Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells
Abstract
1. Introduction
2. Results
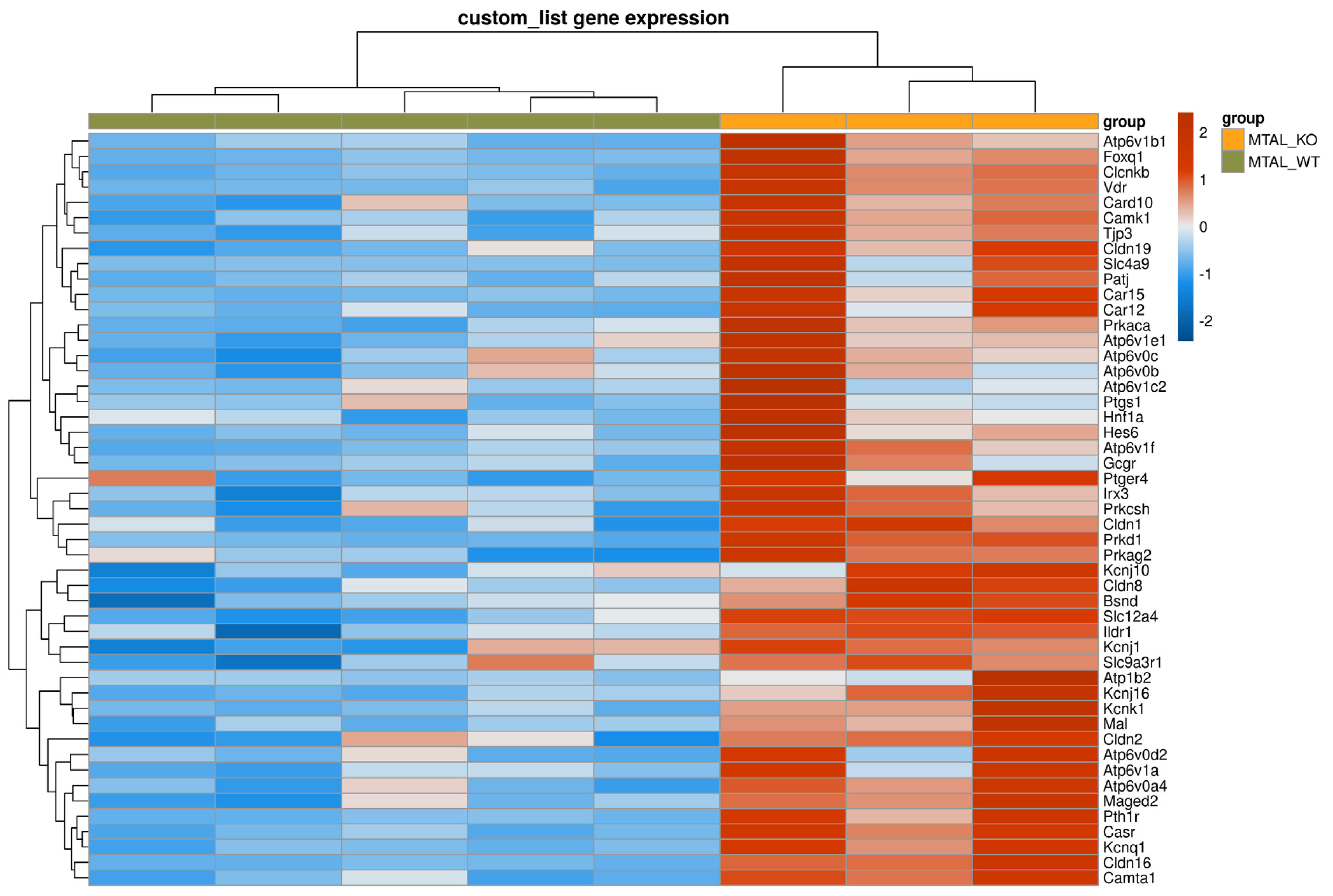
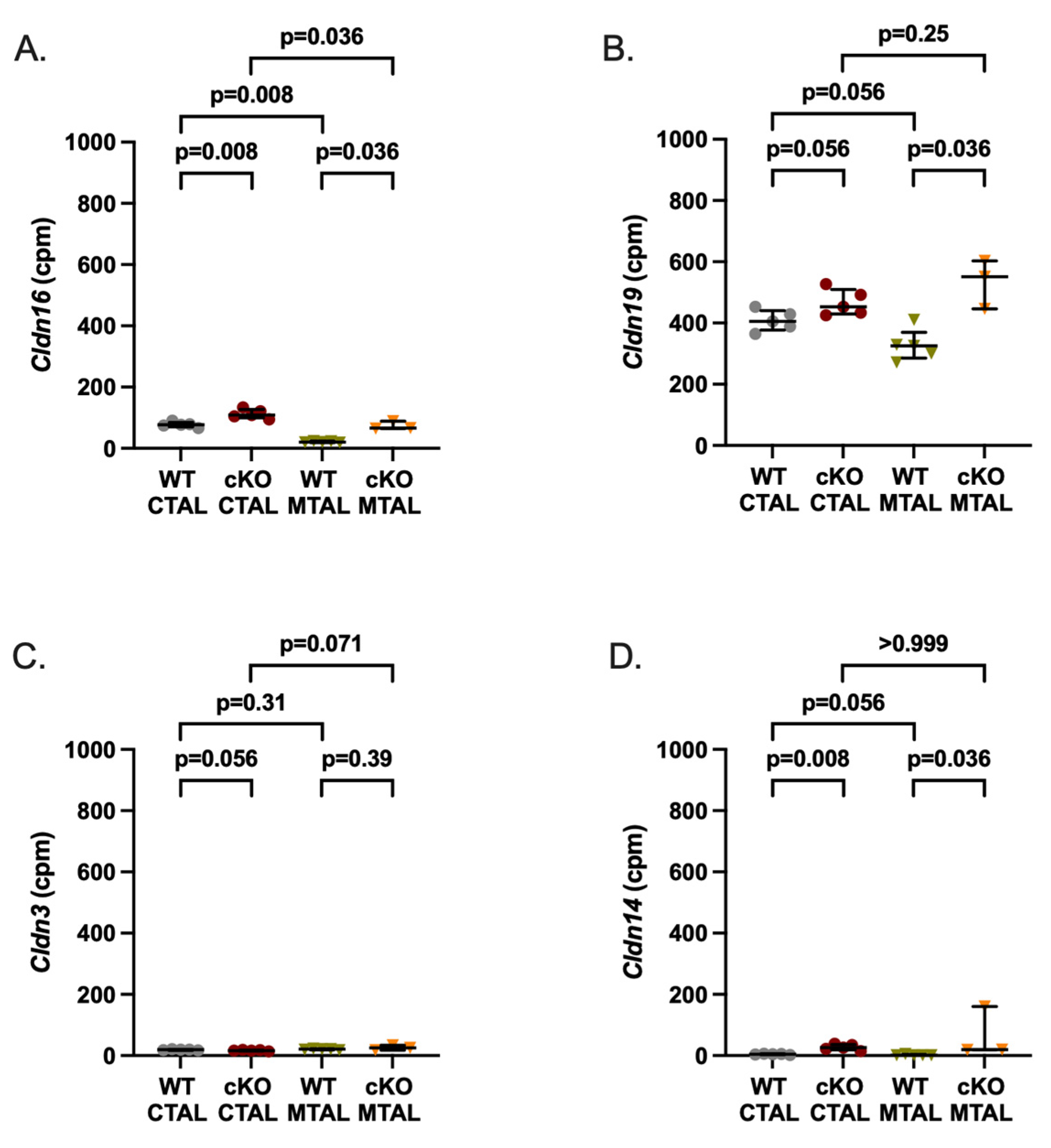
2.1. Differences between cKO MTAL and WT MTAL Transcriptomes
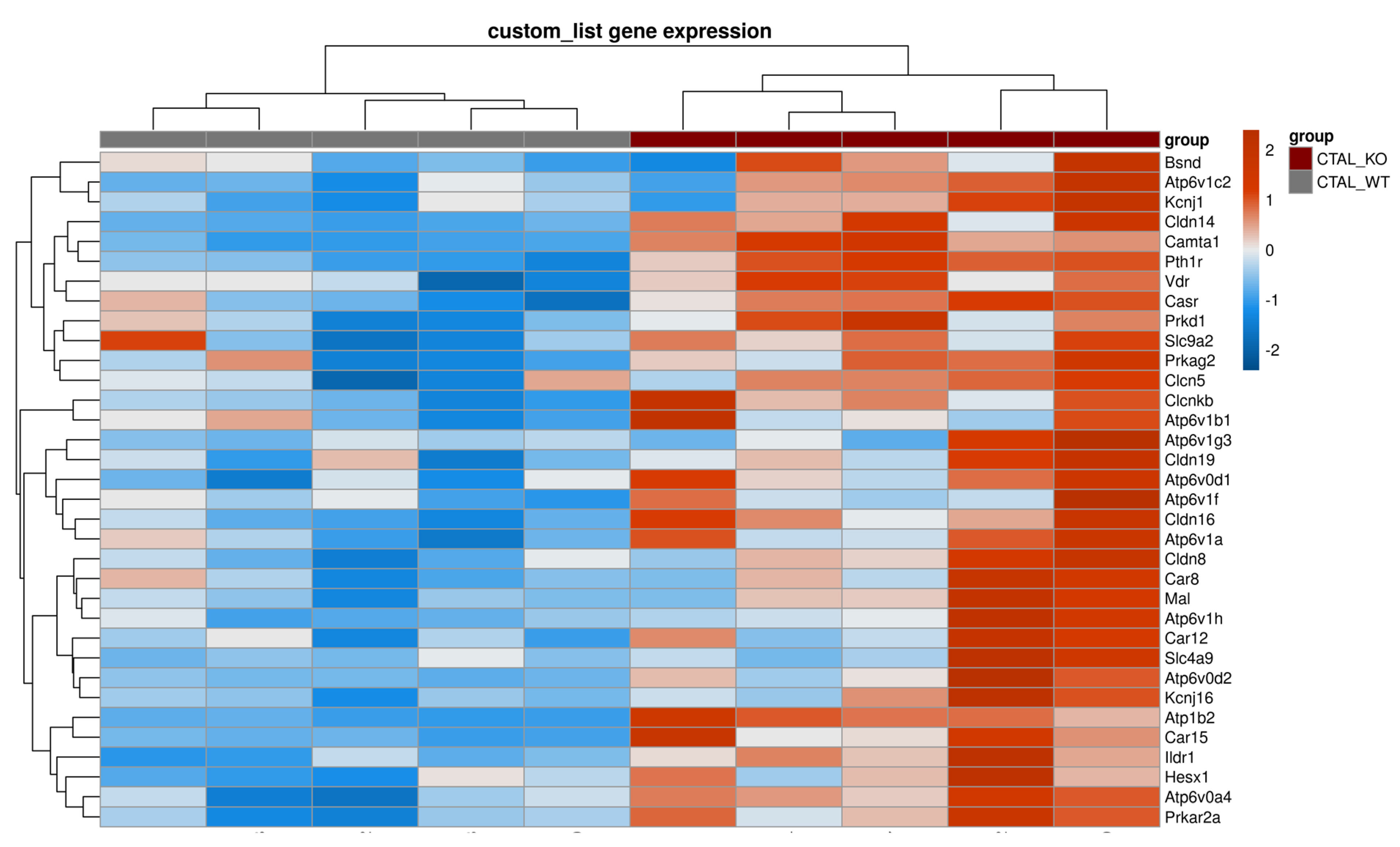
2.2. Differences between cKO CTALs and WT CTALs Transcriptomes
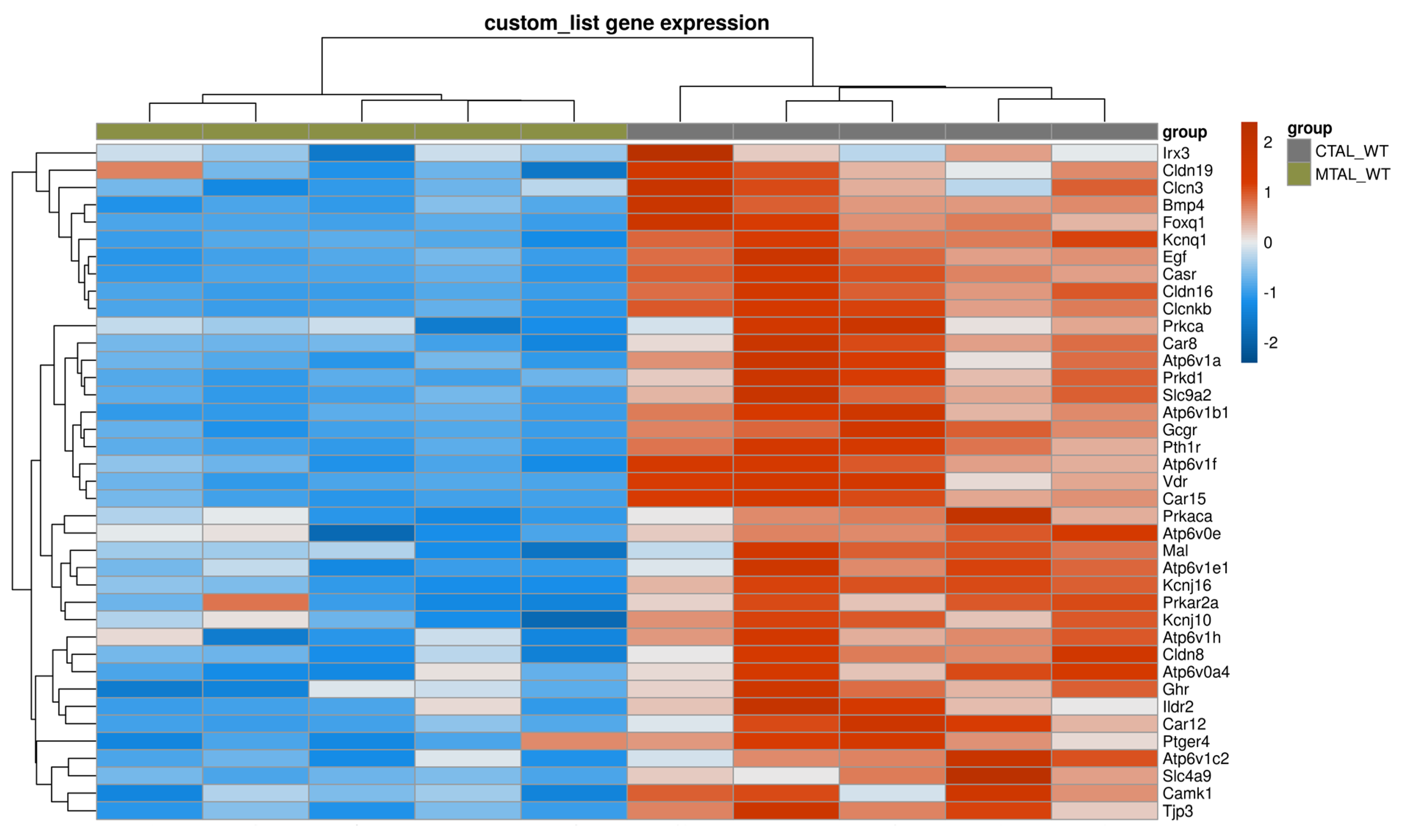
2.3. Differences between WT MTAL and WT CTAL Transcriptomes
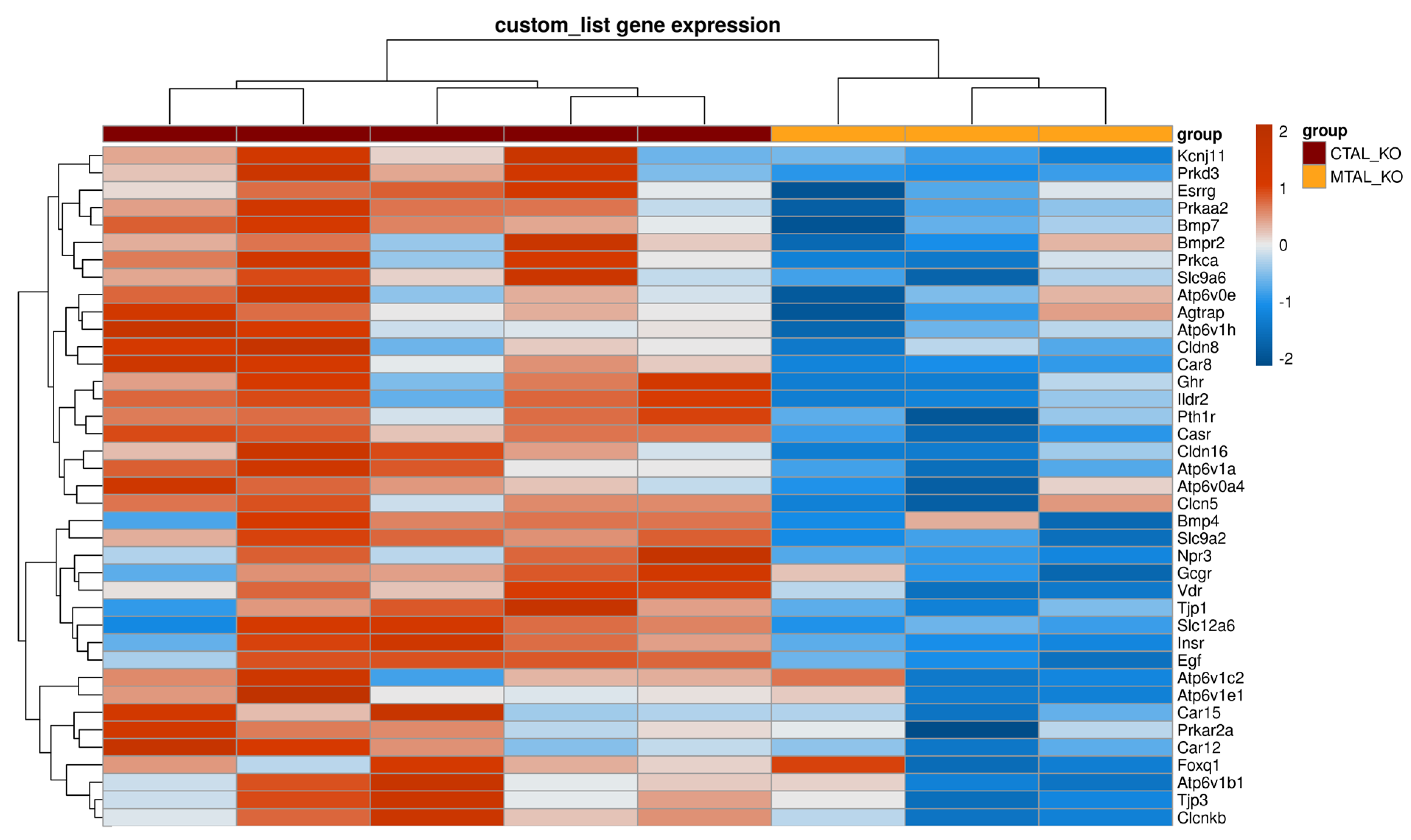
2.4. Differences between cKO MTAL and cKO CTAL Transcriptomes
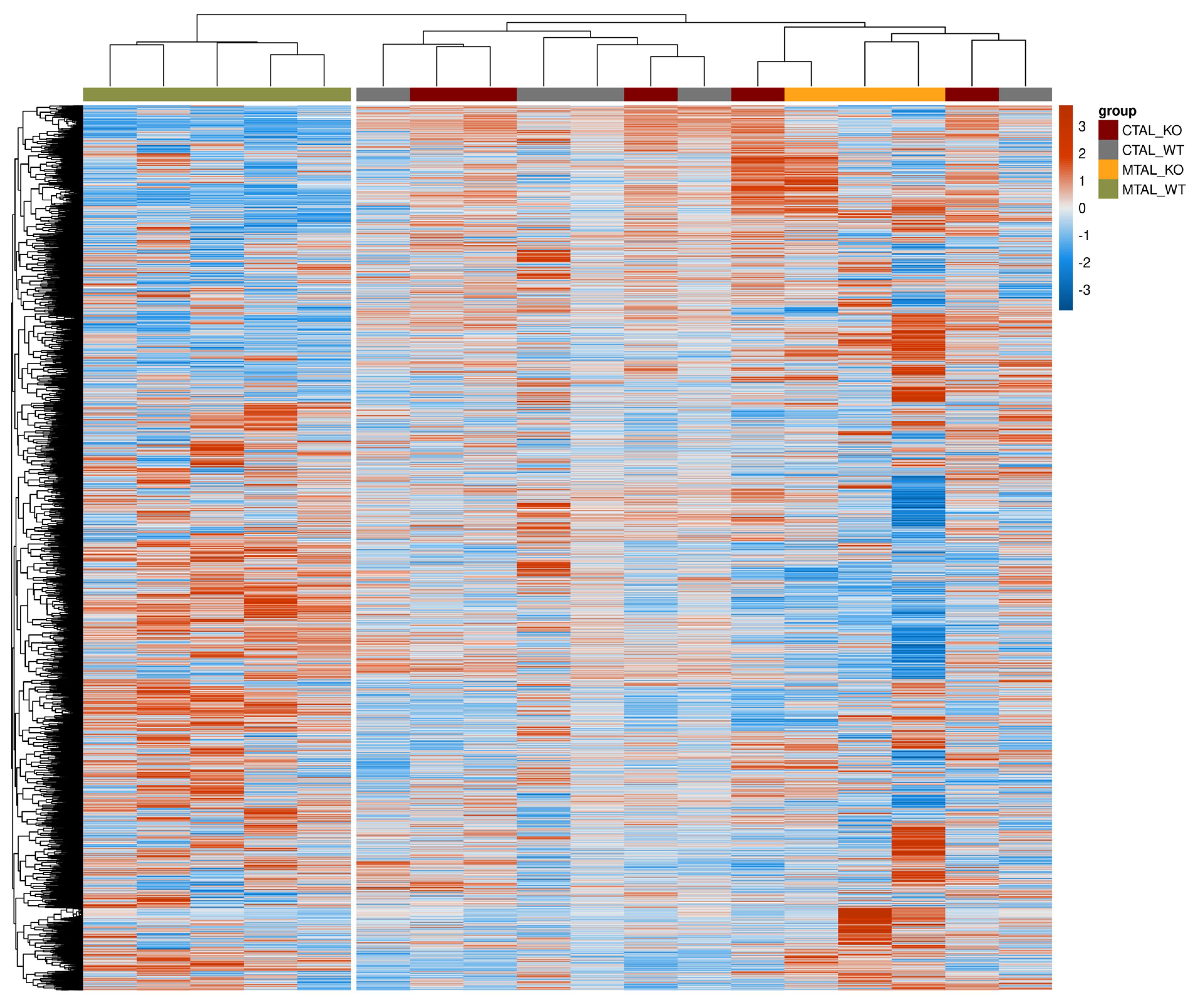
2.5. Differential Transcriptomes of cKO MTAL, WT MTAL, cKO CTAL and WT CTAL
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Microdissection and RNA Sequencing
4.3. Whole-Transcriptome RNA Sequencing
4.4. RNA-Seq Analyses
4.5. Other Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bankir, L.; Figueres, L.; Prot-Bertoye, C.; Bouby, N.; Crambert, G.; Pratt, J.H.; Houillier, P. Medullary and cortical thick ascending limb: Similarities and differences. Am. J. Physiol. Renal. Physiol. 2020, 318, F422–F442. [Google Scholar] [CrossRef]
- Dimke, H.; Schnermann, J. Axial and cellular heterogeneity in electrolyte transport pathways along the thick ascending limb. Acta Physiol. 2018, 223, e13057. [Google Scholar] [CrossRef]
