Novel Tetracyclic Azaphenothiazines with the Quinoline Ring as New Anticancer and Antibacterial Derivatives of Chlorpromazine
Abstract
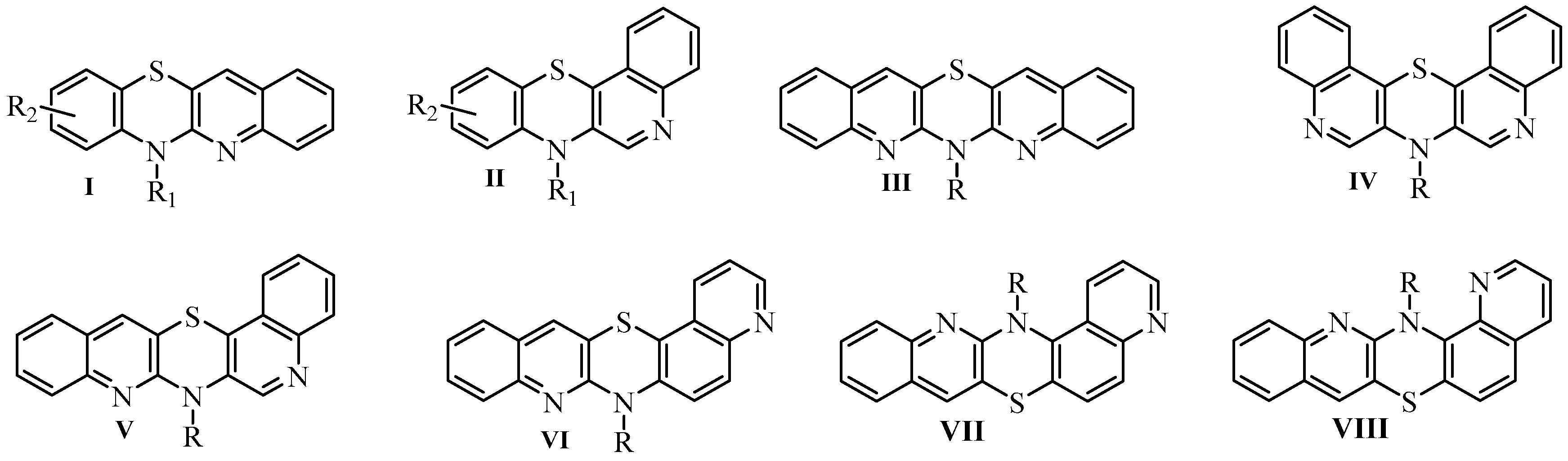
1. Introduction
2. Results and Discussion
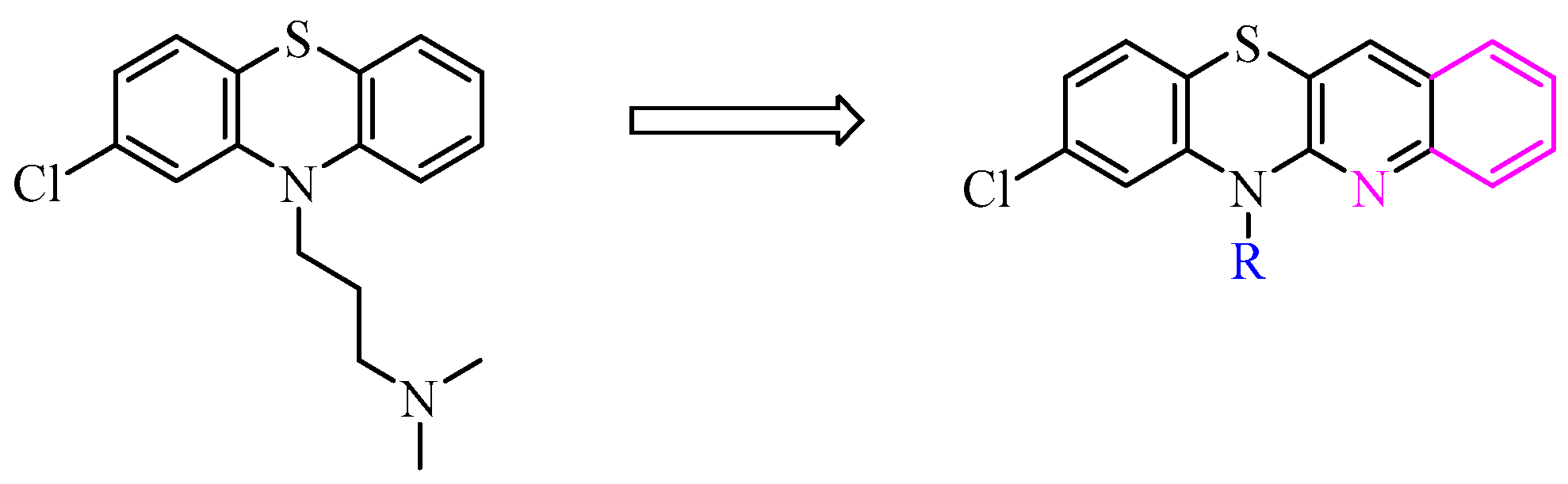
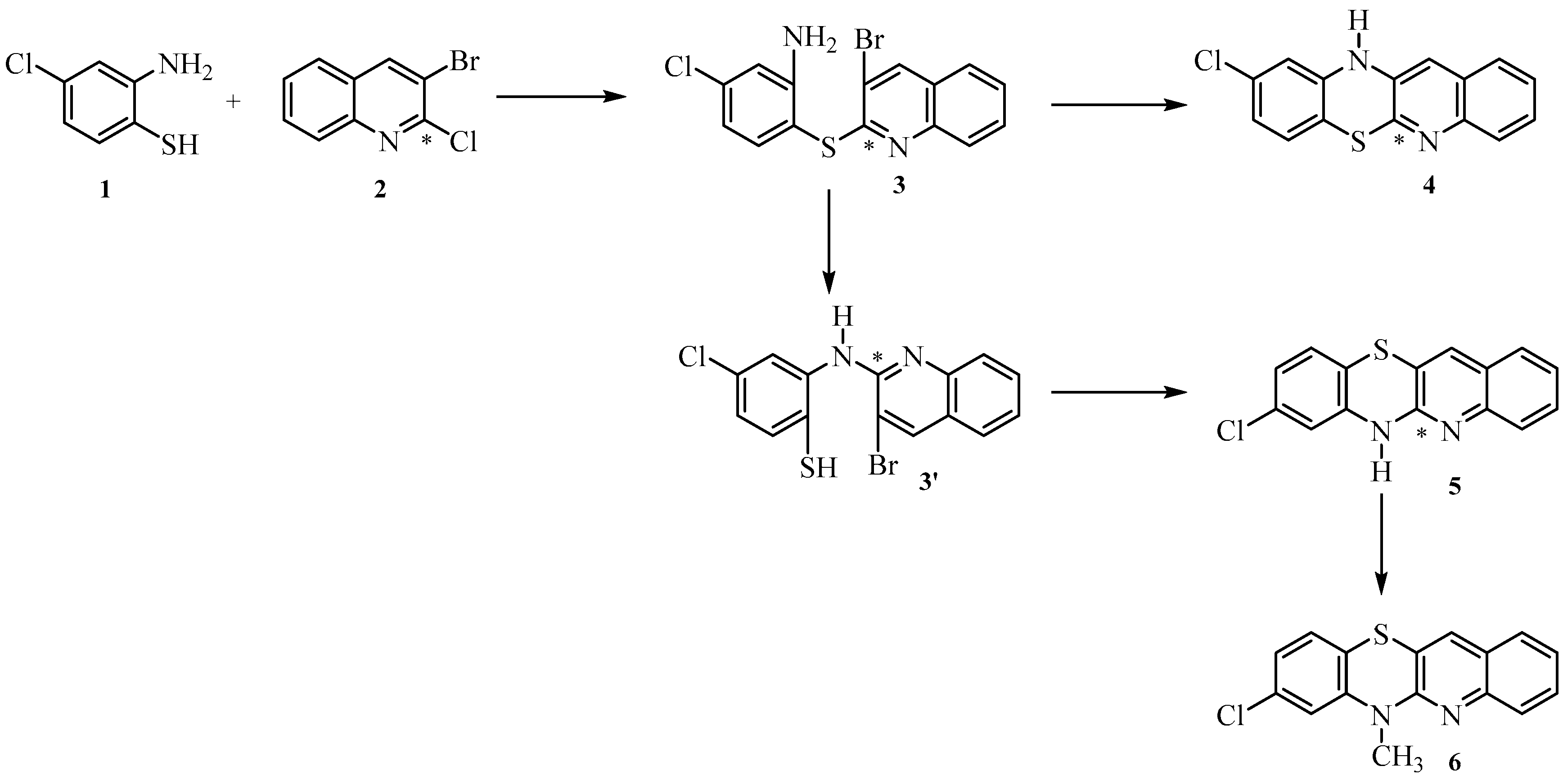
2.1. Synthesis
