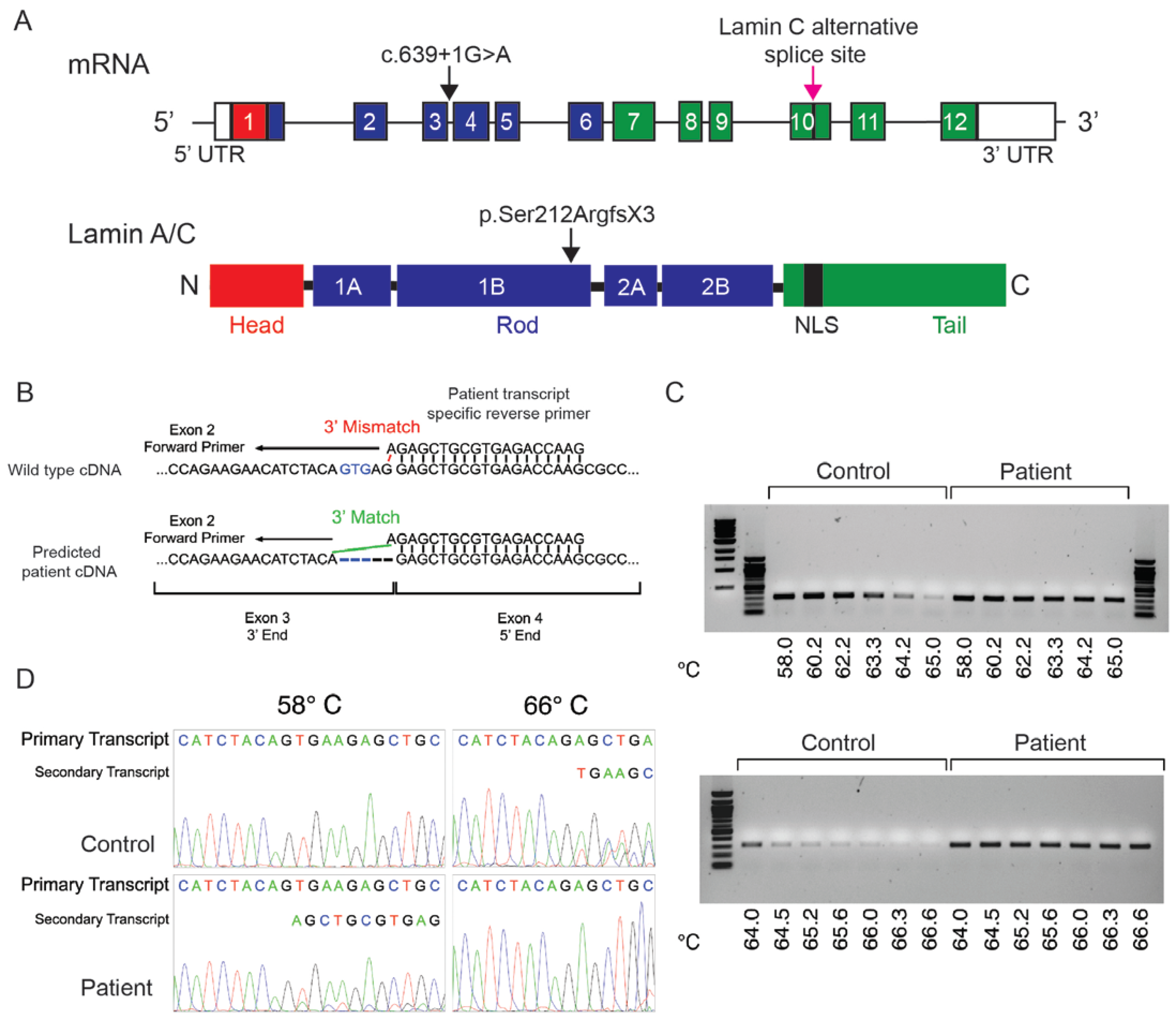
2.3. A Novel LMNA Variant Was Identified as Causal of These Phenotypes
2.4. Muscle Cores Are Present in a Subset of the LGMD1B-Affected Individuals
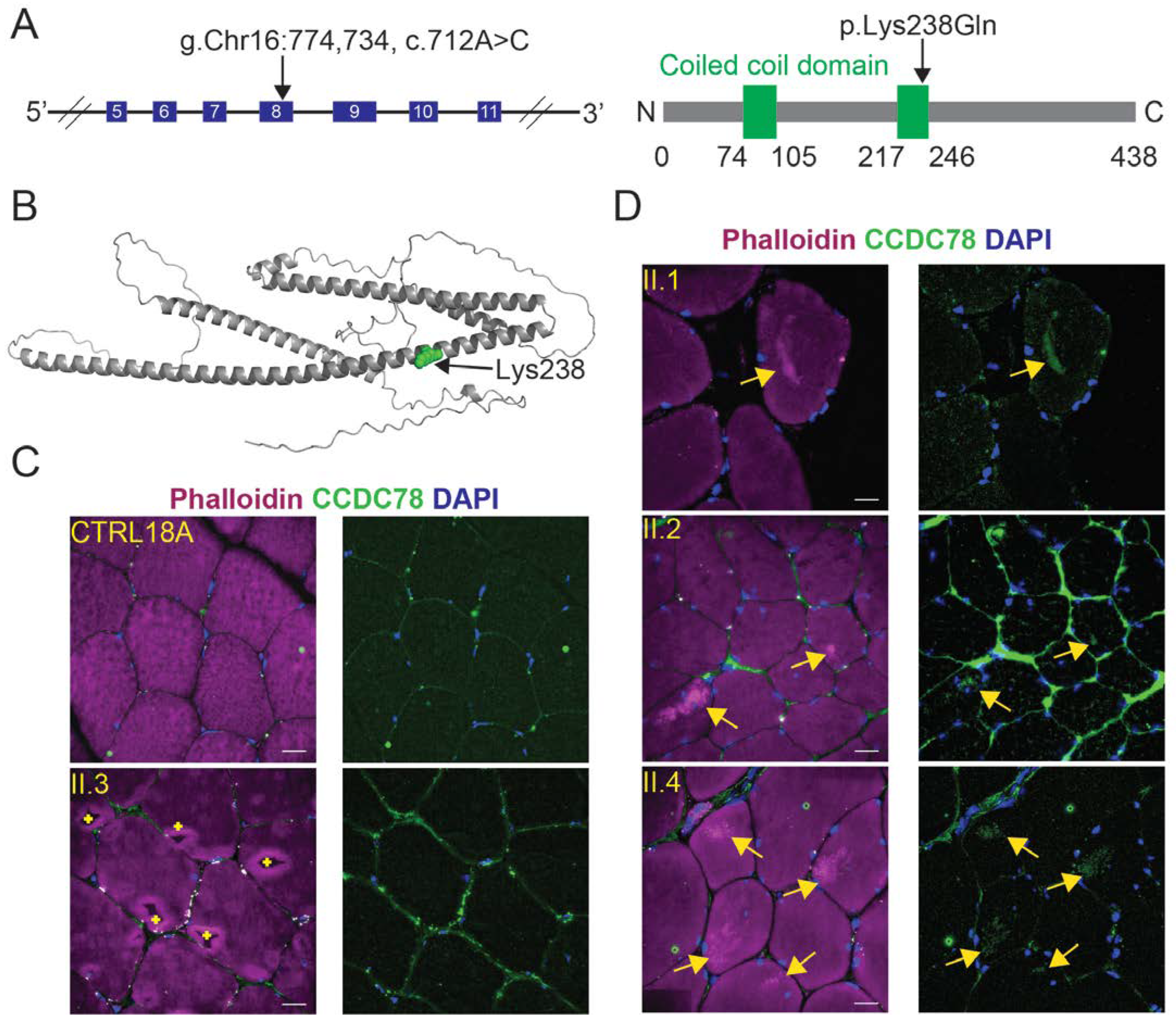
2.5. A Relatively Common Variant in CCDC78 Was Identified in Family Members with Muscle Cores
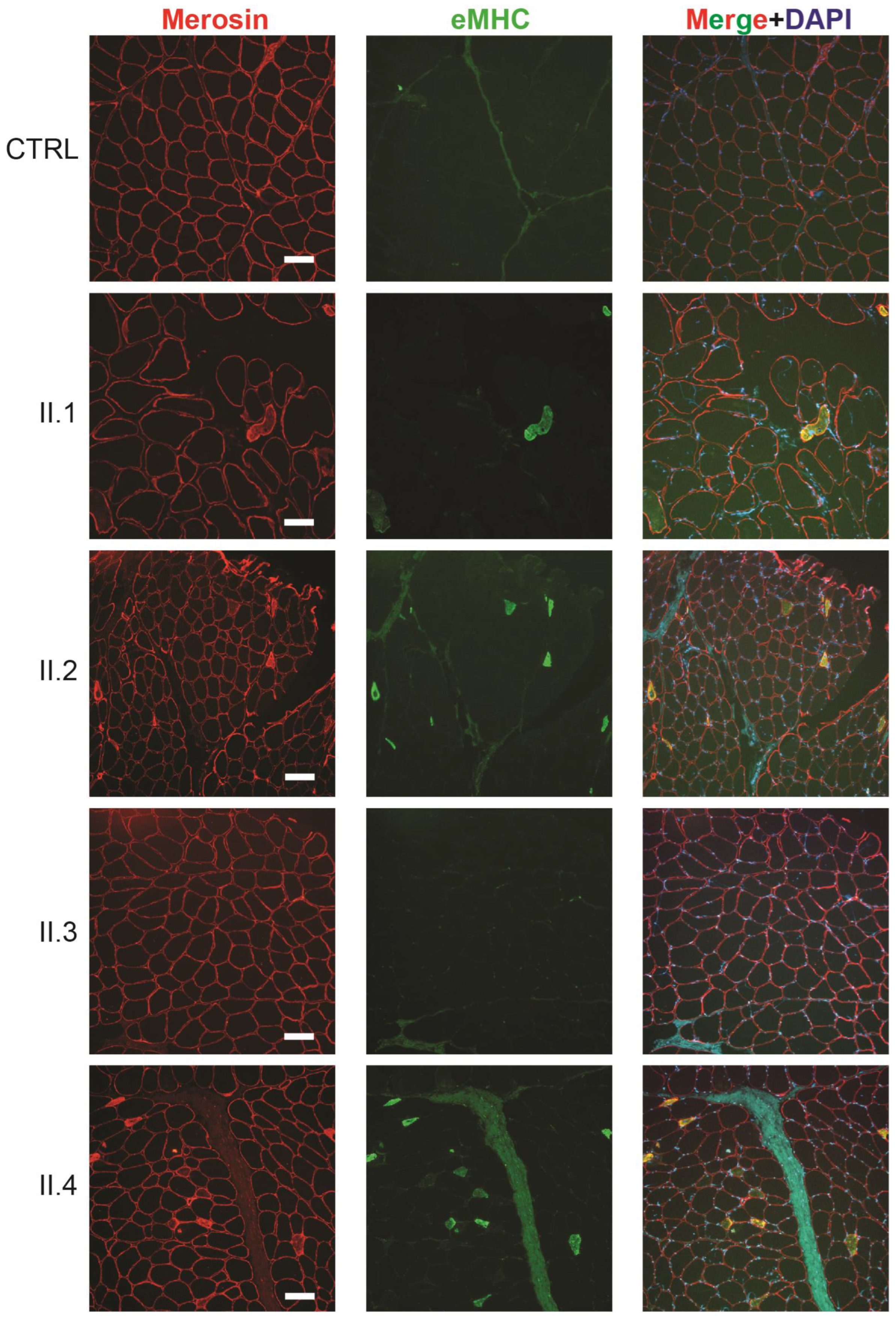
2.6. Individuals with the CCDC78 Variant Show Mislocalization of CCDC78
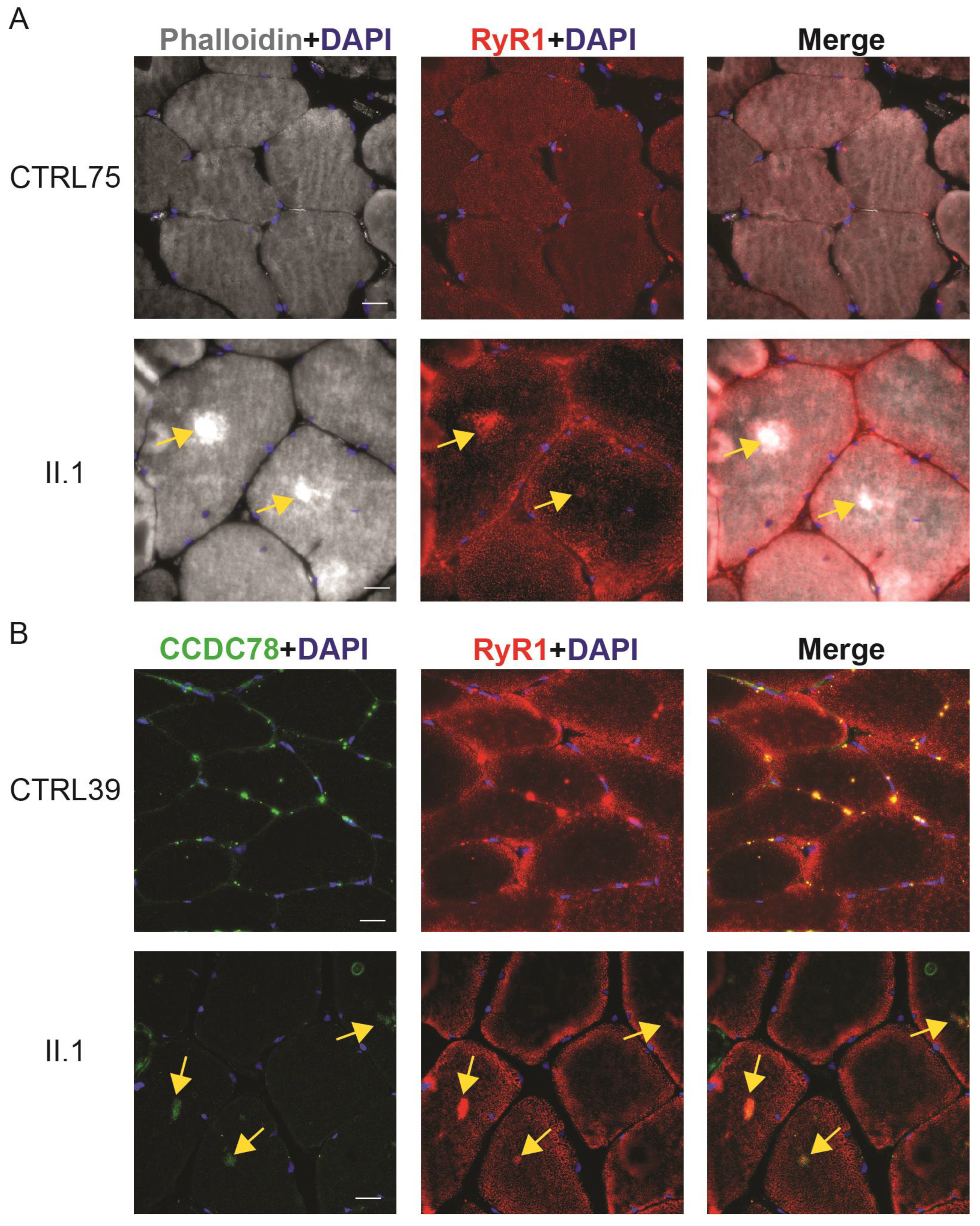
2.7. CCDC78 Aggregates Colocalize with RYR1 Aggregates in Muscle Cores
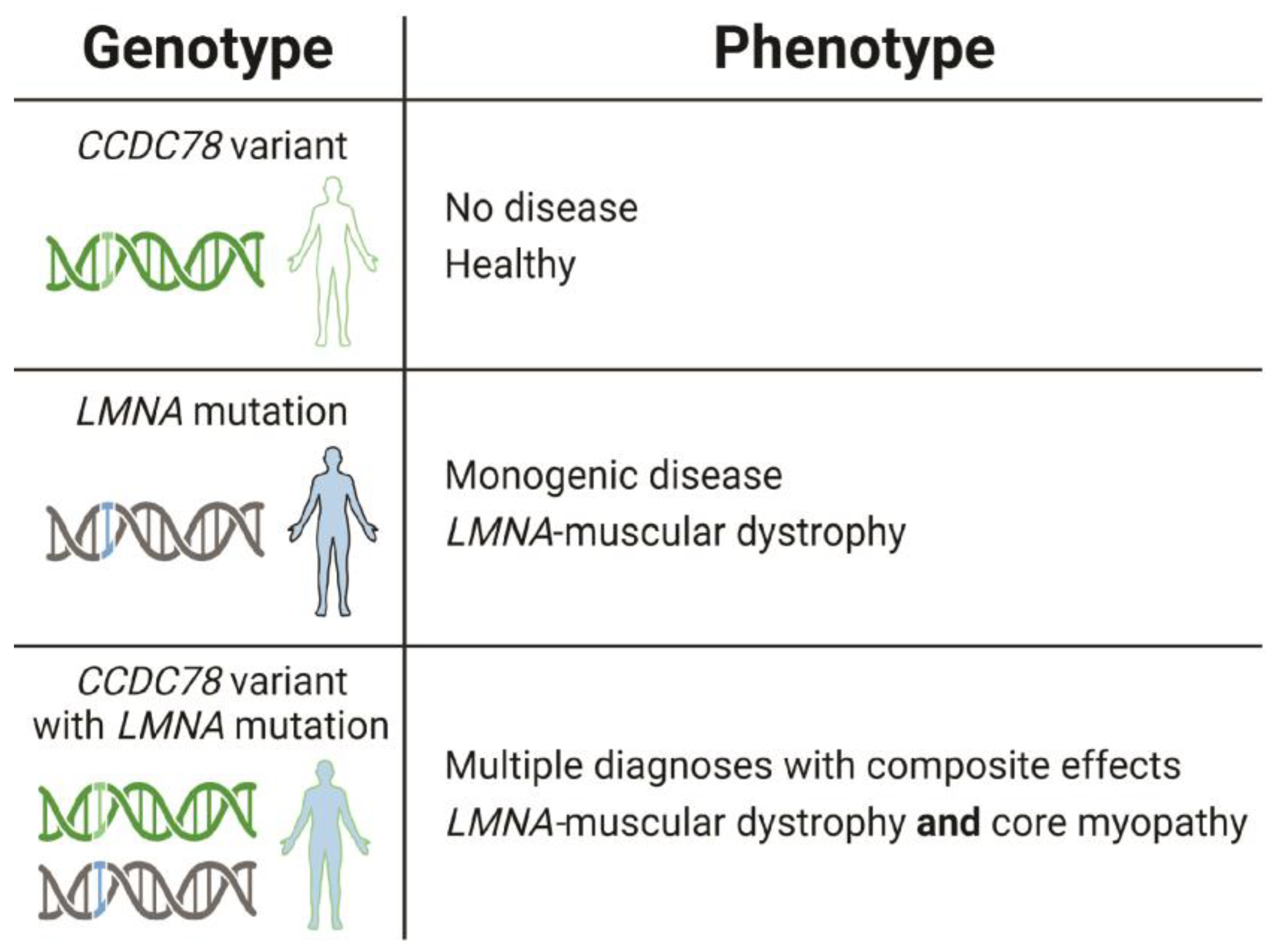
3. Discussion
4. Materials and Methods
4.1. Patient Recruitment
4.2. Tissue Collection and Analysis
4.3. Identification of the LMNA Variant
4.4. In Silico Transcript Analysis
4.5. RNA Isolation and RT-PCR Analysis
4.6. Whole Genome Sequencing and Bioinformatic Analysis
4.7. Immunostaining of Skeletal Muscle Tissue
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahn, J.; Jo, I.; Kang, S.M.; Hong, S.; Kim, S.; Jeong, S.; Kim, Y.H.; Park, B.J.; Ha, N.C. Structural basis for lamin assembly at the molecular level. Nat. Commun. 2019, 10, 3757. [Google Scholar] [CrossRef]
- Tenga, R.; Medalia, O. Structure and unique mechanical aspects of nuclear lamin filaments. Curr. Opin. Struct. Biol. 2020, 64, 152–159. [Google Scholar] [CrossRef]
- Turgay, Y.; Medalia, O. The structure of lamin filaments in somatic cells as revealed by cryo-electron tomography. Nucleus 2017, 8, 475–481. [Google Scholar] [CrossRef]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Margalit, A.; Goldman, R.D.; Shumaker, D.K.; Wilson, K.L. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 2005, 6, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Sobo, J.M.; Alagna, N.S.; Sun, S.X.; Wilson, K.L.; Reddy, K.L. Lamins: The backbone of the nucleocytoskeleton interface. Curr. Opin. Cell Biol. 2024, 86, 102313. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Worman, H.J. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993, 268, 16321–16326. [Google Scholar] [CrossRef]
- Lin, F.; Worman, H.J. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics 1995, 27, 230–236. [Google Scholar] [CrossRef]
- Wydner, K.L.; McNeil, J.A.; Lin, F.; Worman, H.J.; Lawrence, J.B. Chromosomal assignment of human nuclear envelope protein genes LMNA, LMNB1, and LBR by fluorescence in situ hybridization. Genomics 1996, 32, 474–478. [Google Scholar] [CrossRef]
- Worman, H.J. Nuclear lamins and laminopathies. J. Pathol. 2012, 226, 316–325. [Google Scholar] [CrossRef]
- Worman, H.J.; Courvalin, J.C. How do mutations in lamins A and C cause disease? J. Clin. Investig. 2004, 113, 349–351. [Google Scholar] [CrossRef]
- Worman, H.J.; Ostlund, C.; Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000760. [Google Scholar] [CrossRef]
- Crasto, S.; My, I.; Di Pasquale, E. The Broad Spectrum of LMNA Cardiac Diseases: From Molecular Mechanisms to Clinical Phenotype. Front. Physiol. 2020, 11, 761. [Google Scholar] [CrossRef]
- Lu, J.T.; Muchir, A.; Nagy, P.L.; Worman, H.J. LMNA cardiomyopathy: Cell biology and genetics meet clinical medicine. Dis. Model. Mech. 2011, 4, 562–568. [Google Scholar] [CrossRef]
- Worman, H.J.; Bonne, G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007, 313, 2121–2133. [Google Scholar] [CrossRef]
- Hinz, B.E.; Walker, S.G.; Xiong, A.; Gogal, R.A.; Schnieders, M.J.; Wallrath, L.L. In Silico and In Vivo Analysis of Amino Acid Substitutions That Cause Laminopathies. Int. J. Mol. Sci. 2021, 22, 11226. [Google Scholar] [CrossRef]
