Nitrogen Sources Reprogram Carbon and Nitrogen Metabolism to Promote Andrographolide Biosynthesis in Andrographis paniculata (Burm.f.) Nees Seedlings
Abstract
1. Introduction
2. Results
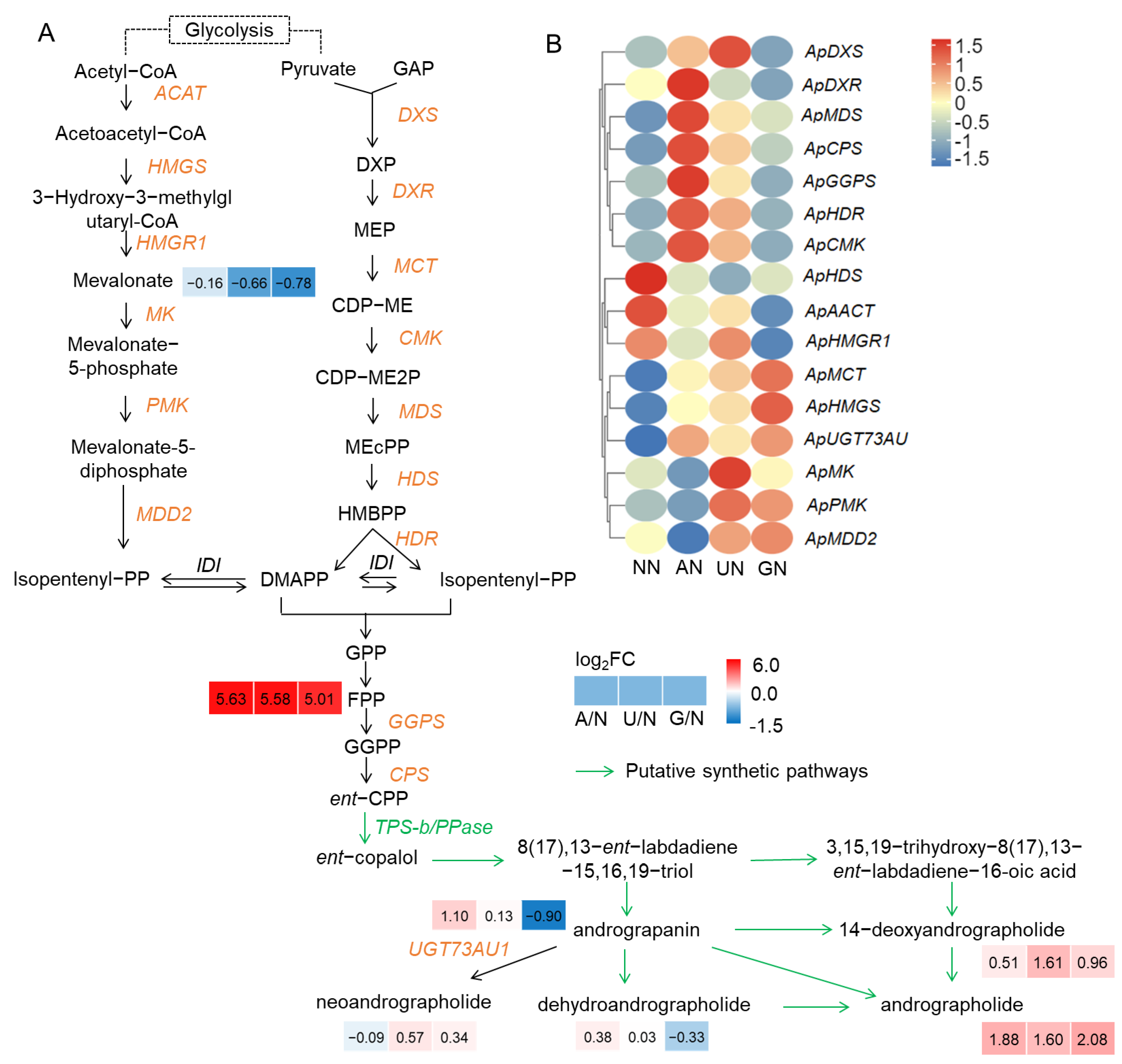
2.1. Diterpenoid Lactone Compounds Affected by N Sources
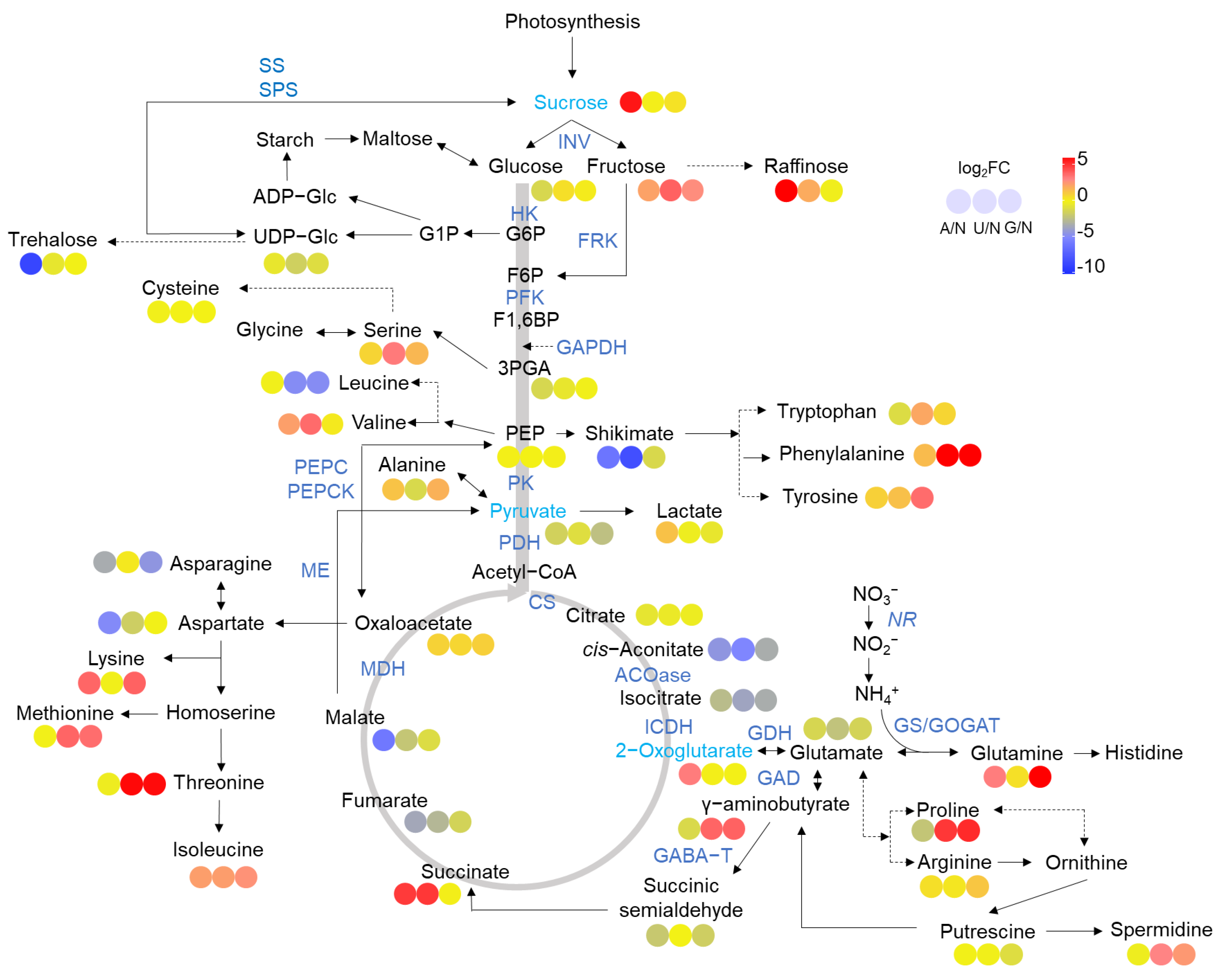
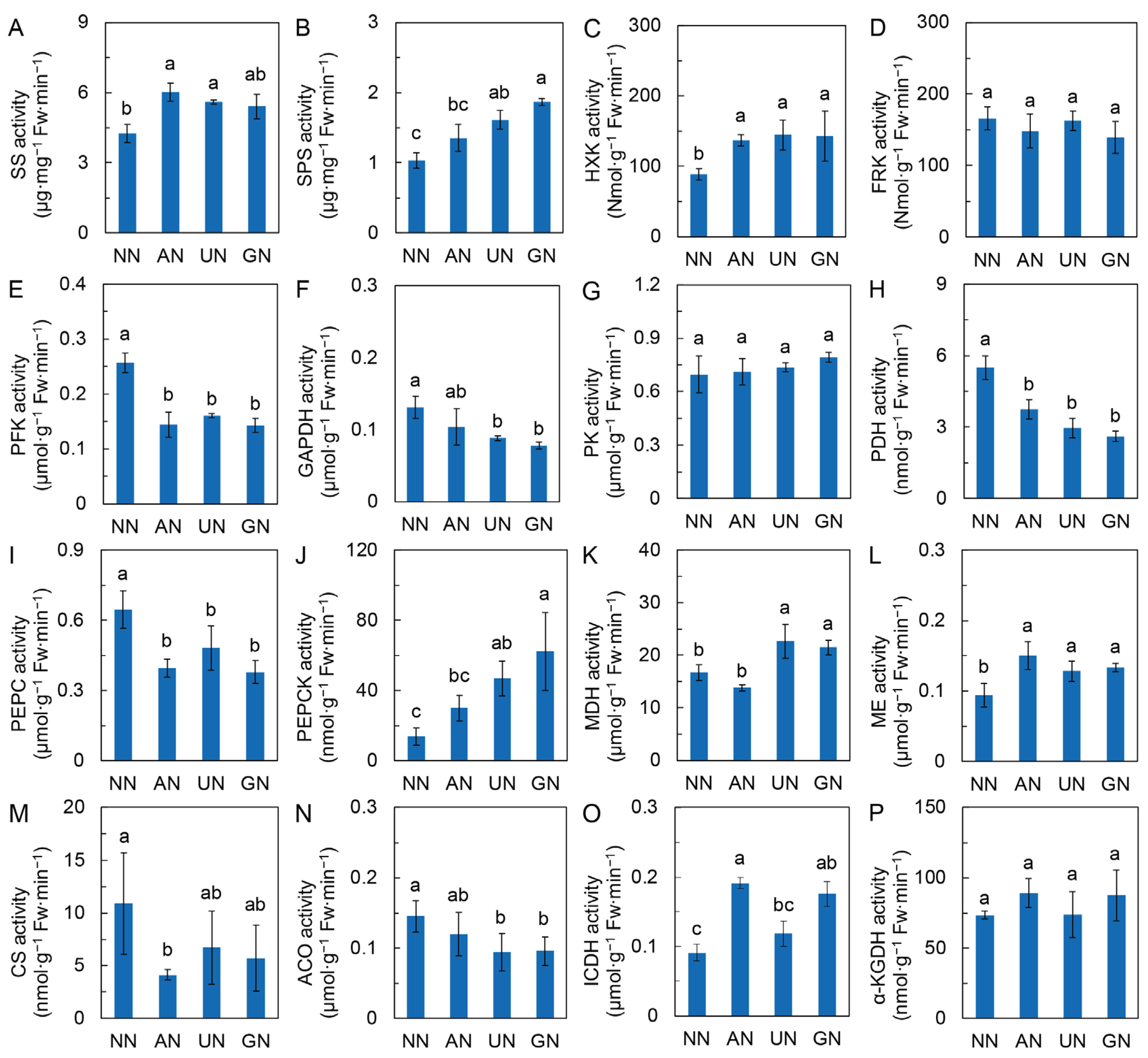
2.2. Glycolysis and the TCA Cycle
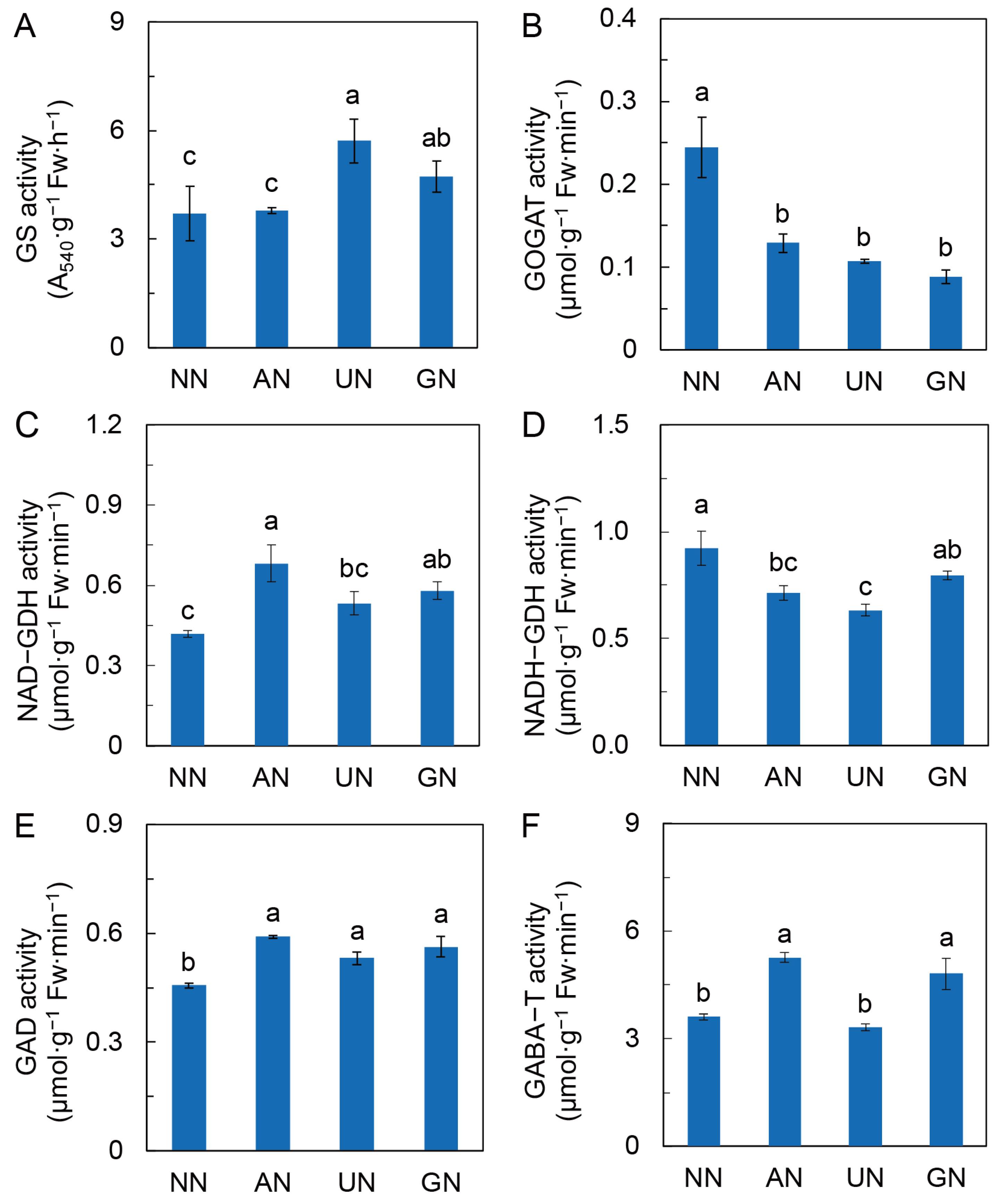
2.3. N Assimilation and Amino Acids Metabolism
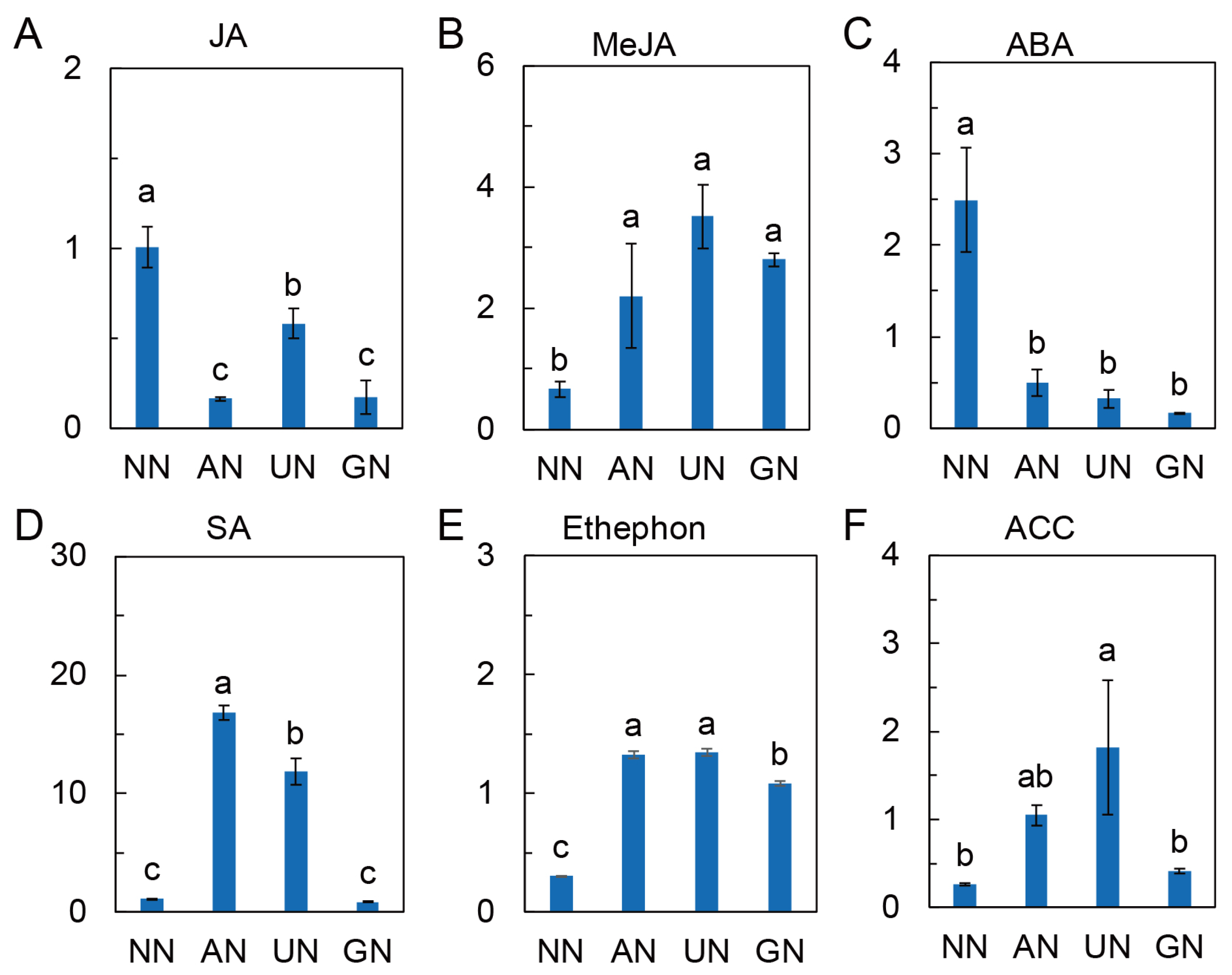
2.4. Phytohormone Levels Affected by N Sources
3. Discussion
3.1. N Sources Affect Diterpenoid Lactone Components Accumulation
3.2. N Sources Reprogram C and N Metabolism
3.3. The Center Metabolic Pathway in Mediating Reprogramming of C Metabolism
3.4. Plant Hormones Involve in N Source Regulated Andrographolide Synthesis
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Photosynthesis and Leaf Nitrogen Allocation Measurements
4.3. Metabolites Analysis
4.4. Enzymes Assays
4.5. RNA Extraction, cDNA Synthesis, and q-PCR Profiling
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Li, Y.; Kong, D.; Fu, Y.; Sussman, M.R.; Wu, H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020, 148, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H.; Takei, K.; Hirose, N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006, 11, 440–448. [Google Scholar] [CrossRef]
- Nchu, F.; Matanzima, Y.; Laubscher, C.P. Prospects of N fertilization in medicinal plants cultivation. In Nitrogen in Agriculture—Updates; Amanullah, A., Fahad, S., Eds.; IntechOpen: London, UK, 2018; pp. 209–222. [Google Scholar] [CrossRef]
- Ye, J.Y.; Tian, W.H.; Jin, C.W. Nitrogen in plants: From nutrition to the modulation of abiotic stress adaptation. Stress Biol. 2022, 2, 4. [Google Scholar] [CrossRef]
- Farzadfar, S.; Knight, J.D.; Congreves, K.A. Soil organic nitrogen: An overlooked but potentially significant contribution to crop nutrition. Plant Soil 2021, 462, 7–23. [Google Scholar] [CrossRef]
- Li, X.; Yan, X.F.; Liu, J.F. Effect of nitrogen forms on berberine, jatrorrhizine and palmatine content in corktree seedlings. Acta Ecol. Sin. 2005, 25, 2159–2164. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, T.; Xu, X.; Yang, Y.; Tong, H.; Mur, L.A.J.; Yuan, H. Transcriptomic characterization of nitrate-enhanced stevioside glycoside synthesis in stevia (Stevia rebaudiana Bertoni). Int. J. Mol. Sci. 2021, 22, 8549. [Google Scholar] [CrossRef]