2.2. Biological Evaluation
2.2.1. Cytotoxic Activity
2.2.2. In Vitro Antibacterial Activity
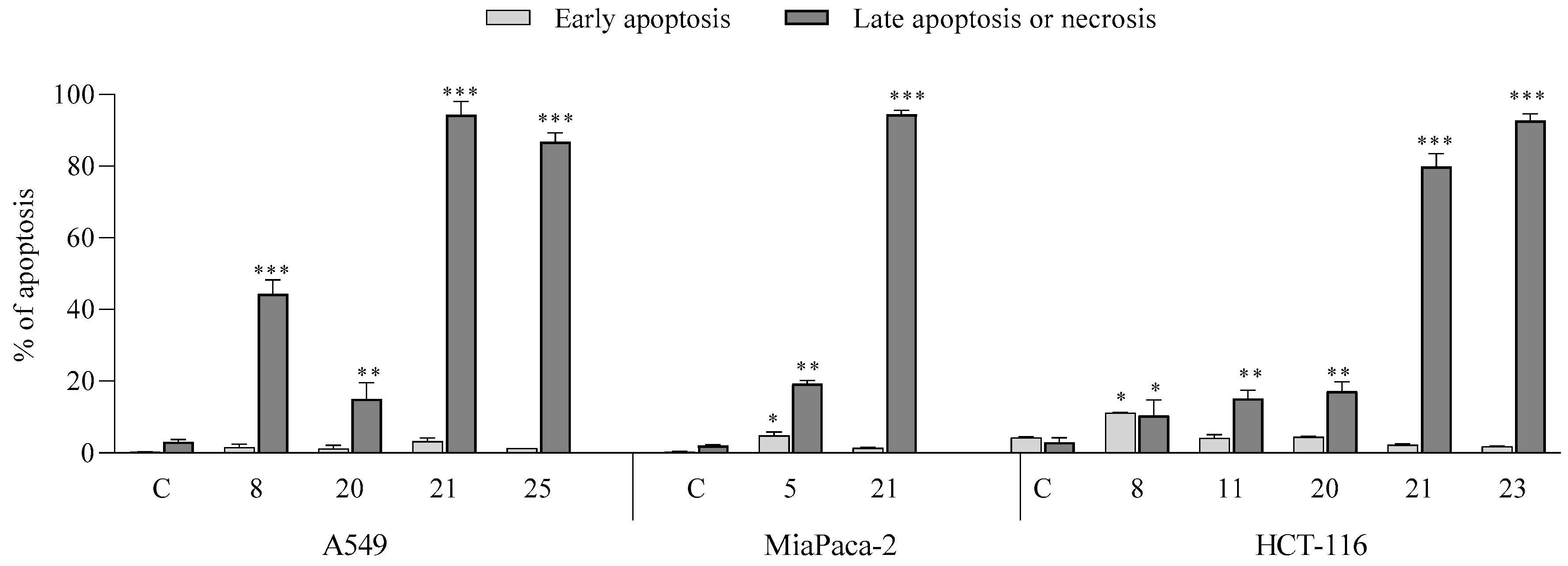
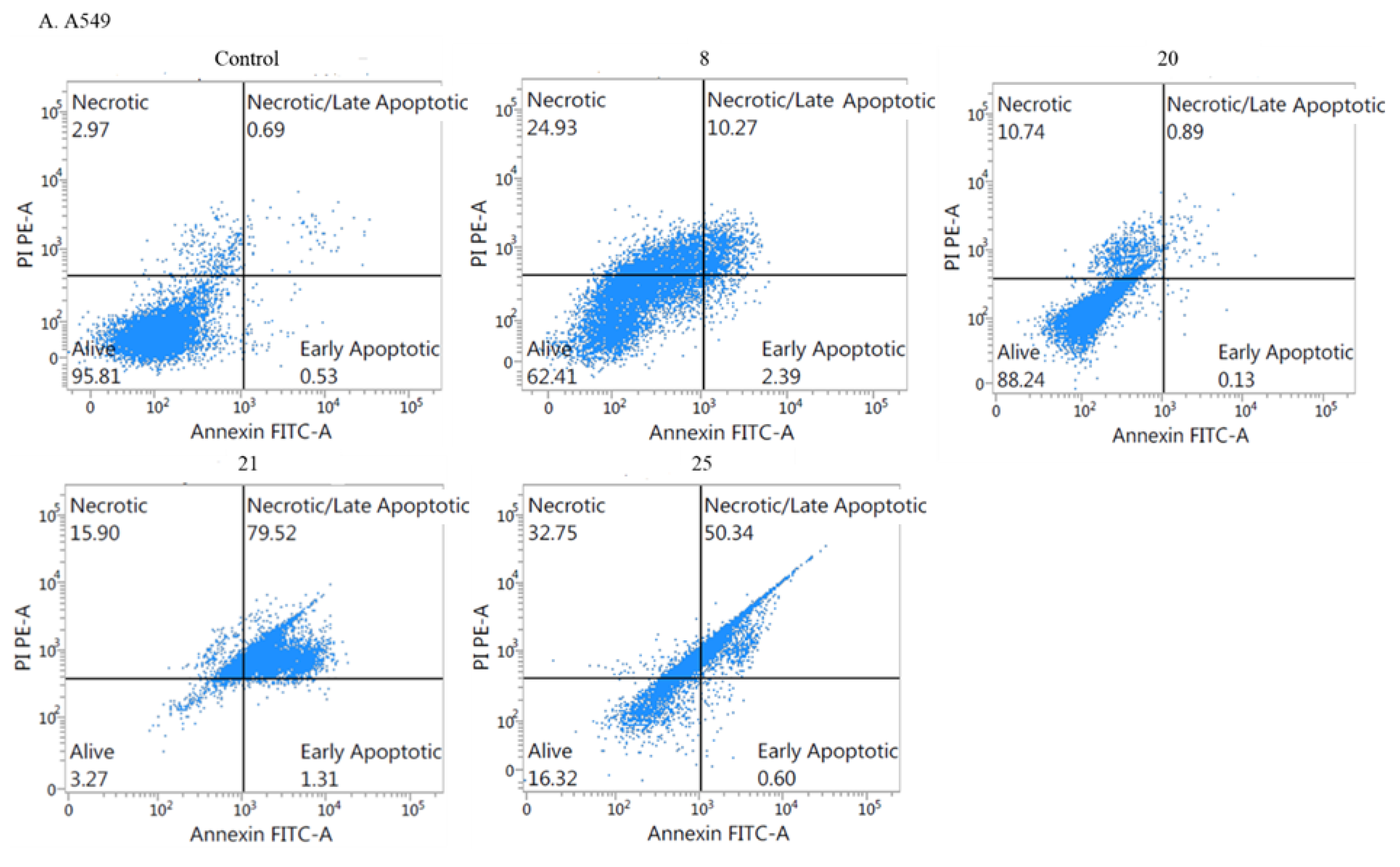
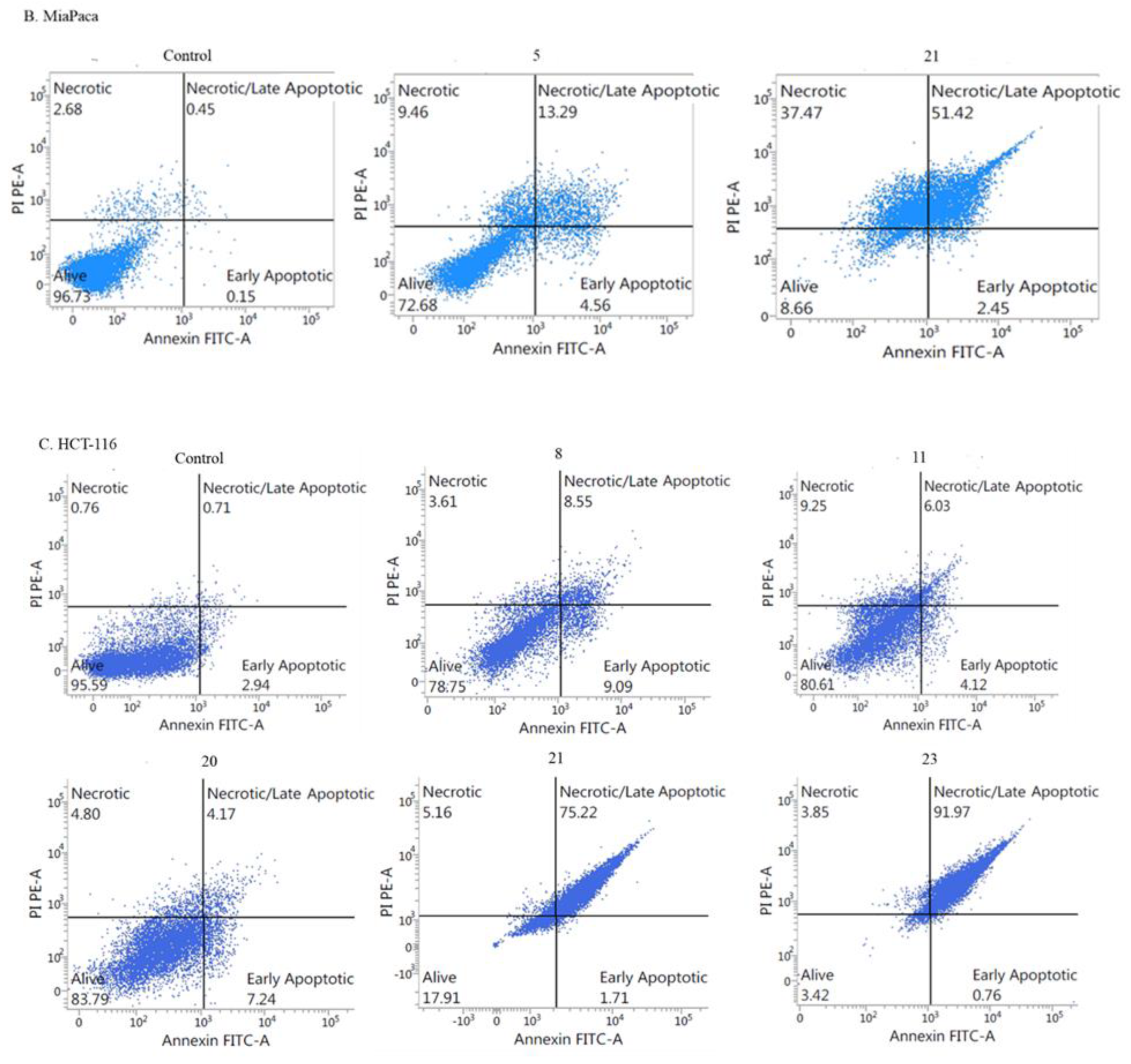
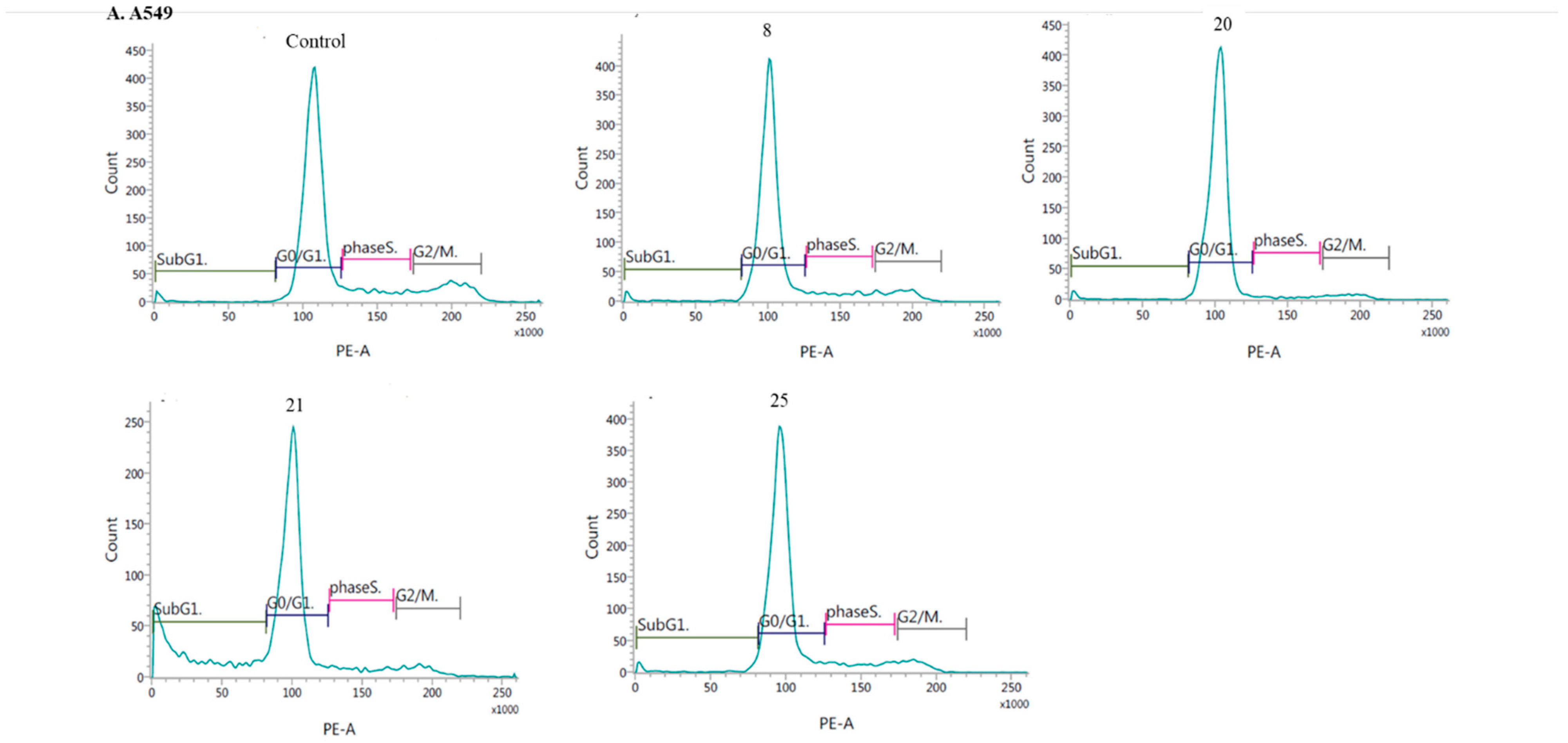
2.2.3. Mechanism of Cytotoxicity of Newly Synthesized Derivatives