- Ben Yaou, R.; Yun, P.; Dabaj, I.; Norato, G.; Donkervoort, S.; Xiong, H.; Nascimento, A.; Maggi, L.; Sarkozy, A.; Monges, S.; et al. International retrospective natural history study of LMNA-related congenital muscular dystrophy. Brain Commun. 2021, 3, fcab075. [Google Scholar] [CrossRef]
- Mounkes, L.; Kozlov, S.; Burke, B.; Stewart, C.L. The laminopathies: Nuclear structure meets disease. Curr. Opin. Genet. Dev. 2003, 13, 223–230. [Google Scholar] [CrossRef]
- Cao, H.; Hegele, R.A. LMNA is mutated in Hutchinson-Gilford progeria (MIM 176670) but not in Wiedemann-Rautenstrauch progeroid syndrome (MIM 264090). J. Hum. Genet. 2003, 48, 271–274. [Google Scholar] [CrossRef]
- De Sandre-Giovannoli, A.; Bernard, R.; Cau, P.; Navarro, C.; Amiel, J.; Boccaccio, I.; Lyonnet, S.; Stewart, C.L.; Munnich, A.; Le Merrer, M.; et al. Lamin a truncation in Hutchinson-Gilford progeria. Science 2003, 300, 2055. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.Y.; Worman, H.J. Molecular Pathology of Laminopathies. Annu. Rev. Pathol. 2022, 17, 159–180. [Google Scholar] [CrossRef]
- Bonne, G.; Di Barletta, M.R.; Varnous, S.; Becane, H.M.; Hammouda, E.H.; Merlini, L.; Muntoni, F.; Greenberg, C.R.; Gary, F.; Urtizberea, J.A.; et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999, 21, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Carboni, N.; Mateddu, A.; Marrosu, G.; Cocco, E.; Marrosu, M.G. Genetic and clinical characteristics of skeletal and cardiac muscle in patients with lamin A/C gene mutations. Muscle Nerve 2013, 48, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Bonne, G.; van der Kooi, A.J.; van Meegen, M.; Baas, F.; Bolhuis, P.A.; de Visser, M.; Schwartz, K. Identification of mutations in the gene encoding lamins A/C in autosomal dominant limb girdle muscular dystrophy with atrioventricular conduction disturbances (LGMD1B). Hum. Mol. Genet. 2000, 9, 1453–1459. [Google Scholar] [CrossRef] [PubMed]
- Wicklund, M.P.; Kissel, J.T. The limb-girdle muscular dystrophies. Neurol. Clin. 2014, 32, 729–749, ix. [Google Scholar] [CrossRef]
- Mathews, K.D.; Moore, S.A. Limb-girdle muscular dystrophy. Curr. Neurol. Neurosci. Rep. 2003, 3, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Angelini, C. LGMD. Identification, description and classification. Acta Myol. 2020, 39, 207–217. [Google Scholar] [CrossRef]
- Groh, W.J. Arrhythmias in the muscular dystrophies. Heart Rhythm. 2012, 9, 1890–1895. [Google Scholar] [CrossRef]
- Petillo, R.; D’Ambrosio, P.; Torella, A.; Taglia, A.; Picillo, E.; Testori, A.; Ergoli, M.; Nigro, G.; Piluso, G.; Nigro, V.; et al. Novel mutations in LMNA A/C gene and associated phenotypes. Acta Myol. 2015, 34, 116–119. [Google Scholar] [PubMed]
- Brodsky, G.L.; Muntoni, F.; Miocic, S.; Sinagra, G.; Sewry, C.; Mestroni, L. Lamin A/C gene mutation associated with dilated cardiomyopathy with variable skeletal muscle involvement. Circulation 2000, 101, 473–476. [Google Scholar] [CrossRef]
- Carboni, N.; Mura, M.; Marrosu, G.; Cocco, E.; Ahmad, M.; Solla, E.; Mateddu, A.; Maioli, M.A.; Marini, S.; Nissardi, V.; et al. Muscle MRI findings in patients with an apparently exclusive cardiac phenotype due to a novel LMNA gene mutation. Neuromuscul. Disord. 2008, 18, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.L.; Langer, E.R.; Routes, T.C.; McWilliams, S.F.; Bereslavskyy, I.; Kamp, T.J.; Eckhardt, L.L. Most myopathic lamin variants aggregate: A functional genomics approach for assessing variants of uncertain significance. NPJ Genom. Med. 2021, 6, 103. [Google Scholar] [CrossRef]