- Konrad, M.; Nijenhuis, T.; Ariceta, G.; Bertholet-Thomas, A.; Calo, L.A.; Capasso, G.; Emma, F.; Schlingmann, K.P.; Singh, M.; Trepiccione, F.; et al. Diagnosis and management of Bartter syndrome: Executive summary of the consensus and recommendations from the European Rare Kidney Disease Reference Network Working Group for Tubular Disorders. Kidney Int. 2021, 99, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Bongers, E.; Shelton, L.M.; Milatz, S.; Verkaart, S.; Bech, A.P.; Schoots, J.; Cornelissen, E.A.M.; Bleich, M.; Hoenderop, J.G.J.; Wetzels, J.F.M.; et al. A Novel Hypokalemic-Alkalotic Salt-Losing Tubulopathy in Patients with CLDN10 Mutations. J. Am. Soc. Nephrol. 2017, 28, 3118–3128. [Google Scholar] [CrossRef]
- Klar, J.; Piontek, J.; Milatz, S.; Tariq, M.; Jameel, M.; Breiderhoff, T.; Schuster, J.; Fatima, A.; Asif, M.; Sher, M.; et al. Altered paracellular cation permeability due to a rare CLDN10B variant causes anhidrosis and kidney damage. PLoS Genet. 2017, 13, e1006897. [Google Scholar] [CrossRef]
- Hadj-Rabia, S.; Brideau, G.; Al-Sarraj, Y.; Maroun, R.C.; Figueres, M.L.; Leclerc-Mercier, S.; Olinger, E.; Baron, S.; Chaussain, C.; Nochy, D.; et al. Multiplex epithelium dysfunction due to CLDN10 mutation: The HELIX syndrome. Genet. Med. 2018, 20, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Meyers, N.; Nelson-Williams, C.; Malaga-Dieguez, L.; Kaufmann, H.; Loring, E.; Knight, J.; Lifton, R.P.; Trachtman, H. Hypokalemia Associated With a Claudin 10 Mutation: A Case Report. Am. J. Kidney Dis. 2019, 73, 425–428. [Google Scholar] [CrossRef]