- Apoptotic activity:
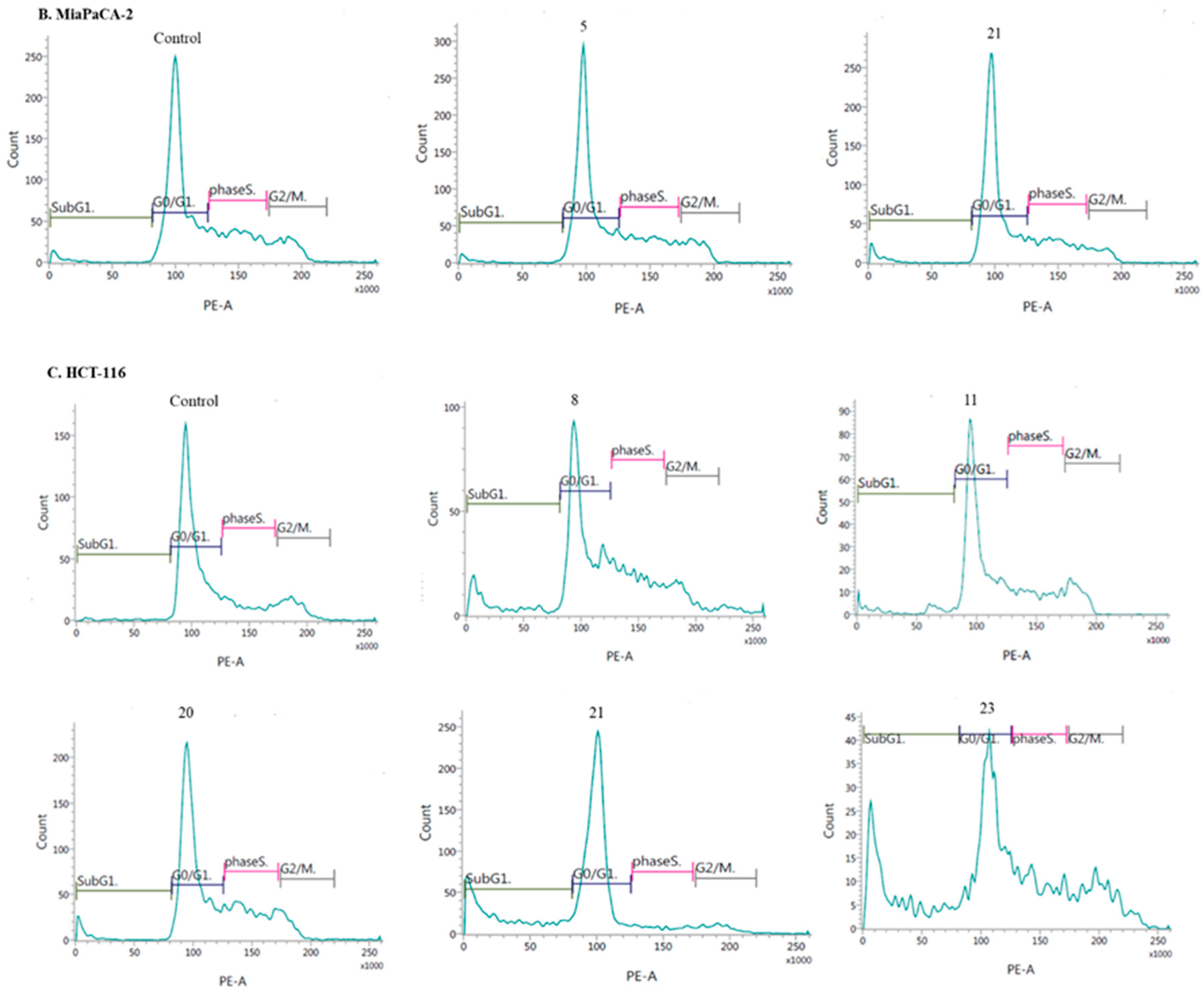
- Induction of cell cycle arrest
3. Methods and Materials
3.1. Synthesis of Compounds
General Procedure for the Synthesis of 8-Chloroquinobenzothiazine Derivatives
- Synthesis of 6H-8-chloroquino[3,2-b]benzo[1,4]thiazine 5.