- Biesiada, A.; Woloszczak, E.; Sokolletowska, A.; Kucharska, A.Z.; Nawirskaolszanska, A. The effect of nitrogen form and dose on yield, chemical composition and antioxidant activity of stinging nettle (Urtica dioica L.). Herba Pol. 2009, 55, 84–93. [Google Scholar]
- Sui, L.; Wang, K.; Yi, J.; Ta, H.; Wei, L.; Wei, H. Effect of different nitrogen forms on growth and quality of Perilla Frutescens (L.) Britt. Chin. J. Soil Sci. 2018, 49, 667–672. [Google Scholar] [CrossRef]
- Zhong, C.; Jian, S.F.; Chen, D.L.; Huang, X.J.; Miao, J.H. Organic nitrogen sources promote andrographolide biosynthesis by reducing nitrogen metabolism and increasing carbon accumulation in Andrographis paniculata. Plant Physiol. Biochem. 2021, 164, 82–91. [Google Scholar] [CrossRef]
- Kováčik, J.; Klejdus, B. Induction of phenolic metabolites and physiological changes in chamomile plants in relation to nitrogen nutrition. Food Chem. 2014, 142, 334–341. [Google Scholar] [CrossRef]
- Bruce, S.O.; Onyegbule, F.A. Biosynthesis of natural products. In Bioactive Compounds—Biosynthesis, Characterization and Applications; Zepka, L.Q., Nascimento, T.C.d., Jacob-Lopes, E., Eds.; IntechOpen: London, UK, 2021. [Google Scholar] [CrossRef]
- Chen, Y.; Bai, Y.; Zhang, Z.; Zhang, Y.; Jiang, Y.; Wang, S.; Wang, Y.; Sun, Z. Transcriptomics and metabolomics reveal the primary and secondary metabolism changes in Glycyrrhiza uralensis with different forms of nitrogen utilization. Front. Plant Sci. 2023, 14, 1229253. [Google Scholar] [CrossRef]
- Feng, H.; Fan, X.; Miller, A.J.; Xu, G. Plant nitrogen uptake and assimilation: Regulation of cellular pH homeostasis. J. Exp. Bot. 2020, 71, 4380–4392. [Google Scholar] [CrossRef]
- Hachiya, T.; Inaba, J.; Wakazaki, M.; Sato, M.; Toyooka, K.; Miyagi, A.; Kawai-Yamada, M.; Sugiura, D.; Nakagawa, T.; Kiba, T.; et al. Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 2021, 12, 4944. [Google Scholar] [CrossRef]
- Sakano, K. Metabolic regulation of pH in plant cells: Role of cytoplasmic pH in defense reaction and secondary metabolism. Int. Rev. Cytol. 2001, 206, 1–44. [Google Scholar] [CrossRef]
- Fritz, C.; Palacios-Rojas, N.; Feil, R.; Stitt, M. Regulation of secondary metabolism by the carbon–nitrogenstatus in tobacco: Nitrate inhibits large sectors of phenylpropanoid metabolism. Plant J. 2006, 46, 533–548. [Google Scholar] [CrossRef]
- Xu, X.J.; Li, Q.Y.; Song, X.H.; Shen, Q.R.; Dong, C.X. Dynamic regulation of nitrogen and organic acid metabolism of cherry tomato fruit as affected by different nitrogen forms. Pedosphere 2012, 22, 67–78. [Google Scholar] [CrossRef]
- Huang, H.; Yao, Q.; Xia, E.; Gao, L. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J. Agric. Food Chem. 2018, 66, 9828–9838. [Google Scholar] [CrossRef]