- Alzahrani, A.S.; Hussein, M.; Alswailem, M.; Mouna, A.; Albalawi, L.; Moria, Y.; Jabbar, M.A.; Shi, Y.; Gunzel, D.; Dasouki, M. A novel claudin-10 mutation with a unique mechanism in two unrelated families with HELIX syndrome. Kidney Int. 2021, 100, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Breiderhoff, T.; Himmerkus, N.; Stuiver, M.; Mutig, K.; Will, C.; Meij, I.C.; Bachmann, S.; Bleich, M.; Willnow, T.E.; Muller, D. Deletion of claudin-10 (Cldn10) in the thick ascending limb impairs paracellular sodium permeability and leads to hypermagnesemia and nephrocalcinosis. Proc. Natl. Acad. Sci. USA 2012, 109, 14241–14246. [Google Scholar] [CrossRef]
- Breiderhoff, T.; Himmerkus, N.; Drewell, H.; Plain, A.; Gunzel, D.; Mutig, K.; Willnow, T.E.; Muller, D.; Bleich, M. Deletion of claudin-10 rescues claudin-16-deficient mice from hypomagnesemia and hypercalciuria. Kidney Int. 2018, 93, 580–588. [Google Scholar] [CrossRef]
- Blanchard, A.; Jeunemaitre, X.; Coudol, P.; Dechaux, M.; Froissart, M.; May, A.; Demontis, R.; Fournier, A.; Paillard, M.; Houillier, P. Paracellin-1 is critical for magnesium and calcium reabsorption in the human thick ascending limb of Henle. Kidney Int. 2001, 59, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.B.; Lu, Y.; Choate, K.A.; Velazquez, H.; Al-Sabban, E.; Praga, M.; Casari, G.; Bettinelli, A.; Colussi, G.; Rodriguez-Soriano, J.; et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science 1999, 285, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Prot-Bertoye, C.; Houillier, P. Claudins in Renal Physiology and Pathology. Genes 2020, 11, 290. [Google Scholar] [CrossRef]