- Lazarte, J.; Jurgens, S.J.; Choi, S.H.; Khurshid, S.; Morrill, V.N.; Weng, L.C.; Nauffal, V.; Pirruccello, J.P.; Halford, J.L.; Hegele, R.A.; et al. LMNA Variants and Risk of Adult-Onset Cardiac Disease. J. Am. Coll. Cardiol. 2022, 80, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.W.; Brady, G.F.; Kwan, R.; Nesvizhskii, A.I.; Omary, M.B. Genotype-phenotype analysis of LMNA-related diseases predicts phenotype-selective alterations in lamin phosphorylation. FASEB J. 2020, 34, 9051–9073. [Google Scholar] [CrossRef]
- Scharner, J.; Lu, H.C.; Fraternali, F.; Ellis, J.A.; Zammit, P.S. Mapping disease-related missense mutations in the immunoglobulin-like fold domain of lamin A/C reveals novel genotype-phenotype associations for laminopathies. Proteins 2014, 82, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Storey, E.C.; Fuller, H.R. Genotype-Phenotype Correlations in Human Diseases Caused by Mutations of LINC Complex-Associated Genes: A Systematic Review and Meta-Summary. Cells 2022, 11, 4065. [Google Scholar] [CrossRef]
- Bonne, G.; Mercuri, E.; Muchir, A.; Urtizberea, A.; Becane, H.M.; Recan, D.; Merlini, L.; Wehnert, M.; Boor, R.; Reuner, U.; et al. Clinical and molecular genetic spectrum of autosomal dominant Emery-Dreifuss muscular dystrophy due to mutations of the lamin A/C gene. Ann. Neurol. 2000, 48, 170–180. [Google Scholar] [CrossRef] [PubMed]
- Mercuri, E.; Poppe, M.; Quinlivan, R.; Messina, S.; Kinali, M.; Demay, L.; Bourke, J.; Richard, P.; Sewry, C.; Pike, M.; et al. Extreme variability of phenotype in patients with an identical missense mutation in the lamin A/C gene: From congenital onset with severe phenotype to milder classic Emery-Dreifuss variant. Arch. Neurol. 2004, 61, 690–694. [Google Scholar] [CrossRef]
- Rankin, J.; Auer-Grumbach, M.; Bagg, W.; Colclough, K.; Nguyen, T.D.; Fenton-May, J.; Hattersley, A.; Hudson, J.; Jardine, P.; Josifova, D.; et al. Extreme phenotypic diversity and nonpenetrance in families with the LMNA gene mutation R644C. Am. J. Med. Genet. Part A 2008, 146A, 1530–1542. [Google Scholar] [CrossRef] [PubMed]
- Cesar, S.; Coll, M.; Fiol, V.; Fernandez-Falgueras, A.; Cruzalegui, J.; Iglesias, A.; Moll, I.; Perez-Serra, A.; Martinez-Barrios, E.; Ferrer-Costa, C.; et al. LMNA-related muscular dystrophy: Identification of variants in alternative genes and personalized clinical translation. Front. Genet. 2023, 14, 1135438. [Google Scholar] [CrossRef] [PubMed]
- Meinke, P.; Mattioli, E.; Haque, F.; Antoku, S.; Columbaro, M.; Straatman, K.R.; Worman, H.J.; Gundersen, G.G.; Lattanzi, G.; Wehnert, M.; et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014, 10, e1004605. [Google Scholar] [CrossRef] [PubMed]
- Granger, B.; Gueneau, L.; Drouin-Garraud, V.; Pedergnana, V.; Gagnon, F.; Ben Yaou, R.; Tezenas du Montcel, S.; Bonne, G. Modifier locus of the skeletal muscle involvement in Emery-Dreifuss muscular dystrophy. Hum. Genet. 2011, 129, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Lamar, K.M.; McNally, E.M. Genetic Modifiers for Neuromuscular Diseases. J. Neuromuscul. Dis. 2014, 1, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Rahit, K.; Tarailo-Graovac, M. Genetic Modifiers and Rare Mendelian Disease. Genes 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Blauwendraat, C.; Reed, X.; Krohn, L.; Heilbron, K.; Bandres-Ciga, S.; Tan, M.; Gibbs, J.R.; Hernandez, D.G.; Kumaran, R.; Langston, R.; et al. Genetic modifiers of risk and age at onset in GBA associated Parkinson’s disease and Lewy body dementia. Brain 2020, 143, 234–248. [Google Scholar] [CrossRef]