- Coleto, I.; Vega-Mas, I.; Glauser, G.; González-Moro, M.B.; Marino, D.; Ariz, I. New insights on Arabidopsis thaliana root adaption to ammonium nutrition by the use of a quantitative proteomic approach. Int. J. Mol. Sci. 2019, 20, 814. [Google Scholar] [CrossRef]
- Sun, Y.; Hou, M.; Mur, L.A.J.; Yang, Y.; Zhang, T.; Xu, X.; Huang, S.; Tong, H. Nitrogen drives plant growth to the detriment of leaf sugar and steviol glycosides metabolisms in stevia (Stevia rebaudiana Bertoni). Plant Physiol. Biochem. 2019, 141, 240–249. [Google Scholar] [CrossRef]
- Ibrahim, M.H.; Jaafar, H.Z. The relationship of nitrogen and C/N ratio with secondary metabolites levels and antioxidant activities in three varieties of Malaysian kacip Fatimah (Labisia pumila Blume). Molecules 2011, 16, 5514–5526. [Google Scholar] [CrossRef]
- Ibrahim, M.H.; Jaafar, H.Z. Involvement of carbohydrate, protein and phenylanine ammonia lyase in up-regulation of secondary metabolites in Labisia pumila under various CO2 and N2 levels. Molecules 2011, 16, 4172–4190. [Google Scholar] [CrossRef]
- Morkunas, I.; Ratajczak, L. The role of sugar signaling in plant defense responses against fungal pathogens. Acta Physiol. Plant. 2014, 36, 1607–1619. [Google Scholar] [CrossRef]
- Jian, S.F.; Huang, X.J.; Yang, X.N.; Zhong, C.; Miao, J.H. Sulfur regulates the trade-off between growth and andrographolide accumulation via nitrogen metabolism in Andrographis paniculata. Front. Plant Sci. 2021, 12, 687954. [Google Scholar] [CrossRef]
- Sun, J.; Guo, H.; Liu, M.; Chen, M.; Zhu, M.; Liu, D.; Tao, J. Histology and transcriptomic profiling reveal the dynamics of seed coat and endosperm formation in tree peony (Paeonia ostii). Hortic. Res. 2022, 9, uhac106. [Google Scholar] [CrossRef]
- Subramanian, R.; Asmawi, M.Z.; Sadikun, A. A bitter plant with a sweet future? A comprehensive review of an oriental medicinal plant: Andrographis paniculata. Phytochem. Rev. 2012, 11, 39–75. [Google Scholar] [CrossRef]
- Talei, D.; Valdiani, A.; Rafii, M.Y.; Maziah, M. Proteomic analysis of the salt-responsive leaf and root proteins in the anticancer plant Andrographis paniculata Nees. PLoS ONE 2014, 9, e112907. [Google Scholar] [CrossRef]
- Valdiani, A.; Talei, D.; Tan, S.G.; Kadir, M.A.; Maziah, M.; Rafii, M.Y.; Sagineedu, S.R. A classical genetic solution to enhance the biosynthesis of anticancer phytochemicals in Andrographis paniculata Nees. PLoS ONE 2014, 9, e87034. [Google Scholar] [CrossRef]