- Synthesis of 8-chloro-6-methylquino[3,2-b]benzo[1,4]thiazine 6.
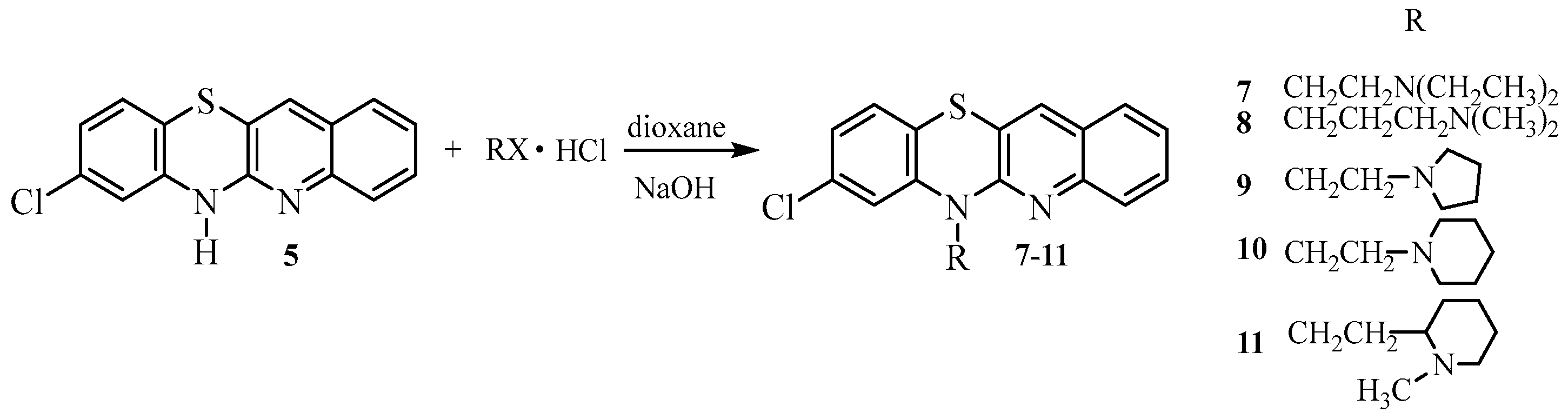
- Synthesis of 8-chloro-6-dialkylaminoalkylquinobenzothiazines 7–11.
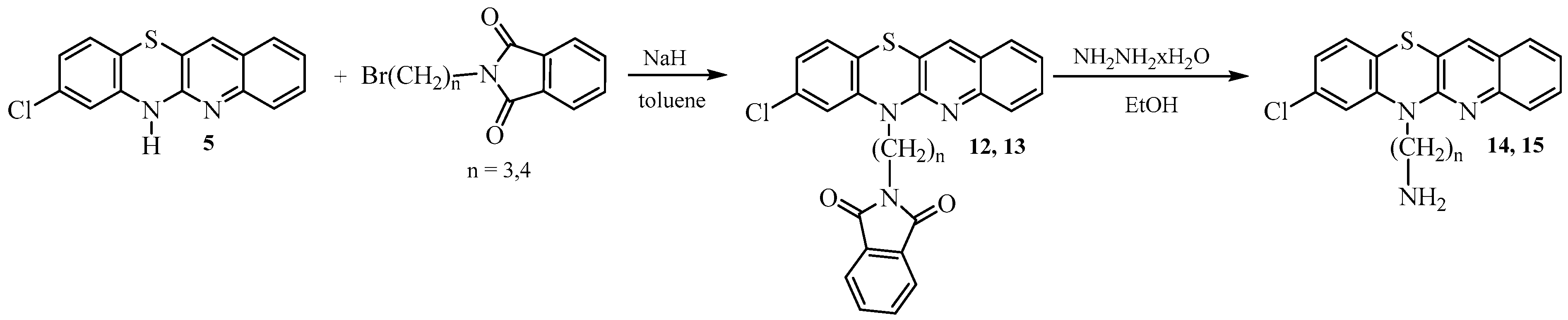
- Synthesis of 8-chloro-6-phthalimidoalkylquinobenzothiazines 12 and 13.
- Synthesis of 8-chloro-6-aminoalkylquinobenzothiazines 14 and 15.
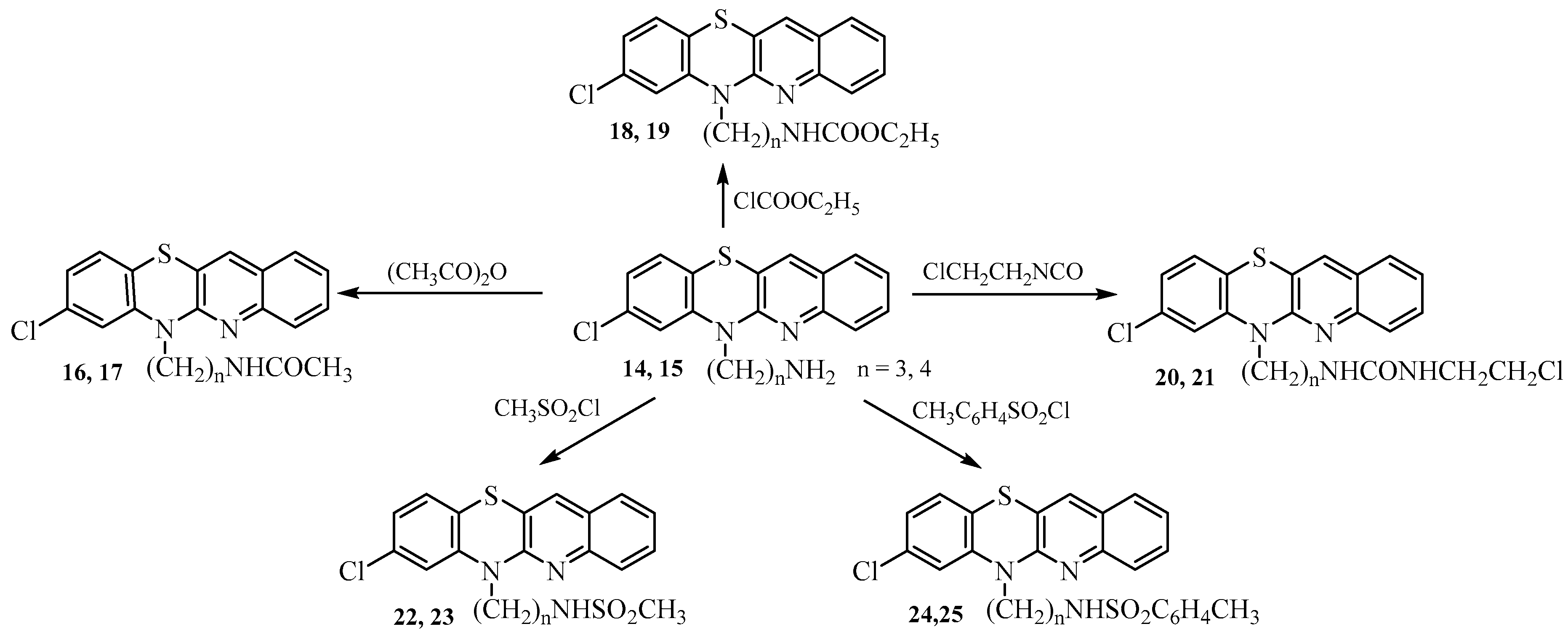
- Synthesis of 8-chloro-6-acetylaminoalkylquinobenzothiazines 16–25.