- Konrad, M.; Schaller, A.; Seelow, D.; Pandey, A.V.; Waldegger, S.; Lesslauer, A.; Vitzthum, H.; Suzuki, Y.; Luk, J.M.; Becker, C.; et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am. J. Hum. Genet. 2006, 79, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Will, C.; Breiderhoff, T.; Thumfart, J.; Stuiver, M.; Kopplin, K.; Sommer, K.; Gunzel, D.; Querfeld, U.; Meij, I.C.; Shan, Q.; et al. Targeted deletion of murine Cldn16 identifies extra- and intrarenal compensatory mechanisms of Ca2+ and Mg2+ wasting. Am. J. Physiol. Renal. Physiol. 2010, 298, F1152–F1161. [Google Scholar] [CrossRef] [PubMed]
- Van Itallie, C.M.; Rogan, S.; Yu, A.; Vidal, L.S.; Holmes, J.; Anderson, J.M. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. Am. J. Physiol. Renal. Physiol. 2006, 291, F1288–F1299. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.B.; Baekgaard, A.; Hall, A.E.; Plata, C.; Xu, J.; Beier, D.R.; Gamba, G.; Hebert, S.C. Isoforms of the Na-K-2Cl cotransporter in murine TAL I. Molecular characterization and intrarenal localization. Am. J. Physiol. 1999, 276, F347–F358. [Google Scholar] [CrossRef]
- Payne, J.A.; Forbush, B., 3rd. Alternatively spliced isoforms of the putative renal Na-K-Cl cotransporter are differentially distributed within the rabbit kidney. Proc. Natl. Acad. Sci. USA 1994, 91, 4544–4548. [Google Scholar] [CrossRef]
- Kobayashi, K.; Uchida, S.; Mizutani, S.; Sasaki, S.; Marumo, F. Intrarenal and cellular localization of CLC-K2 protein in the mouse kidney. J. Am. Soc. Nephrol. 2001, 12, 1327–1334. [Google Scholar] [CrossRef]