- Shi, T.H.; Huang, Y.L.; Chen, C.C.; Pi, W.C.; Hsu, Y.L.; Lo, L.C.; Chen, W.Y.; Fu, S.L.; Lin, C.H. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochem. Biophys. Res. Commun. 2020, 533, 467–473. [Google Scholar] [CrossRef]
- Intharuksa, A.; Arunotayanun, W.; Yooin, W.; Sirisa-Ard, P. A comprehensive review of Andrographis paniculata (Burm. f.) Nees and its constituents as potential lead compounds for COVID-19 drug discovery. Molecules 2022, 27, 4479. [Google Scholar] [CrossRef]
- Tanuja; Parani, M. Whole transcriptome analysis identifies full-length genes for neoandrographolide biosynthesis from Andrographis alata, an alternate source for antiviral compounds. Gene 2023, 851, 146981. [Google Scholar] [CrossRef]
- Sun, W.; Leng, L.; Yin, Q.; Xu, M.; Huang, M.; Xu, Z.; Zhang, Y.; Yao, H.; Wang, C.; Xiong, C.; et al. The genome of the medicinal plant Andrographis paniculata provides insight into the biosynthesis of the bioactive diterpenoid neoandrographolide. Plant J. 2019, 97, 841–857. [Google Scholar] [CrossRef]
- Liang, Y.; Chen, S.; Wei, K.; Yang, Z.; Duan, S.; Du, Y.; Qu, P.; Miao, J.; Chen, W.; Dong, Y. Chromosome level genome assembly of Andrographis paniculata. Front. Genet. 2020, 11, 701. [Google Scholar] [CrossRef]
- Sharma, S.N.; Jha, Z.; Sinha, R.K.; Geda, A.K. Jasmonate-induced biosynthesis of andrographolide in Andrographis paniculata. Physiol. Plant. 2015, 153, 221–229. [Google Scholar] [CrossRef]
- Srivastava, N.; Akhila, A. Biosynthesis of andrographolide in Andrographis paniculata. Phytochemistry 2010, 71, 1298–1304. [Google Scholar] [CrossRef]
- Sun, M.; Xu, S.; Li, J.; Gu, Y.; Mei, Y.; Zhang, W.; Zhou, F.; Wang, J. Transcriptome analysis and discovery of genes related to andrographolide synthesis of Andrographis paniculata under UV-C treatment. J. South. Agric. 2022, 53, 618–627. [Google Scholar] [CrossRef]
- Garg, A.; Agrawal, L.; Misra, R.C.; Sharma, S.; Ghosh, S. Andrographis paniculata transcriptome provides molecular insights into tissue-specific accumulation of medicinal diterpenes. BMC Genom. 2015, 16, 659. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Ahammed, G.J.; Li, Z.X.; Wei, J.P.; Shen, C.; Yan, P.; Zhang, L.P.; Han, W.Y. Stimulation in primary and secondary metabolism by elevated carbon dioxide alters green tea quality in Camellia sinensis L. Sci. Rep. 2017, 7, 7937. [Google Scholar] [CrossRef]
- He, Z.; Webster, S.; He, S.Y. Growth-defense trade-offs in plants. Curr. Biol. 2022, 32, R634–R639. [Google Scholar] [CrossRef]