- Ryan, N.M.; Lihm, J.; Kramer, M.; McCarthy, S.; Morris, S.W.; Arnau-Soler, A.; Davies, G.; Duff, B.; Ghiban, E.; Hayward, C.; et al. DNA sequence-level analyses reveal potential phenotypic modifiers in a large family with psychiatric disorders. Mol. Psychiatry 2018, 23, 2254–2265. [Google Scholar] [CrossRef]
- Wilk, M.A.; Braun, A.T.; Farrell, P.M.; Laxova, A.; Brown, D.M.; Holt, J.M.; Birch, C.L.; Sosonkina, N.; Wilk, B.M.; Worthey, E.A. Applying whole-genome sequencing in relation to phenotype and outcomes in siblings with cystic fibrosis. Mol. Case Stud. 2020, 6, a004531. [Google Scholar] [CrossRef]
- Ogasawara, M.; Nishino, I. A review of core myopathy: Central core disease, multiminicore disease, dusty core disease, and core-rod myopathy. Neuromuscul. Disord. 2021, 31, 968–977. [Google Scholar] [CrossRef]
- Topaloglu, H. Core myopathies—A short review. Acta Myol. 2020, 39, 266–273. [Google Scholar] [CrossRef]
- Mathews, K.D.; Moore, S.A. Multiminicore myopathy, central core disease, malignant hyperthermia susceptibility, and RYR1 mutations: One disease with many faces? Arch. Neurol. 2004, 61, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.A.; Shilling, C.J.; Westra, S.; Wall, C.; Wicklund, M.P.; Stolle, C.; Brown, C.A.; Michele, D.E.; Piccolo, F.; Winder, T.L.; et al. Limb-girdle muscular dystrophy in the United States. J. Neuropathol. Exp. Neurol. 2006, 65, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- van Rijsingen, I.A.; Nannenberg, E.A.; Arbustini, E.; Elliott, P.M.; Mogensen, J.; Hermans-van Ast, J.F.; van der Kooi, A.J.; van Tintelen, J.P.; van den Berg, M.P.; Grasso, M.; et al. Gender-specific differences in major cardiac events and mortality in lamin A/C mutation carriers. Eur. J. Hear. Fail. 2013, 15, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Darbro, B.; Cox, E.M.; Stence, A.A.; Cabuay, B.M.; Mathews, K.D.; Nagy, P.L.; Moore, S.A. In silico and experimental analysis of a novel intronic LMNA DNA variation implicated in Limb Girdle Muscular Dystrophy Type 1B and Dilated Cardiomyopathy. In Proceedings of the 60th Annual Meeting of the American Society of Human Genetics, Washington, DC, USA, 2–6 November 2010. Abstract 847F. [Google Scholar]
- Hartmann, L.; Theiss, S.; Niederacher, D.; Schaal, H. Diagnostics of pathogenic splicing mutations: Does bioinformatics cover all bases? Front. Biosci. 2008, 13, 3252–3272. [Google Scholar] [CrossRef] [PubMed]
- Joyce, N.C.; Oskarsson, B.; Jin, L.W. Muscle biopsy evaluation in neuromuscular disorders. Phys. Med. Rehabil. Clin. N. Am. 2012, 23, 609–631. [Google Scholar] [CrossRef] [PubMed]
- Nix, J.S.; Moore, S.A. What Every Neuropathologist Needs to Know: The Muscle Biopsy. J. Neuropathol. Exp. Neurol. 2020, 79, 719–733. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Miller, R.G.; Brownell, A.K. Central core disease: Ultrastructure of the sarcoplasmic reticulum and T-tubules. Muscle Nerve 1989, 12, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Palmucci, L.; Schiffer, D.; Monga, G.; Mollo, F.; de Marchi, M. Central core disease: Histochemical and ultrastructural study of muscle biopsies of father and daughter. J. Neurol. 1978, 218, 55–62. [Google Scholar] [CrossRef]