- Synthesis of 8-chloro-6-etoxycarbonylaminoalkylquinobenzothiazines 18 and 19.
- Synthesis of 8-chloro-6-chloroethylureidoalkylquinobenzothiazines 20 and 21.
- Synthesis of 8-chloro 6-methanesulfonylaminoalkylquinobenzothiazines 22 and 23.
- Synthesis of 8-chloro-6-p-toluenesulfonylaminolkylquinobenzothiazines 24 and 25.
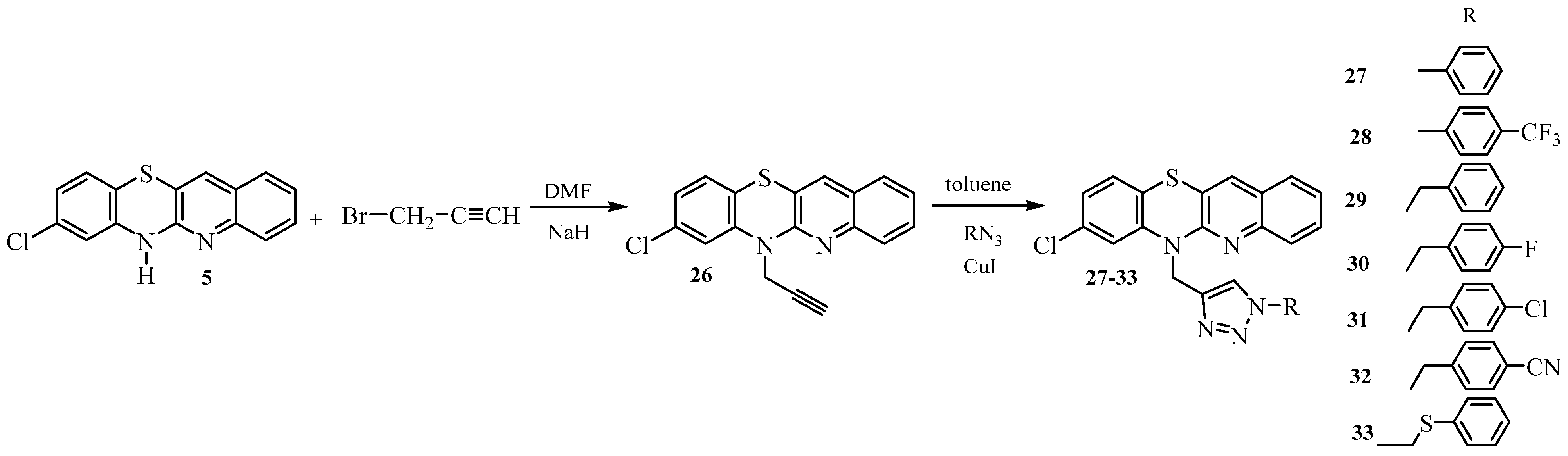
- Synthesis of 8-chloroquinobenzothiazines with triazole substituents 27–33.
3.2. Biological Assays
3.2.1. Cell Line and Culture
3.2.2. MTT Assay
3.2.3. Apoptosis and Cell Cycle Analysis by Flow Cytometry (FCM)
- Cell cycle analysis:
3.2.4. In Vitro Antibacterial Studies
3.2.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vitaku, E.; Smith, D.T.; Njardarson, J.T. Analysis of the structural diversity, substitution patterns, and frequency of nitrogen heterocycles among U.S. FDA approved pharmaceuticals. J. Med. Chem. 2014, 57, 10257–10274. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Sinhmar, A.; Sharma, A.; Rajak, H.; Pathak, D.P. Medicinal significance of benzothiazole scaffold: An insight view. J. Enzym. Inhib. Med. Chem. 2013, 28, 240–266. [Google Scholar] [CrossRef]
- Chhabria, M.T.; Patel, S.; Modi, P.; Brahmkshatriya, P.S. Thiazole: A review on chemistry, synthesis and therapeutic importance of its derivatives. Curr. Top. Med. Chem. 2016, 16, 2841–2862. [Google Scholar] [CrossRef]
- Mitchell, S.C. Phenothiazine: The parent molecule. Curr. Drug Targets 2006, 7, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, J.E.; Dastidar, S.G.; Palchoudhuri, S.; Roy, D.S.; Das, S.; Hendricks, O.; Christensen, J.B. Phenothiazines as a solution for multidrug resistant tuberculosis: From the origin to the present. Int. Microbiol. 2015, 18, 1–12. [Google Scholar] [CrossRef]
- Remington, G.; Hahn, M.K.; Agarwal, S.M.; Chintoh, A.; Agid, O. Schizophrenia: Antipsychotics and drug development. Behav. Brain Res. 2021, 414, 113507. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.E.; Awad, G.A.; Rathbone, J.; Thornley, B.; Soares-Weiser, K. Chlorpromazine versus placebo for schizophrenia. Cochrane Database Syst. Rev. 2014, 1, CD00028. [Google Scholar] [CrossRef]
- Boyd-Kimball, D.; Gonczy, K.; Lewis, B.; Mason, T.; Siliko, N.; Wolfe, J. Classics in chemical neuroscience: Chlorpromazine. ACS Chem. Neurosci. 2019, 10, 79–88. [Google Scholar] [CrossRef]
- Rai, S.; Tanaka, H.; Suzuki, M.; Espinoza, J.L.; Kumode, T.; Tanimura, A.; Yokota, T.; Oritani, K.; Watanabe, T.; Kanakura, Y.; et al. Chlorpromazine eliminates acute myeloid leukemia cells by altering the subcellular localisation of FLT3-ITD and KIT-D816V. Nat. Commun. 2020, 11, 4147. [Google Scholar] [CrossRef]
- Lidsky, T.; Yablonsky-Alter, E.; Zuck, L.; Banerjee, S. Antipsychotic drug effects on glutamatergic activity. Brain Res. 1997, 764, 46–52. [Google Scholar] [CrossRef]
- López-Muñoz, F.; Alamo, C.; Cuenca, E.; Shen, W.; Clervoy, P.; Rubio, G. History of the discovery and clinical introduction of chlorpromazine. Ann. Clin. Psychiatry 2005, 17, 113–135. [Google Scholar] [CrossRef] [PubMed]
- Otręba, M.; Kośmider, L. In vitro anticancer activity of fluphenazine, perphenazine and prochlorperazine. A review. J. Appl. Toxicol. 2021, 41, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Kamgar-Dayhoff, P.; Brelidze, T.I. Multifaceted effect of chlorpromazine in cancer: Implications for cancer treatment. Oncotarget 2021, 12, 1406–1426. [Google Scholar] [CrossRef] [PubMed]
- Hendouei, N.; Saghafi, F.; Shadfar, F.; Hosseinimehr, S.J. Molecular mechanisms of antipsychotic drugs for improvement of cancer treatment. Eur. J. Pharmacol. 2019, 856, 172402. [Google Scholar] [CrossRef] [PubMed]
- Csatary, L.K. Chlorpromazines and cancer. Lancet 1972, 300, 338–339. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, P.B. Neuroleptic treatment and other factors modifying cancer risk in schizophrenic patients. Acta Psychiatr. Scand. 1987, 75, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Yde, C.W.; Clausen, M.P.; Bennetzen, M.V.; Lykkesfeldt, A.E.; Mouritsen, O.G.; Guerra, B. The antipsychotic drug chlorpromazine enhances the cytotoxic effect of tamoxifen in tamoxifen-sensitive and tamoxifen-resistant human breast cancer cells. Anti-Cancer Drugs 2009, 20, 723–735. [Google Scholar] [CrossRef]
- Yang, C.E.; Lee, W.Y.; Cheng, H.W.; Chung, C.H.; Mi, F.L.; Lin, C.W. The antipsychotic chlorpromazine suppresses YAP signaling, stemness properties, and drug resistance in breast cancer cells. Chem. Biol. Interact. 2019, 302, 28–35. [Google Scholar] [CrossRef]
- Lee, W.Y.; Lee, W.T.; Cheng, C.H.; Chen, K.C.; Chou, C.M.; Chung, C.H.; Sun, M.S.; Cheng, H.W.; Ho, M.N.; Lin, C.W. Repositioning antipsychotic chlorpromazine for treating colorectal cancer by inhibiting sirtuin 1. Oncotarget 2015, 6, 27580–27595. [Google Scholar] [CrossRef]
- Lee, M.S.; Johansen, L.; Zhang, Y.; Wilson, A.; Keegan, M.; Avery, W.; Elliott, P.; Borisy, A.A.; Keith, C.T. The novel combination of chlorpromazine and pentamidine exerts synergistic antiproliferative effects through dual mitotic action. Cancer Res. 2007, 67, 11359–11367. [Google Scholar] [CrossRef]