- Larbat, R.; Robin, C.; Lillo, C.; Drengstig, T.; Ruoff, P. Modeling the diversion of primary carbon flux into secondary metabolism under variable nitrate and light/dark conditions. J. Theor. Biol. 2016, 402, 144–157. [Google Scholar] [CrossRef]
- Si, C.; Shi, C.; Liu, H.; Zhan, X.; Liu, Y. Effects of nitrogen forms on carbohydrate metabolism and storage-root formation of sweet potato. J. Plant Nutr. Soil Sci. 2018, 181, 419–428. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, C.; Jia, R.; Yang, N.; Jin, L.; Zhu, L.; Ma, B.; Yao, Y.X.; Ma, F.; Li, M. Malate metabolism mediated by the cytoplasmic malate dehydrogenase gene MdcyMDH affects sucrose synthesis in apple fruit. Hortic. Res. 2022, 9, uhac194. [Google Scholar] [CrossRef]
- Nunes-Nesi, A.; Fernie, A.R.; Stitt, M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 2010, 3, 973–996. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G.; Hodges, M. Respiration and nitrogen assimilation: Targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 2011, 62, 1467–1482. [Google Scholar] [CrossRef]
- Medeiros, D.B.; Aarabi, F.; Rivas, F.J.M.; Fernie, A.R. The knowns and unknowns of intercellular partitioning of carbon and nitrogen, with focus on the organic acid-mediated interplay between mitochondrion and chloroplast. J. Plant Physiol. 2021, 266, 153521. [Google Scholar] [CrossRef]
- Vega-Mas, I.; Cukier, C.; Coleto, I.; González-Murua, C.; Limami, A.M.; González-Moro, M.B.; Marino, D. Isotopic labelling reveals the efficient adaptation of wheat root TCA cycle flux modes to match carbon demand under ammonium nutrition. Sci. Rep. 2019, 9, 8925. [Google Scholar] [CrossRef]
- Nomura, M.; Mai, H.T.; Fujii, M.; Hata, S.; Izui, K.; Tajima, S. Phosphoenolpyruvate carboxylase plays a crucial role in limiting nitrogen fixation in Lotus japonicus nodules. Plant Cell Physiol. 2006, 47, 613–621. [Google Scholar] [CrossRef]
- Foyer, C.H.; Parry, M.; Noctor, G. Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 2003, 54, 585–593. [Google Scholar] [CrossRef]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef]
- Huergo, L.F.; Chandra, G.; Merrick, M. PII signal transduction proteins: Nitrogen regulation and beyond. FEMS Microbiol. Rev. 2013, 37, 251–283. [Google Scholar] [CrossRef]
- Gent, L.; Forde, B.G. How do plants sense their nitrogen status? J. Exp. Bot. 2017, 68, 2531–2539. [Google Scholar] [CrossRef]
- Chellamuthu, V.R.; Ermilova, E.; Lapina, T.; Lüddecke, J.; Minaeva, E.; Herrmann, C.; Hartmann, M.D.; Forchhammer, K. A widespread glutamine-sensing mechanism in the plant kingdom. Cell 2014, 159, 1188–1199. [Google Scholar] [CrossRef]