- Lawal, T.A.; Todd, J.J.; Meilleur, K.G. Ryanodine Receptor 1-Related Myopathies: Diagnostic and Therapeutic Approaches. Neurotherapeutics 2018, 15, 885–899. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; Program, N.C.S.; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Rodelsperger, C.; Schuelke, M.; Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 2010, 7, 575–576. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.; Edwards, K.J.; Day, I.N.; Gaunt, T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Shihab, H.A.; Rogers, M.F.; Gough, J.; Mort, M.; Cooper, D.N.; Day, I.N.; Gaunt, T.R.; Campbell, C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics 2015, 31, 1536–1543. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Singleton, M.V.; Guthery, S.L.; Voelkerding, K.V.; Chen, K.; Kennedy, B.; Margraf, R.L.; Durtschi, J.; Eilbeck, K.; Reese, M.G.; Jorde, L.B.; et al. Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am. J. Hum. Genet. 2014, 94, 599–610. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Majczenko, K.; Davidson, A.E.; Camelo-Piragua, S.; Agrawal, P.B.; Manfready, R.A.; Li, X.; Joshi, S.; Xu, J.; Peng, W.; Beggs, A.H.; et al. Dominant mutation of CCDC78 in a unique congenital myopathy with prominent internal nuclei and atypical cores. Am. J. Hum. Genet. 2012, 91, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Katsuragi, Y.; Ichimura, Y.; Komatsu, M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015, 282, 4672–4678. [Google Scholar] [CrossRef]
- Jeong, S.J.; Zhang, X.; Rodriguez-Velez, A.; Evans, T.D.; Razani, B. p62/SQSTM1 and Selective Autophagy in Cardiometabolic Diseases. Antioxid. Redox Signal. 2019, 31, 458–471. [Google Scholar] [CrossRef]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays Biochem. 2017, 61, 609–624. [Google Scholar] [CrossRef]
- Sedaghat-Hamedani, F.; Rebs, S.; Kayvanpour, E.; Zhu, C.; Amr, A.; Muller, M.; Haas, J.; Wu, J.; Steinmetz, L.M.; Ehlermann, P.; et al. Genotype Complements the Phenotype: Identification of the Pathogenicity of an LMNA Splice Variant by Nanopore Long-Read Sequencing in a Large DCM Family. Int. J. Mol. Sci. 2022, 23, 12230. [Google Scholar] [CrossRef]
- Rogozhina, Y.; Mironovich, S.; Shestak, A.; Adyan, T.; Polyakov, A.; Podolyak, D.; Bakulina, A.; Dzemeshkevich, S.; Zaklyazminskaya, E. New intronic splicing mutation in the LMNA gene causing progressive cardiac conduction defects and variable myopathy. Gene 2016, 595, 202–206. [Google Scholar] [CrossRef]
- Widyastuti, H.P.; Norden-Krichmar, T.M.; Grosberg, A.; Zaragoza, M.V. Gene expression profiling of fibroblasts in a family with LMNA-related cardiomyopathy reveals molecular pathways implicated in disease pathogenesis. BMC Med. Genet. 2020, 21, 152. [Google Scholar] [CrossRef]
- Zaragoza, M.V.; Fung, L.; Jensen, E.; Oh, F.; Cung, K.; McCarthy, L.A.; Tran, C.K.; Hoang, V.; Hakim, S.A.; Grosberg, A. Exome Sequencing Identifies a Novel LMNA Splice-Site Mutation and Multigenic Heterozygosity of Potential Modifiers in a Family with Sick Sinus Syndrome, Dilated Cardiomyopathy, and Sudden Cardiac Death. PLoS ONE 2016, 11, e0155421. [Google Scholar] [CrossRef]
- Ito, K.; Patel, P.N.; Gorham, J.M.; McDonough, B.; DePalma, S.R.; Adler, E.E.; Lam, L.; MacRae, C.A.; Mohiuddin, S.M.; Fatkin, D.; et al. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc. Natl. Acad. Sci. USA 2017, 114, 7689–7694. [Google Scholar] [CrossRef]
- Al-Saaidi, R.; Rasmussen, T.B.; Palmfeldt, J.; Nissen, P.H.; Beqqali, A.; Hansen, J.; Pinto, Y.M.; Boesen, T.; Mogensen, J.; Bross, P. The LMNA mutation p.Arg321Ter associated with dilated cardiomyopathy leads to reduced expression and a skewed ratio of lamin A and lamin C proteins. Exp. Cell Res. 2013, 319, 3010–3019. [Google Scholar] [CrossRef]