- Mandon, B.; Siga, E.; Roinel, N.; de Rouffignac, C. Ca2+, Mg2+ and K+ transport in the cortical and medullary thick ascending limb of the rat nephron: Influence of transepithelial voltage. Pflügers Arch. 1993, 424, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.; Roinel, N.; de Rouffignac, C.; Wittner, M. Transepithelial Ca2+ and Mg2+ transport in the cortical thick ascending limb of Henle’s loop of the mouse is a voltage-dependent process. Ren. Physiol. Biochem. 1993, 16, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.; Wittner, M.; Nitschke, R.; Braitsch, R.; Greger, R.; Bailly, C.; Amiel, C.; Roinel, N.; de Rouffignac, C. Effects of parathyroid hormone and calcitonin on Na+, Cl−, K+, Mg2+ and Ca2+ transport in cortical and medullary thick ascending limbs of mouse kidney. Pflugers Arch. 1990, 417, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chou, C.L.; Knepper, M.A. A Comprehensive Map of mRNAs and Their Isoforms across All 14 Renal Tubule Segments of Mouse. J. Am. Soc. Nephrol. 2021, 32, 897–912. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Chou, C.L.; Knepper, M.A. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J. Am. Soc. Nephrol. 2015, 26, 2669–2677. [Google Scholar] [CrossRef] [PubMed]
- Limbutara, K.; Chou, C.L.; Knepper, M.A. Quantitative Proteomics of All 14 Renal Tubule Segments in Rat. J. Am. Soc. Nephrol. 2020, 31, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Muto, Y.; Wilson, P.C.; Ledru, N.; Wu, H.; Dimke, H.; Waikar, S.S.; Humphreys, B.D. Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat. Commun. 2021, 12, 2190. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chou, C.L.; Knepper, M.A. Targeted Single-Cell RNA-seq Identifies Minority Cell Types of Kidney Distal Nephron. J. Am. Soc. Nephrol. 2021, 32, 886–896. [Google Scholar] [CrossRef]
- Milatz, S.; Himmerkus, N.; Wulfmeyer, V.C.; Drewell, H.; Mutig, K.; Hou, J.; Breiderhoff, T.; Muller, D.; Fromm, M.; Bleich, M.; et al. Mosaic expression of claudins in thick ascending limbs of Henle results in spatial separation of paracellular Na+ and Mg2+ transport. Proc. Natl. Acad. Sci. USA 2017, 114, E219–E227. [Google Scholar] [CrossRef]
- Prot-Bertoye, C.; Griveau, C.; Skjodt, K.; Cheval, L.; Brideau, G.; Lievre, L.; Ferriere, E.; Arbaretaz, F.; Garbin, K.; Zamani, R.; et al. Differential localization patterns of claudin 10, 16, and 19 in human, mouse, and rat renal tubular epithelia. Am. J. Physiol. Renal. Physiol. 2021, 321, F207–F224. [Google Scholar] [CrossRef]
- Plain, A.; Wulfmeyer, V.C.; Milatz, S.; Klietz, A.; Hou, J.; Bleich, M.; Himmerkus, N. Corticomedullary difference in the effects of dietary Ca(2)(+) on tight junction properties in thick ascending limbs of Henle’s loop. Pflugers Arch. 2016, 468, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Loupy, A.; Ramakrishnan, S.K.; Wootla, B.; Chambrey, R.; de la Faille, R.; Bourgeois, S.; Bruneval, P.; Mandet, C.; Christensen, E.I.; Faure, H.; et al. PTH-independent regulation of blood calcium concentration by the calcium-sensing receptor. J. Clin. Investig. 2012, 122, 3355–3367. [Google Scholar] [CrossRef] [PubMed]