- Liu, C.; Xue, Z.; Tang, D.; Shen, Y.; Shi, W.; Ren, L.; Du, G.; Li, Y.; Cheng, Z. Ornithine δ-aminotransferase is critical for floret development and seed setting through mediating nitrogen reutilization in rice. Plant J. 2018, 96, 842–854. [Google Scholar] [CrossRef]
- Khan, M.I.R.; Jalil, S.U.; Chopra, P.; Chhillar, H.; Ferrante, A.; Khan, N.A.; Ansari, M.I. Role of GABA in plant growth, development and senescence. Plant Gene 2021, 26, 100283. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Huang, X.; Cui, J.; Shabala, L.; Zhou, M.; Yu, M.; Shabala, S. Hypoxia-induced increase in GABA content is essential for restoration of membrane potential and preventing ROS-induced disturbance to ion homeostasis. Plant Commun. 2021, 2, 100188. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Tyerman, S.D.; Gilliham, M.; Xu, B. γ-Aminobutyric acid (GABA) signalling in plants. Cell. Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar] [CrossRef]
- Ma, W.; Xu, L.; Gao, S.; Lyu, X.; Cao, X.; Yao, Y. Melatonin alters the secondary metabolite profile of grape berry skin by promoting VvMYB14-mediated ethylene biosynthesis. Hortic. Res. 2021, 8, 43. [Google Scholar] [CrossRef]
- Guan, R.; Xu, S.; Lu, Z.; Su, L.; Zhang, L.; Sun, W.; Zhang, Y.; Jiang, C.; Liu, Z.; Duan, L.; et al. Genomic characterization of bZIP transcription factors related to andrographolide biosynthesis in Andrographis paniculata. Int. J. Biol. Macromol. 2022, 223, 1619–1631. [Google Scholar] [CrossRef]
- Nandy, S.; Das, T.; Dey, A. Role of jasmonic acid and salicylic acid signaling in secondary metabolite production. In Jasmonates and Salicylates Signaling in Plants. Signaling and Communication in Plants; Aftab, T., Yusuf, M., Eds.; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Ali, B. Salicylic acid: An efficient elicitor of secondary metabolite production in plants. Biocatal. Agric. Biotechnol. 2021, 31, 101884. [Google Scholar] [CrossRef]
- Ding, M.; Zhang, B.; Zhang, S.; Hao, R.R.; Xia, Y.; Ma, P.; Dong, J. The SmNPR4-SmTGA5 module regulates SA-mediated phenolic acid biosynthesis in Salvia miltiorrhiza hairy roots. Hortic. Res. 2023, 10, uhad066. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Bhuiyan, T.F.; Anee, T.I.; Inafuku, M.; Oku, H.; Fujita, M. Salicylic acid: An all-rounder in regulating abiotic stress responses in plants. In Phytohormones—Signaling Mechanisms and Crosstalk in Plant Development and Stress Responses; El-Esawi, M., Ed.; InTech: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef]