- Kurita, J.; Hirao, Y.; Nakano, H.; Fukunishi, Y.; Nishimura, Y. Sertraline, chlorprothixene and chlorpromazine characteristically interact with the REST-binding site of the corepressor mSin3, showing inhibitory activities of medulloblastoma cell growth. Sci. Rep. 2018, 8, 13763. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Zhang, W.; Langford, C.; Suto, M.J.; Griguer, C.E. Repositioning chlorpromazine for treating chemoresistant glioma through the inhibition of cytochrome c oxidase bearing the COX4-1 regulatory subunit. Oncotarget 2017, 8, 37568–37583. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Kim, C.; Kim, S.; Kim, S.H.; Kim, Y.S.; Lim, Y.; Lee, Y.H. Chlorpromazine activates p21Waf1/Cip1 gene transcription via early growth response-1 (Egr-1) in C6 glioma cells. Exp. Mol. Med. 2010, 42, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Lee, K.S.; Choi, Y.K.; Lim, H.J.; Lee, H.G.; Lim, Y.; Lee, Y.H. The antipsychotic agent chlorpromazine induces autophagic cell death by inhibiting the Akt/mTOR pathway in human U-87MG glioma cells. Carcinogenesis 2013, 34, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Gil-Ad, I.; Shtaif, B.; Levkovitz, Y.; Nordenberg, J.; Taler, M.; Korov, I.; Weizman, A. Phenothiazines induce apoptosis in a B16 mouse melanoma cell line and attenuate in vivo melanoma tumor growth. Oncol. Rep. 2006, 15, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Zhelev, Z.; Ohba, H.; Bakalova, R.; Hadjimitova, V.; Ishikawa, M.; Shinohara, Y.; Baba, Y. Phenothiazines suppress proliferation and induce apoptosis in cultured leukemic cells without any influence on the viability of normal lymphocytes. Cancer Chemother. Pharmacol. 2004, 53, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Zong, D.; Zielinska-Chomej, K.; Juntti, T.; Mörk, B.; Lewensohn, R.; Hååg, P.; Viktorsson, K. Harnessing the lysosome-dependent antitumor activity of phenothiazines in human small cell lung cancer. Cell Death Dis. 2014, 5, 1111. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, S.; Giehl, K.; Henis, Y.I.; Ehrlich, M. Differential interference of chlorpromazine with the membrane interactions of oncogenic K-Ras and its effects on cell growth. J. Biol. Chem. 2008, 283, 27279–27288. [Google Scholar] [CrossRef] [PubMed]
- Jhou, A.J.; Chang, H.C.; Hung, C.C.; Lin, H.C.; Lee, Y.C.; Liu, W.; Lee, C.H. Chlorpromazine, an antipsychotic agent, induces G2/M phase arrest and apoptosis via regulation of the PI3K/AKT/mTOR-mediated autophagy pathways in human oral cancer. Biochem. Pharmacol. 2021, 184, 114403. [Google Scholar] [CrossRef]
- Abbruzzese, C.; Matteoni, S.; Persico, M.; Villani, V.; Paggi, M.G. Repurposing chlorpromazine in the treatment of glioblastoma multiforme: Analysis of literature and forthcoming steps. J. Exp. Clin. Cancer Res. 2020, 39, 26. [Google Scholar] [CrossRef]
- Xu, F.; Xi, H.; Liao, M.; Zhang, Y.; Ma, H.; Wu, M.; Xue, Q.; Sun, H.; Zhang, Y.; Xia, Y. Repurposed antipsychotic chlorpromazine inhibits colorectal cancer and pulmonary metastasis by inducing G2/M cell cycle arrest, apoptosis, and autophagy. Cancer Chemother. Pharmacol. 2022, 89, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Matteoni, S.; Matarrese, P.; Ascione, B.; Buccarelli, M.; Ricci-Vitiani, L.; Pallini, R.; Villani, V.; Pace, A.; Paggi, M.G.; Abbruzzese, C. Anticancer properties of the antipsychotic drug chlorpromazine and its synergism with temozolomide in restraining human glioblastoma proliferation in vitro. Front Oncol. 2021, 11, 635472. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.C.; Ekins, S.; Williams, A.J.; Tropsha, A. A bibliometric review of drug repurposing. Drug Discov. Today 2018, 23, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Varga, B.; Csonka, Á.; Csonka, A.; Molnár, J.; Amaral, L.; Spengler, G. Possible biological and clinical applications of phenothiazines. Anticancer Res. 2017, 37, 5983–5993. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.Y.; Tay, W.L.; Chui, W.K.; Chan, A. Clinically relevant drug interactions between anticancer drugs and psychotropic agents. Eur. J. Cancer Care 2011, 20, 6–32. [Google Scholar] [CrossRef] [PubMed]
- Lialiaris, T.S.; Papachristou, F.; Mourelatos, C.; Simopoulou, M. Antineoplastic and cytogenetic effects of chlorpromazine on human lymphocytes in vitro and on Ehrlich ascites tumor cells in vivo. Anticancer Drugs. 2009, 20, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Pluta, K.; Jeleń, M.; Morak-Młodawska, B.; Zimecki, M.; Artym, J.; Kocięba, M.; Zaczyńska, E. Azaphenothiazines—Promising phenothiazine derivatives. An insight into nomenclature, synthesis, structure elucidation and biological properties. Eur. J. Med. Chem. 2017, 138, 774–806. [Google Scholar] [CrossRef] [PubMed]
- Pluta, K.; Morak-Młodawska, B.; Jeleń, M. Recent progress in biological activities of synthesized phenothiazines. Eur. J. Med. Chem. 2011, 46, 3179–3189. [Google Scholar] [CrossRef]
- Empel, A.; Bak, A.; Kozik, V.; Latocha, M.; Cizek, A.; Jampilek, J.; Suwinska, K.; Sochanik, A.; Zieba, A. Towards Property Profiling: SYNTHESIS and SAR Probing of New Tetracyclic Diazaphenothiazine Analogues. Int. J. Mol. Sci. 2021, 22, 12826. [Google Scholar] [CrossRef]
- Jeleń, M.; Morak-Młodawska, B.; Korlacki, R. Anticancer activities of tetra-, penta-, and hexacyclic phenothiazines modified with quinoline moiety. J. Mol. Struct. 2023, 1287, 135700. [Google Scholar] [CrossRef]
- Kumar, A.; Vigato, C.; Boschi, D.; Lolli, M.L.; Kumar, D. Phenothiazines as anti-cancer agents: SAR overview and synthetic strategies. Eur. J. Med. Chem. 2023, 254, 115337. [Google Scholar] [CrossRef]
- Posso, M.C.; Domingues, F.C.; Ferreira, S.; Silvestre, S. Development of phenothiazine hybrids with potential medicinal interest: A review. Molecules 2022, 27, 276. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, M.; Pluta, K.; Szmielew, M.; Morak-Młodawska, B.; Herman, K.; Giercuszkiewicz, K.; Kasprzycka, A.; Skonieczna, M. 14-Substituted diquinothiazines as a new group of anticancer agents. Molecules 2023, 28, 3248. [Google Scholar] [CrossRef] [PubMed]