- Shareghi, G.R.; Agus, Z.S. Magnesium transport in the cortical thick ascending limb of Henle’s loop of the rabbit. J. Clin. Investig. 1982, 69, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Prot-Bertoye, C.; Lievre, L.; Houillier, P. The importance of kidney calcium handling in the homeostasis of extracellular fluid calcium. Pflugers Arch. 2022, 474, 885–900. [Google Scholar] [CrossRef]
- Yamamoto, M.; Kawanobe, Y.; Takahashi, H.; Shimazawa, E.; Kimura, S.; Ogata, E. Vitamin D deficiency and renal calcium transport in the rat. J. Clin. Investig. 1984, 74, 507–513. [Google Scholar] [CrossRef]
- Frische, S.; Alexander, R.T.; Ferreira, P.; Tan, R.S.G.; Wang, W.; Svenningsen, P.; Skjodt, K.; Dimke, H. Localization and regulation of claudin-14 in experimental models of hypercalcemia. Am. J. Physiol. Renal. Physiol. 2021, 320, F74–F86. [Google Scholar] [CrossRef] [PubMed]
- Dimke, H.; Desai, P.; Borovac, J.; Lau, A.; Pan, W.; Alexander, R.T. Activation of the Ca(2+)-sensing receptor increases renal claudin-14 expression and urinary Ca(2+) excretion. Am. J. Physiol. Renal. Physiol. 2013, 304, F761–F769. [Google Scholar] [CrossRef]
- Holmes, J.L.; Van Itallie, C.M.; Rasmussen, J.E.; Anderson, J.M. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Exp. Patterns 2006, 6, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, V.; Quiros, M.; Nusrat, A. Intestinal epithelial claudins: Expression and regulation in homeostasis and inflammation. Ann. N. Y. Acad. Sci. 2017, 1397, 66–79. [Google Scholar] [CrossRef]