- Ding, S.; Shao, X.; Li, J.; Ahammed, G.J.; Yao, Y.; Ding, J.; Hu, Z.; Yu, J.; Shi, K. Nitrogen forms and metabolism affect plant defence to foliar and root pathogens in tomato. Plant Cell Environ. 2021, 44, 1596–1610. [Google Scholar] [CrossRef]
- Fang, R.; Zhao, H.; Liao, Y.; Tang, C.; Wu, F.; Zhu, Y.; Pang, Y.; Lu, G.; Wang, X.; Yang, R.; et al. Dual regulating effects of ethylene on the formation of plant secondary metabolites. Chin. Bull. Bot. 2014, 49, 626–639. [Google Scholar] [CrossRef]
- Baharudin, N.F.; Osman, N.I. Plant development, stress responses, and secondary metabolism under ethylene regulation. Plant Stress 2023, 7, 100146. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Pollmann, S.; Müller, M. Ethylene and jasmonates signaling network mediating secondary metabolites under abiotic stress. Int. J. Mol. Sci. 2023, 24, 5990. [Google Scholar] [CrossRef] [PubMed]
- Jian, S.; Liao, Q.; Song, H.; Liu, Q.; Lepo, J.E.; Guan, C.; Zhang, J.; Ismail, A.M.; Zhang, Z. NRT1.1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 2018, 178, 1473–1488. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Q.; Wang, X.; Zhang, J.; Ismail, A.M.; Zhang, Z. Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell Environ. 2021, 44, 3576–3588. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Ma, T.; Xian, W.; Hu, B.; Chu, C. Interplay between ethylene and nitrogen nutrition: How ethylene orchestrates nitrogen responses in plants. J. Integr. Plant Biol. 2023, 65, 399–407. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Liu, C.; Cui, G.; Qu, L.; Bao, M.; Wang, J.; Chan, Z.; Wang, Y. Integrating physiological and metabolites analysis to identify ethylene involvement in petal senescence in Tulipa gesneriana. Plant Physiol. Biochem. 2020, 149, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Wang, Y.; Zhang, Z.H.; Zu, Y.G.; Efferth, T.; Tang, Z.H. The combined effects of ethylene and MeJA on metabolic profiling of phenolic compounds in Catharanthus roseus revealed by metabolomics analysis. Front. Physiol. 2016, 7, 217. [Google Scholar] [CrossRef] [PubMed]
- De Boer, K.; Tilleman, S.; Pauwels, L.; Vanden Bossche, R.; De Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef]
- Farquhar, G.D.; von Caemmerer, S.; Berry, J.A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 1980, 149, 78–90. [Google Scholar] [CrossRef]
- Long, S.P.; Bernacchi, C.J. Gas exchange measurements, what can they tell us about the underlying limitations to photosynthesis? Procedures and sources of error. J. Exp. Bot. 2003, 54, 2393–2401. [Google Scholar] [CrossRef] [PubMed]