- Carmosino, M.; Gerbino, A.; Schena, G.; Procino, G.; Miglionico, R.; Forleo, C.; Favale, S.; Svelto, M. The expression of Lamin A mutant R321X leads to endoplasmic reticulum stress with aberrant Ca(2+) handling. J. Cell. Mol. Med. 2016, 20, 2194–2207. [Google Scholar] [CrossRef]
- Geiger, S.K.; Bar, H.; Ehlermann, P.; Walde, S.; Rutschow, D.; Zeller, R.; Ivandic, B.T.; Zentgraf, H.; Katus, H.A.; Herrmann, H.; et al. Incomplete nonsense-mediated decay of mutant lamin A/C mRNA provokes dilated cardiomyopathy and ventricular tachycardia. J. Mol. Med. 2008, 86, 281–289. [Google Scholar] [CrossRef]
- Kato, K.; Ohno, S.; Sonoda, K.; Fukuyama, M.; Makiyama, T.; Ozawa, T.; Horie, M. LMNA Missense Mutation Causes Nonsense-Mediated mRNA Decay and Severe Dilated Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, 435–443. [Google Scholar] [CrossRef]
- Tao, J.; Duan, J.; Pi, X.; Wang, H.; Li, S. A splicing LMNA mutation causing laminopathies accompanied by aortic valve malformation. J. Clin. Lab. Anal. 2021, 35, e23736. [Google Scholar] [CrossRef] [PubMed]
- Shaw, N.M.; Rios-Monterrosa, J.L.; Fedorchak, G.R.; Ketterer, M.R.; Coombs, G.S.; Lammerding, J.; Wallrath, L.L. Effects of mutant lamins on nucleo-cytoskeletal coupling in Drosophila models of LMNA muscular dystrophy. Front. Cell Dev. Biol. 2022, 10, 934586. [Google Scholar] [CrossRef]
- Dialynas, G.; Shrestha, O.K.; Ponce, J.M.; Zwerger, M.; Thiemann, D.A.; Young, G.H.; Moore, S.A.; Yu, L.; Lammerding, J.; Wallrath, L.L. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 2015, 11, e1005231. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Flannery, K.M.; Zirbel, L.N.; Nagy, P.L.; Mathews, K.D.; Moore, S.A.; Wallrath, L.L. LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle. Hum. Mol. Genet. 2012, 21, 1544–1556. [Google Scholar] [CrossRef]
- Coombs, G.S.; Rios-Monterrosa, J.L.; Lai, S.; Dai, Q.; Goll, A.C.; Ketterer, M.R.; Valdes, M.F.; Uche, N.; Benjamin, I.J.; Wallrath, L.L. Modulation of muscle redox and protein aggregation rescues lethality caused by mutant lamins. Redox Biol. 2021, 48, 102196. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.E.; Hayashi, Y.K.; Goto, K.; Komaki, H.; Hayashi, Y.; Inuzuka, T.; Noguchi, S.; Nonaka, I.; Nishino, I. Nuclear changes in skeletal muscle extend to satellite cells in autosomal dominant Emery-Dreifuss muscular dystrophy/limb-girdle muscular dystrophy 1B. Neuromuscul. Disord. 2009, 19, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.; Carpenter, D.; Shaw, M.A.; Halsall, J.; Hopkins, P. Mutations in RYR1 in malignant hyperthermia and central core disease. Hum. Mutat. 2006, 27, 977–989. [Google Scholar] [CrossRef]
- Dowling, J.J.; Lawlor, M.W.; Dirksen, R.T. Triadopathies: An emerging class of skeletal muscle diseases. Neurotherapeutics 2014, 11, 773–785. [Google Scholar] [CrossRef]
- Tang, T.K. Centriole biogenesis in multiciliated cells. Nat. Cell Biol. 2013, 15, 1400–1402. [Google Scholar] [CrossRef]
- Kim, S.K.; Brotslaw, E.; Thome, V.; Mitchell, J.; Ventrella, R.; Collins, C.; Mitchell, B. A role for Cep70 in centriole amplification in multiciliated cells. Dev. Biol. 2021, 471, 10–17. [Google Scholar] [CrossRef]
- Klos Dehring, D.A.; Vladar, E.K.; Werner, M.E.; Mitchell, J.W.; Hwang, P.; Mitchell, B.J. Deuterosome-mediated centriole biogenesis. Dev. Cell 2013, 27, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Truebestein, L.; Leonard, T.A. Coiled-coils: The long and short of it. Bioessays 2016, 38, 903–916. [Google Scholar] [CrossRef]
- Hartmann, M.D. Functional and Structural Roles of Coiled Coils. Fibrous Proteins Struct. Mech. 2017, 82, 63–93. [Google Scholar] [CrossRef]