- Cheval, L.; Pierrat, F.; Rajerison, R.; Piquemal, D.; Doucet, A. Of mice and men: Divergence of gene expression patterns in kidney. PLoS ONE 2012, 7, e46876. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 September 2023).
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brideau, G.; Cheval, L.; Griveau, C.; Ling, W.-M.E.; Lievre, L.; Crambert, G.; Müller, D.; Broćić, J.; Cherchame, E.; Houillier, P.; et al. Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells. Int. J. Mol. Sci. 2024, 25, 4008. https://doi.org/10.3390/ijms25074008
Brideau G, Cheval L, Griveau C, Ling W-ME, Lievre L, Crambert G, Müller D, Broćić J, Cherchame E, Houillier P, et al. Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells. International Journal of Molecular Sciences. 2024; 25(7):4008. https://doi.org/10.3390/ijms25074008
Chicago/Turabian StyleBrideau, Gaelle, Lydie Cheval, Camille Griveau, Wung-Man Evelyne Ling, Loïc Lievre, Gilles Crambert, Dominik Müller, Jovana Broćić, Emeline Cherchame, Pascal Houillier, and et al. 2024. "Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells" International Journal of Molecular Sciences 25, no. 7: 4008. https://doi.org/10.3390/ijms25074008
APA StyleBrideau, G., Cheval, L., Griveau, C., Ling, W.-M. E., Lievre, L., Crambert, G., Müller, D., Broćić, J., Cherchame, E., Houillier, P., & Prot-Bertoye, C. (2024). Claudin-10 Expression and the Gene Expression Pattern of Thick Ascending Limb Cells. International Journal of Molecular Sciences, 25(7), 4008. https://doi.org/10.3390/ijms25074008