- Jeleń, M.; Ying, P.; Ting, C.; Hao, Y.J.; Balachandran, A.; Anamalay, K.; Morak-Młodawska, B.; Gaurav, A.; Lavilla, C.A.; Uy, M.M.; et al. In vitro study of antioxidant, antigylycation, sugar hydrolysis enzyme inhibitory effect and molecular in silico docking study of angularly condensed diquinothiazines. J. Mol. Struct. 2024, 1296, 136856. [Google Scholar] [CrossRef]
- Artym, J.; Kocięba, M.; Zaczyńska, E.; Kochanowska, I.; Zimecki, M.; Kałas, W.; Strządała, L.; Zioło, E.; Jeleń, M.; Morak-Młodawska, B.; et al. Prolongation of skin graft survival in mice by an azaphenothiazine derivative. Immunol. Lett. 2019, 208, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, N.L.; Sharma, A.K.; Jamwal, R.S.; Atal, C.K.; Torres, T. A convenient and single-step synthesis of substituted 12H-quinoxalino [2,3-b][1,4]benzothiazines and -benzoxazines. Liebigs Ann. Chem. 1987, 1987, 921–925. [Google Scholar] [CrossRef]
- Yoneda, F.; Ohtaka, T.; Nitta, Y. Pyridazine derivatives. VII. Syntheses of 2,3-diazaphenothiazine derivatives. Yakugaku Zasshi 1966, 86, 887–890. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yoneda, F.; Ohtaka, T.; Nitta, Y. 10H-Benzo[b]pyridazino [3,4-e][1,4]thiazin und 5H-Benz[b]pyridazino [4,3-e][1,4]thiazin. Chem. Pharm. Bull. 1963, 11, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Nagashima, H.; Oda, H. Regiocontrolled cyclization to the isomeric dipyridazino [4,5-b;4′,5′-e][1,4]thiazines-competence and performance of benzylamino and thioether functions on the 3(2H)-pyridazinones as regiocontrolling elements. Heterocycles 1984, 22, 675–679. [Google Scholar] [CrossRef]
- Jeleń, M.; Pluta, K. Synthesis of quinobenzo-1,4-thiazines from diquino-1,4-dithiin and 2,2′-dichloro-3,3′-diquinolinyl disulfide. Heterocycles 2009, 78, 2325–2336. [Google Scholar] [CrossRef]
- Matin, M.M.; Matin, P.; Rahman, M.R.; Hadda, T.B.; Almalki, F.A.; Mahmud, S.; Ghoneim, M.M.; Alruwaily, M.; Alshehri, S. Triazoles and their derivatives: Chemistry, synthesis, and therapeutic applications. Front. Mol. Biosci. 2022, 9, 864286. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Song, Q.; Liu, Z.; Guo, J.; Cao, S.; Long, S. Recent advances in 1,2,3- and 1,2,4-triazole hybrids as antimicrobials and their SAR: A critical review. Eur. J. Med. Chem. 2023, 259, 115603. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.; Lv, Z.; Zhang, G.; Xu, Z. Recent updates on 1,2,3-triazole-containing hybrids with in vivo therapeutic potential against cancers: A mini-review. Eur. J. Med. Chem. 2023, 251, 115254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B. Comprehensive review on the anti-bacterial activity of 1,2,3-triazole hybrids. Eur. J. Med. Chem. 2019, 168, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Bozorov, K.; Zhao, J.; Aisa, H.A. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 2019, 27, 3511–3531. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.M. 1,2,3-Triazole hybrids as anticancer agents: A review. Arch. Pharm. 2022, 355, 2100158. [Google Scholar] [CrossRef]
- Reddyrajula, R.; Dalimba, U.; Kumar, S. Molecular hybridization approach for phenothiazine incorporated 1,2,3-triazole hybrids as promising antimicrobial agents: Design, synthesis, molecular docking and in silico ADME studies. Eur. J. Med. Chem. 2019, 168, 263–282. [Google Scholar] [CrossRef]
- Zhang, J.X.; Guo, J.M.; Zhang, T.T.; Lin, H.J.; Qi, N.S.; Li, Z.G.; Zhou, J.C.; Zhang, Z.Z. Antiproliferative phenothiazine hybrids as novel apoptosis inducers against MCF-7 breast cancer. Molecules. 2018, 23, 1288. [Google Scholar] [CrossRef]
- Liu, N.; Jin, Z.; Zhang, J.; Jin, J. Antitumor evaluation of novel phenothiazine derivatives that inhibit migration and tubulin polymerization against gastric cancer MGC-803 cells. Investig. New Drugs 2019, 37, 188–198. [Google Scholar] [CrossRef]
- Sylvester, P.W. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol. Biol. 2011, 716, 157–168. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; CLSI: Wayne, PA, USA, 2006. [Google Scholar]
- Bonelli, M.; La Monica, S.; Fumarola, C.; Alfieri, R. Multiple effects of CDK4/6 inhibition in cancer: From cell cycle arrest to immunomodulation. Biochem. Pharmacol. 2019, 170, 113676. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Zhang, Y. Downregulation of NIMA-related kinase-7 inhibits cell proliferation by inducing cell cycle arrest in human retinoblastoma cells. Exp. Ther. Med. 2018, 15, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Plati, J.; Bucur, O.; Khosravi-Far, R. Dysregulation of apoptotic signaling in cancer: Molecular mechanisms and therapeutic opportunities. J. Cell. Biochem. 2008, 104, 1124–1149. [Google Scholar] [CrossRef] [PubMed]
- Otto-Ślusarczyk, D.; Mielczarek-Puta, M.; Graboń, W. The real cytotoxic effect of artemisinins on colon cancer cells in a physiological cell culture setting. How composition of the culture medium biases experimental findings. Pharmaceuticals 2021, 14, 976. [Google Scholar] [CrossRef]
- Struga, M.; Roszkowski, P.; Bielenica, A.; Otto-Ślusarczyk, D.; Stępień, K.; Stefańska, J.; Zabost, A.; Augustynowicz-Kopeć, E.; Koliński, M.; Kmiecik, S.; et al. N-Acylated ciprofloxacin derivatives: Synthesis and in vitro biological evaluation as antibacterial and anticancer agents. ACS Omega 2023, 8, 18663–18684. [Google Scholar] [CrossRef]
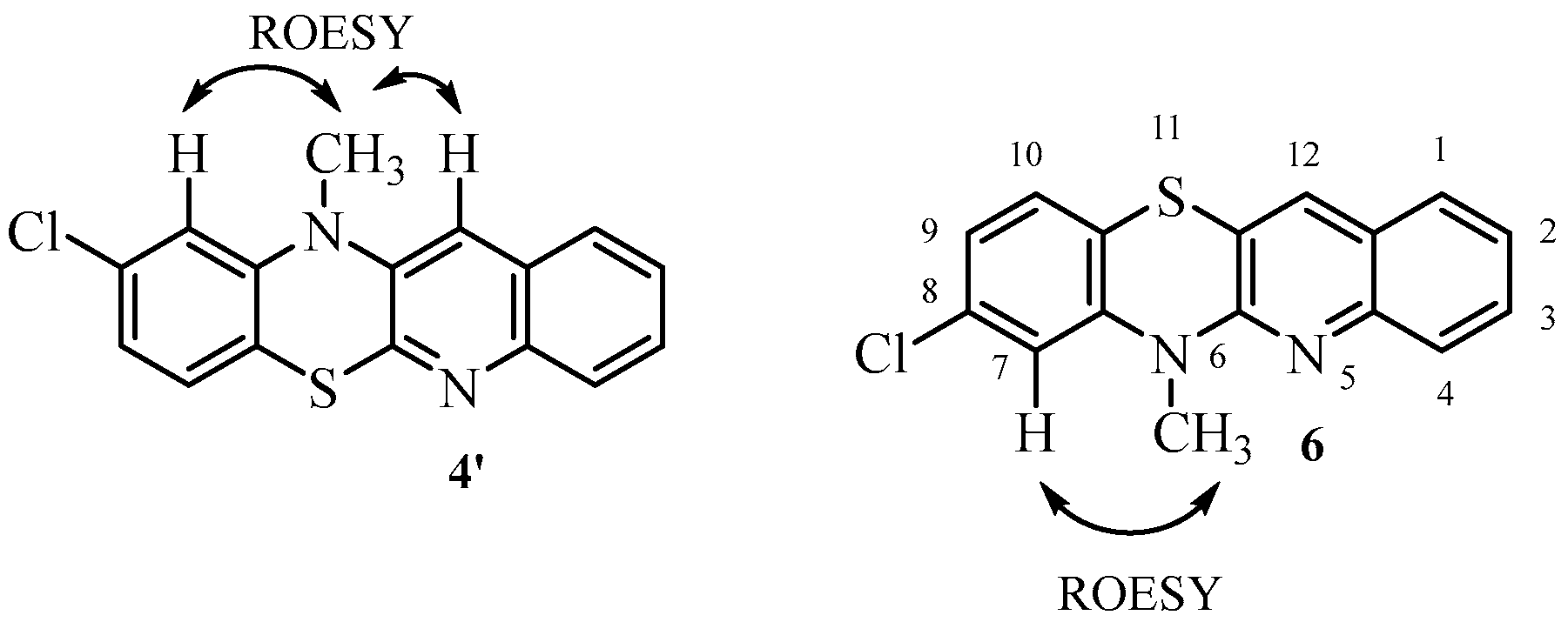

| 1H NMR (ppm) | ROESY | COSY |
|---|---|---|
| 3.61 CH3 | 6.94 | |
| 6.94 H7 | 3.61 | |
| 6.96 H9 | 7.05 | |
| 7.05 H10 | 6.96 | |
| 7.32 H2 | 7.55, 7.56 | |
| 7.55 H3 7.56 H1 | 7.32 | |
| 7.70 H12 | ||
| 7.79 H4 | 7.55, 7.56 |
| 13C NMR | HSQC | HMBC |