- Brunak, S.; Engelbrecht, J.; Knudsen, S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991, 220, 49–65. [Google Scholar] [CrossRef]
- Buratti, E.; Chivers, M.; Kralovicova, J.; Romano, M.; Baralle, M.; Krainer, A.R.; Vorechovsky, I. Aberrant 5′ splice sites in human disease genes: Mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007, 35, 4250–4263. [Google Scholar] [CrossRef]
- Churbanov, A.; Rogozin, I.B.; Deogun, J.S.; Ali, H. Method of predicting splice sites based on signal interactions. Biol. Direct 2006, 1, 10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eng, L.; Coutinho, G.; Nahas, S.; Yeo, G.; Tanouye, R.; Babaei, M.; Dork, T.; Burge, C.; Gatti, R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: Maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004, 23, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Hebsgaard, S.M.; Korning, P.G.; Tolstrup, N.; Engelbrecht, J.; Rouze, P.; Brunak, S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996, 24, 3439–3452. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Lin, X.; Salzberg, S.L. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 2001, 29, 1185–1190. [Google Scholar] [CrossRef]
- Reese, M.G.; Eeckman, F.H.; Kulp, D.; Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 1997, 4, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, K.; Roca, X.; Beiglbock, H.; Callens, T.; Etzler, J.; Rao, A.R.; Krainer, A.R.; Fonatsch, C.; Messiaen, L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5′ splice-site disruption. Hum. Mutat. 2007, 28, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef] [PubMed]
- Burge, C.B.; Karlin, S. Finding the genes in genomic DNA. Curr. Opin. Struct. Biol. 1998, 8, 346–354. [Google Scholar] [CrossRef]
- Churbanov, A.; Vorechovsky, I.; Hicks, C. A method of predicting changes in human gene splicing induced by genetic variants in context of cis-acting elements. BMC Bioinform. 2010, 11, 22. [Google Scholar] [CrossRef]
- Fairbrother, W.G.; Yeh, R.F.; Sharp, P.A.; Burge, C.B. Predictive identification of exonic splicing enhancers in human genes. Science 2002, 297, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Holla, O.L.; Nakken, S.; Mattingsdal, M.; Ranheim, T.; Berge, K.E.; Defesche, J.C.; Leren, T.P. Effects of intronic mutations in the LDLR gene on pre-mRNA splicing: Comparison of wet-lab and bioinformatics analyses. Mol. Genet. Metab. 2009, 96, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Houdayer, C.; Dehainault, C.; Mattler, C.; Michaux, D.; Caux-Moncoutier, V.; Pages-Berhouet, S.; d’Enghien, C.D.; Lauge, A.; Castera, L.; Gauthier-Villars, M.; et al. Evaluation of in silico splice tools for decision-making in molecular diagnosis. Hum. Mutat. 2008, 29, 975–982. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]

| Patient ID (Age at Last Evaluation) | Sex | Age at Onset of Symptomatic Muscle Weakness (Years) | Age at Fulltime Wheelchair Current Motor Ability (Years) | Age at First Abnormal Echocardiogram (Years) | Age of ICD Placement (Years) | CK (U/L) 2 | Skeletal Muscle Pathology (Age at Biopsy; Years) | CCDC78 c.712A>C Variant |
|---|---|---|---|---|---|---|---|---|
| I.2 (deceased at age 31) | Female | N/A | N/A | 27 | Not Done | >600 | Not evaluated | Not tested 5 |
| II.1 (60 years) | Female | mid 20s | 59 | Normal echocardiogram at 59 | 50 | 617 | Mildly dystrophic with cores (34) | Present |
| II.2 (62 years) | Male | 45 | Able to walk short distances at 60 | On initial evaluation at 41 1 | Not Done 1 | 1000 | Mildly dystrophic with cores (29) 3 | Present |