- Santoni, S.; Bonifacio, E.; Zanini, E. Indophenol blue colorimetric method for measuring cation exchange capacity in sandy soils. Commun. Soil Sci. Plant Anal. 2001, 32, 2519–2530. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta (BBA)-Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Zhong, C.; Cao, X.; Hu, J.; Zhu, L.; Zhang, J.; Huang, J.; Jin, Q. Nitrogen metabolism in adaptation of photosynthesis to water stress in rice grown under different nitrogen levels. Front. Plant Sci. 2017, 8, 1079. [Google Scholar] [CrossRef]
- Kulichikhin, K.Y.; Chirkova, T.V.; Fagerstedt, K.V. Activity of biochemical pH-stat enzymes in cereal root tips under oxygen deficiency. Russ. J. Plant Physiol. 2009, 56, 377–388. [Google Scholar] [CrossRef]
- Deewatthanawong, R.; Nock, J.F.; Watkins, C.B. γ-Aminobutyric acid (GABA) accumulation in four strawberry cultivars in response to elevated CO2 storage. Postharvest Biol. Technol. 2010, 57, 92–96. [Google Scholar] [CrossRef]
- Zhong, C.; Bai, Z.G.; Zhu, L.F.; Zhang, J.H.; Zhu, C.Q.; Huang, J.L.; Jin, Q.Y.; Cao, X.C. Nitrogen-mediated alleviation of photosynthetic inhibition under moderate water deficit stress in rice (Oryza sativa L.). Environ. Exp. Bot. 2019, 157, 269–282. [Google Scholar] [CrossRef]
- Walker, R.P.; Chen, Z.H.; Acheson, R.M.; Leegood, R.C. Effects of phosphorylation on phosphoenolpyruvate carboxykinase from the C4 plant Guinea grass. Plant Physiol. 2002, 128, 165–172. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jian, S.; Wan, S.; Lin, Y.; Zhong, C. Nitrogen Sources Reprogram Carbon and Nitrogen Metabolism to Promote Andrographolide Biosynthesis in Andrographis paniculata (Burm.f.) Nees Seedlings. Int. J. Mol. Sci. 2024, 25, 3990. https://doi.org/10.3390/ijms25073990
Jian S, Wan S, Lin Y, Zhong C. Nitrogen Sources Reprogram Carbon and Nitrogen Metabolism to Promote Andrographolide Biosynthesis in Andrographis paniculata (Burm.f.) Nees Seedlings. International Journal of Molecular Sciences. 2024; 25(7):3990. https://doi.org/10.3390/ijms25073990
Chicago/Turabian StyleJian, Shaofen, Si Wan, Yang Lin, and Chu Zhong. 2024. "Nitrogen Sources Reprogram Carbon and Nitrogen Metabolism to Promote Andrographolide Biosynthesis in Andrographis paniculata (Burm.f.) Nees Seedlings" International Journal of Molecular Sciences 25, no. 7: 3990. https://doi.org/10.3390/ijms25073990
APA StyleJian, S., Wan, S., Lin, Y., & Zhong, C. (2024). Nitrogen Sources Reprogram Carbon and Nitrogen Metabolism to Promote Andrographolide Biosynthesis in Andrographis paniculata (Burm.f.) Nees Seedlings. International Journal of Molecular Sciences, 25(7), 3990. https://doi.org/10.3390/ijms25073990