|---|---|---|
| 29.73 | 3.61 | |
| 115.70 | 3.61 and 6.94 C7 | 6.96 |
| 118.19 | 6.96 and 6.94 C8 | |
| 118.95 | 6.94 C4a | |
| 122.48 | 6.96 C9 | |
| 124.53 | 7.32 C2 | |
| 125.99 | 7.32 C11a | |
| 126.30 | 7.54–7.57 C1 | |
| 127.23 | 7.79 C4 | |
| 127.47 | 7.05 C10 | |
| 129.33 | 7.54–7.57 C3 | 7.54–7.57 |
| 132.21 | 7.70 C12 | |
| 133.60 | 6.94 and 7.05 C10a | |
| 144.17 | 7.05 and 3.61 C6a | |
| 145.74 | 7.70 and 7.54–7.57 C12a | |
| 152.77 | 7.70 and 3.61 C5a |
| Compound | Cancer Cells | Normal Cells | |||
|---|---|---|---|---|---|
| A549 d | MDA e | HaCaT f | |||
| IC50 b | SI c | IC50 | SI | IC50 | |
| 5 | 14.9 ± 2.7 | 3.8 | 76.5 ± 0.8 | 0.7 | 56.5 ± 4.5 |
| 7 | 27.2 ± 8.4 | 3.7 | >100 | 1.0 | >100 |
| 8 | 8.2 ± 3.6 | 7.6 | 52.1 ± 2.7 | 1.2 | 62.3 ± 2.5 |
| 9 | 17.4 ± 4.1 | 5.7 | 77.1 ± 1.6 | 1.3 | >100 |
| 10 | 63.8 ± 8.5 | 1.6 | >100 | 1.0 | >100 |
| 11 | 16.0 ± 4.2 | 1 | 16.7 ± 1.1 | 0.9 | 12.7 ± 1.9 |
| 16 | 24.6 ± 6.1 | 4.1 | 76.6 ± 8.9 | 1.3 | >100 |
| 17 | 82.3 ± 6.2 | 1.2 | >100 | 1.0 | >100 |
| 18 | 86.6 ± 0.8 | 1.1 | >100 | 1.0 | >100 |
| 19 | >100 | 1.0 | >100 | 1.0 | >100 |
| 20 | 9.3 ± 1.2 | 10.7 | 48.6 ± 7.3 | 2.0 | >100 |
| 21 | 6.98 ± 1.2 | 0.16 | 7.4 ± 1.2 | 0.15 | 1.1 ± 0.3 |
| 22 | >100 | 1.0 | >100 | 1.0 | >100 |
| 23 | 30.5 ± 8.3 | 3.3 | >100 | 0.2 | >100 |
| 24 | >100 | 1.0 | >100 | 1.0 | >100 |
| 25 | 9.45 ± 1.3 | 10.5 | >100 | 1.0 | <100 |
| 26 | >100 | 1.0 | 95.8 ± 9.2 | 0.7 | 71.7 ± 5.7 |
| 27 | 27.5 ± 6.2 | 2.5 | >100 | 0.7 | 70.2 ± 10.4 |
| 28 | >100 | 1.0 | >100 | 1.0 | >100 |
| 29 | >100 | 1.0 | >100 | 1.0 | >100 |
| 30 | >100 | 1.0 | >100 | 1.0 | >100 |
| 31 | >100 | 1.0 | >100 | 1.0 | >100 |
| 32 | >100 | 1.0 | >100 | 1.0 | >100 |
| 33 | >100 | 1.0 | >100 | 1.0 | >100 |
| DX g | 0.6 ± 0.2 | 0.14 | 0.8 ± 0.1 | 0.15 | 0.3 ± 0.1 |
| Compound | Cancer Cells | Normal Cells | |||||
|---|---|---|---|---|---|---|---|
| MiaPaCa-2 d | PC3 e | HCT116 f | HaCaT g | ||||
| IC50 b | SI c | IC50 | SI | IC50 | SI | IC50 | |
| 5 | 11.1 ± 0.4 | 5.0 | 76.5 ± 8.1 | 0.7 | 33.5 ± 6.8 | 1.7 | 56.5 ± 6.4 |
| 8 | 40.2 ± 0.7 | 1.6 | 52.1 ± 7.1 | 1.2 | 1.6 ± 0.8 | 39 | 62.3 ± 3.5 |
| 9 | 57.4 ± 9.6 | 1.7 | 77.1 ± 9.4 | 1.3 | 17.5 ± 1.4 | 5.7 | >100 |
| 11 | 24.3 ± 3.5 | 0.5 | 16.7 ± 1.8 | 0.7 | 7.7 ± 1.2 | 1.6 | 12.7 ± 2.1 |
| 20 | 23.2 ± 2.7 | 4.3 | 34.8 ± 9.8 | 2.8 | 10.4 ± 1.6 | 8.8 | >100 |
| 21 | 6.4 ± 2.4 | 0.2 | 76.6 ± 9.8 | 0.1 | 11.3 ± 2.2 | 0.1 | 1.1 ± 0.2 |
| 23 | 98.4 ± 5.6 | 1.0 | >100 | 1.0 | 0.7 ± 0.08 | 143 | >100 |
| 25 | >100 | 1.0 | 48.6 ± 4.7 | 2.0 | >100 | 1.0 | >100 |
| 27 | 37.4 ± 5.6 | 1.9 | >100 | 1.0 | 49.6 ± 4.7 | 1.4 | 71.7 ± 7.5 |
| DX h | 0.6 ± 0.2 | 0.14 | 0.8 ± 0.1 | 0.15 | 0.59 ± 0.02 | 0.5 | 0.3 ± 0.1 |
| Compound | Bacterial Strains | |||||||
|---|---|---|---|---|---|---|---|---|
| S. aureus NCTC 4163 | S. aureus ATCC 25923 | S. aureus ATCC 6538 | S. aureus ATCC 29213 | S. epidermidis ATCC 12228 | S. epidermidis ATCC 35984 | E. coli ATCC 25922 | P. aeruginosa ATCC 15442 | |
| 5 | 8 | 8 | 8 | 8 | 8 | 8 | 256 | 256 |
| 7 | 8 | 8 | 8 | 8 | 8 | 8 | >256 | >256 |
| 8 | 8 | 8 | 8 | 8 | 8 | 8 | 64 | 256 |
| 9 | 16 | 16 | 16 | 16 | 16 | 16 | >256 | >256 |
| 10 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 11 | 8 | 128 | 128 | 128 | 256 | 256 | >256 | >256 |
| 16 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 17 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 18 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 19 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 20 | 8 | 8 | 8 | 8 | 8 | 4 | >256 | >256 |
| 21 | 2 | 2 | 2 | 2 | 2 | 2 | 8 | >256 |
| 22 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 23 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 24 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 25 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 26 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 27 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 28 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 29 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 30 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 31 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 32 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| 33 | >256 | >256 | >256 | >256 | >256 | >256 | >256 | >256 |
| control—ciprofloxacin | 0.125 | 0.25 | 0.125 | 0.25 | 0.125 | 0.125 | 0.0075 | 0.125 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeleń, M.; Otto-Ślusarczyk, D.; Morak-Młodawska, B.; Struga, M. Novel Tetracyclic Azaphenothiazines with the Quinoline Ring as New Anticancer and Antibacterial Derivatives of Chlorpromazine. Int. J. Mol. Sci. 2024, 25, 4148. https://doi.org/10.3390/ijms25084148
Jeleń M, Otto-Ślusarczyk D, Morak-Młodawska B, Struga M. Novel Tetracyclic Azaphenothiazines with the Quinoline Ring as New Anticancer and Antibacterial Derivatives of Chlorpromazine. International Journal of Molecular Sciences. 2024; 25(8):4148. https://doi.org/10.3390/ijms25084148
Chicago/Turabian StyleJeleń, Małgorzata, Dagmara Otto-Ślusarczyk, Beata Morak-Młodawska, and Marta Struga. 2024. "Novel Tetracyclic Azaphenothiazines with the Quinoline Ring as New Anticancer and Antibacterial Derivatives of Chlorpromazine" International Journal of Molecular Sciences 25, no. 8: 4148. https://doi.org/10.3390/ijms25084148
APA StyleJeleń, M., Otto-Ślusarczyk, D., Morak-Młodawska, B., & Struga, M. (2024). Novel Tetracyclic Azaphenothiazines with the Quinoline Ring as New Anticancer and Antibacterial Derivatives of Chlorpromazine. International Journal of Molecular Sciences, 25(8), 4148. https://doi.org/10.3390/ijms25084148