| II.3 (60 years, deceased at age 60) | Male | 50–55 | Ambulatory at age 60 with nearly normal strength | 47 4 | 48 | 281 | Type II fiber predominance, rare necrotic fibers (47) 3 | Absent |
| II.4 (65 years) | Female | ~30 | 60 | 68 | 58 | 610 | Mildly dystrophic with cores (31) | Present |
| III.1 (28 years) | Male | N/A | No weakness at 28 | Normal cMRI at 28 | 28, for symptomatic arrhythmia | N/A | No biopsy | Not tested |
| III.2 (32 years) | Male | N/A | No weakness at 32 | Normal echocardiogram at 32 | N/A | N/A | No biopsy | Not tested |
| III.5 (30 years) | Male | N/A | No weakness at 31 | 29 | 30 | 288 | No biopsy | Present |
| III.6 (38 years) | Male | N/A | No weakness at 38 | Normal cMRI at 38 | N/A | 813 | No biopsy | Not tested |
| Gene Symbol | Mutation Type | Transcript Consequence | Protein Consequence | gnmoAD Allele Frequency 1 |
|---|---|---|---|---|
| CCDC78 | Missense | c.712A>C | p.Lys238Gln | 1.11 × 0−2 |
| VPS13A | Splice region | c.385+6_385+15delGAAAACAGTA | Potential splicing effect | 5.39 × 10−3 |
| AXIN1 | Missense | c.1948G>A | p.Gly650Ser | 1.87 × 10−2 |
| TMC4 | Stop retained | c.2120G>A | p.Ter707= | 1.58 × 10−2 |
| PTPRD | Missense | c.2341A>G | p.Thr781Ala | 2.74 × 10−2 |
| TDO2 | Missense | c.685A>C | p.Asn229His | 3.87 × 10−2 |
| SPIN1 | Splice region | c.590-8delT | Potential splicing effect | 2.39 × 10−1 |
| SPIN1 | Splice region | c.590-9_590-8delTT | Potential splicing effect | 1.26 × 10−1 |
| ZNF343 | Missense | c.1373C>T | p.Thr458Met | 5.65 × 10−3 |
| WDR90 | Splice region | c.1380-8G>A | Potential splicing effect | 3.36 × 10−2 |
| FNIP2 | Missense | c.1615G>C | p.Gly539Arg | 1.02 × 10−2 |
| WNK2 | In-frame deletion | c.4190_4204delATGGAGCAGCTCCAG | p.Asp1397_Pro1401del | 2.56 × 10−4 |
| KIF27 | Missense | c.2579G>A | p.Arg860Gln | 3.50 × 10−3 |
| RNF151 | Splice region | c.149+8C>T | Potential splicing effect | 4.71 × 10−3 |
| ZNF75A | Premature start codon gain | c.540C>T | p.Tyr180= | 1.16 × 10−2 |
| RPL3L | Missense | c.784G>A | p.Val262Met | 2.86 × 10−2 |
| SIRPD | Missense | c.206G>A | p.Gly69Glu | 9.42 × 10−5 |
| HEXD | Missense | c.1299C>T | p.Ala433= | Not reported |
| JMJD8 | Splice region | c.323-6C>T | Potential splicing effect | 3.36 × 10−2 |
| PRR35 | Missense | c.1504G>A | p.Gly502Ser | 3.03 × 10−2 |
| TUT7 | Missense | c.1679G>A | p.Arg560Gln | 4.80 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohar, N.P.; Cox, E.M.; Adelizzi, E.; Moore, S.A.; Mathews, K.D.; Darbro, B.W.; Wallrath, L.L. The Influence of a Genetic Variant in CCDC78 on LMNA-Associated Skeletal Muscle Disease. Int. J. Mol. Sci. 2024, 25, 4930. https://doi.org/10.3390/ijms25094930
Mohar NP, Cox EM, Adelizzi E, Moore SA, Mathews KD, Darbro BW, Wallrath LL. The Influence of a Genetic Variant in CCDC78 on LMNA-Associated Skeletal Muscle Disease. International Journal of Molecular Sciences. 2024; 25(9):4930. https://doi.org/10.3390/ijms25094930
Chicago/Turabian StyleMohar, Nathaniel P., Efrem M. Cox, Emily Adelizzi, Steven A. Moore, Katherine D. Mathews, Benjamin W. Darbro, and Lori L. Wallrath. 2024. "The Influence of a Genetic Variant in CCDC78 on LMNA-Associated Skeletal Muscle Disease" International Journal of Molecular Sciences 25, no. 9: 4930. https://doi.org/10.3390/ijms25094930
APA StyleMohar, N. P., Cox, E. M., Adelizzi, E., Moore, S. A., Mathews, K. D., Darbro, B. W., & Wallrath, L. L. (2024). The Influence of a Genetic Variant in CCDC78 on LMNA-Associated Skeletal Muscle Disease. International Journal of Molecular Sciences, 25(9), 4930. https://doi.org/10.3390/ijms25094930






