mRNA-LNP COVID-19 Vaccine Lipids Induce Complement Activation and Production of Proinflammatory Cytokines: Mechanisms, Effects of Complement Inhibitors, and Relevance to Adverse Reactions
Abstract
1. Introduction
2. Results
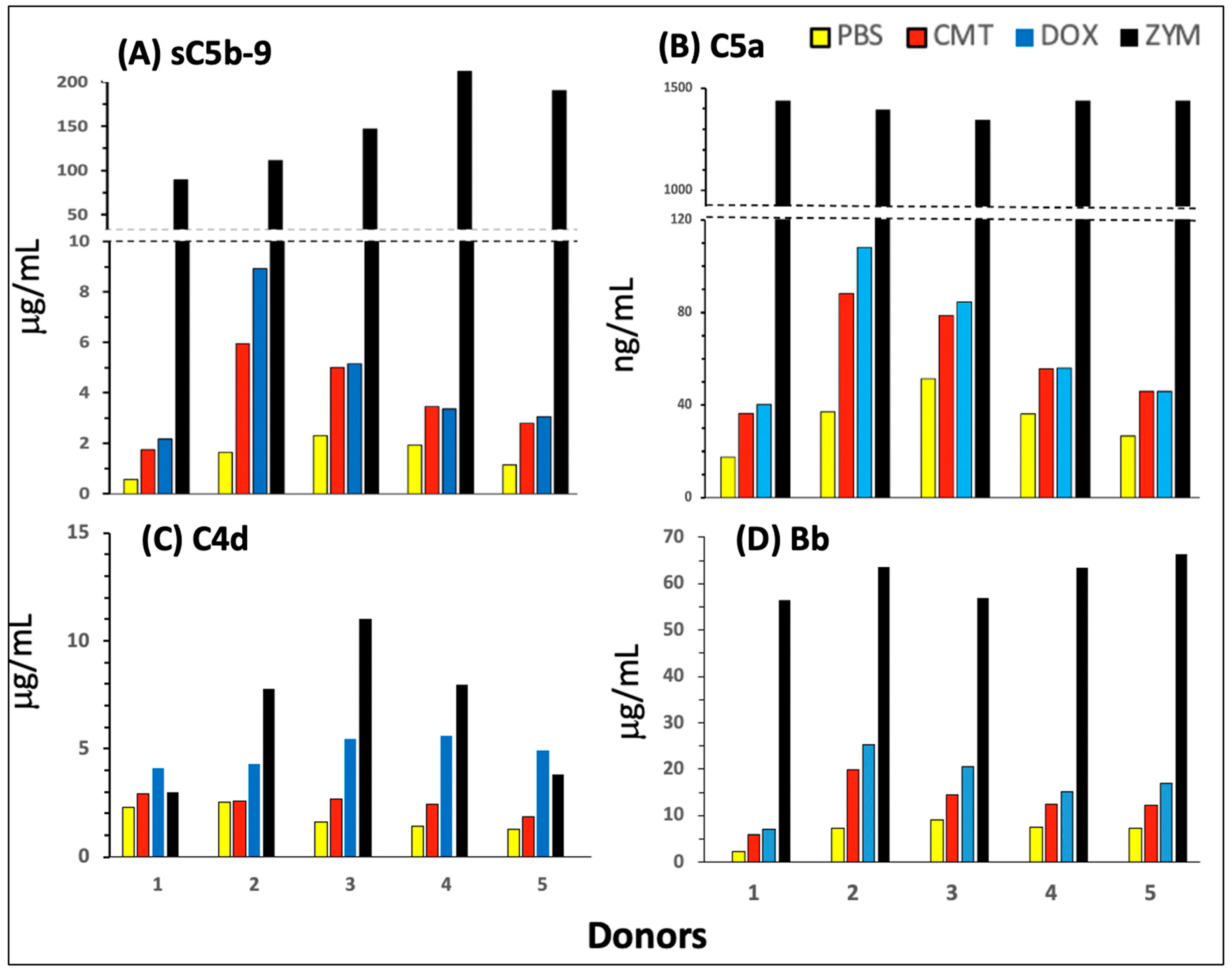
2.1. Complement Activation by Comirnaty in PBMC Culture
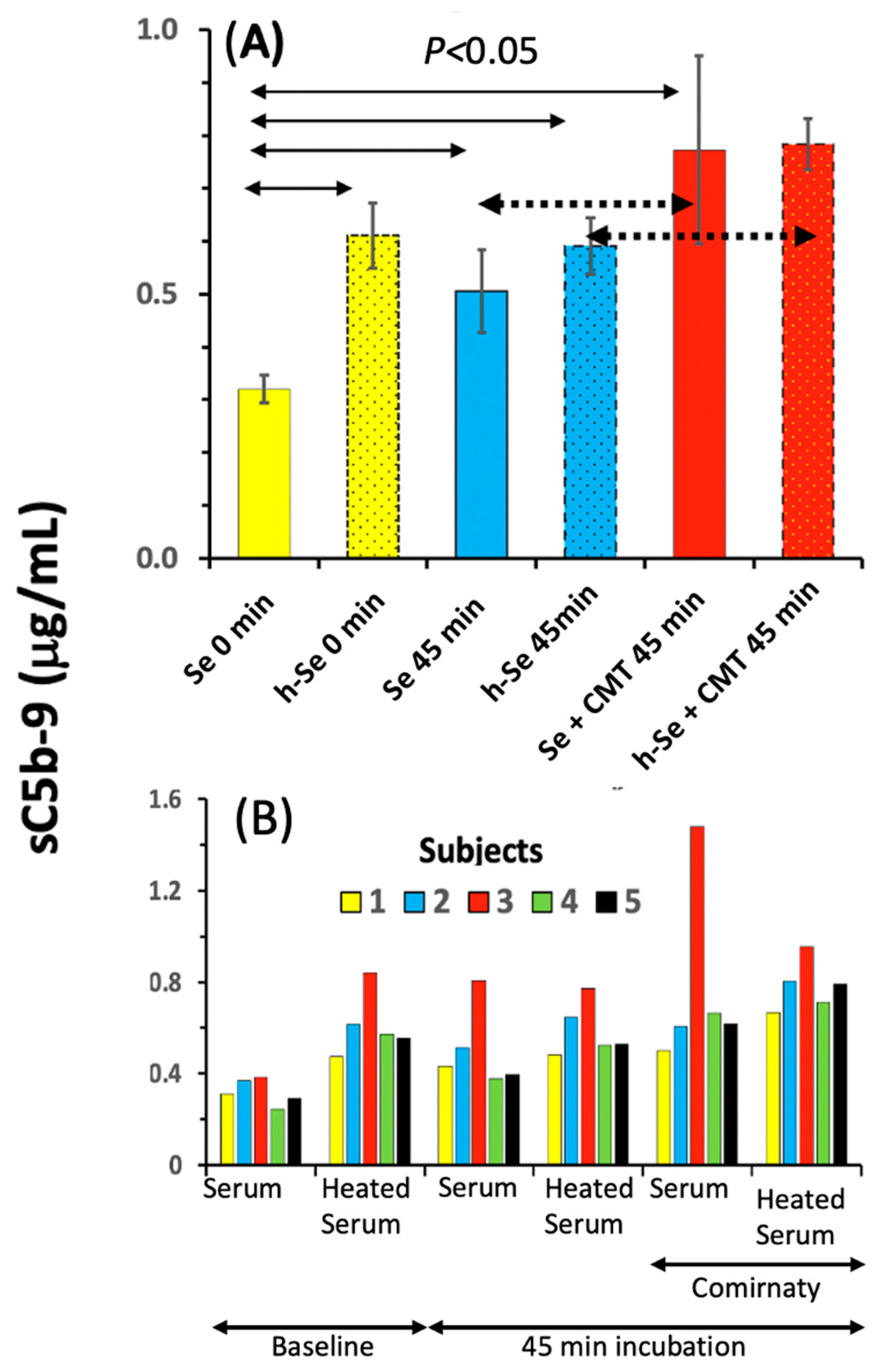
2.2. Features of C Activation by Comirnaty in 75% Human Serum
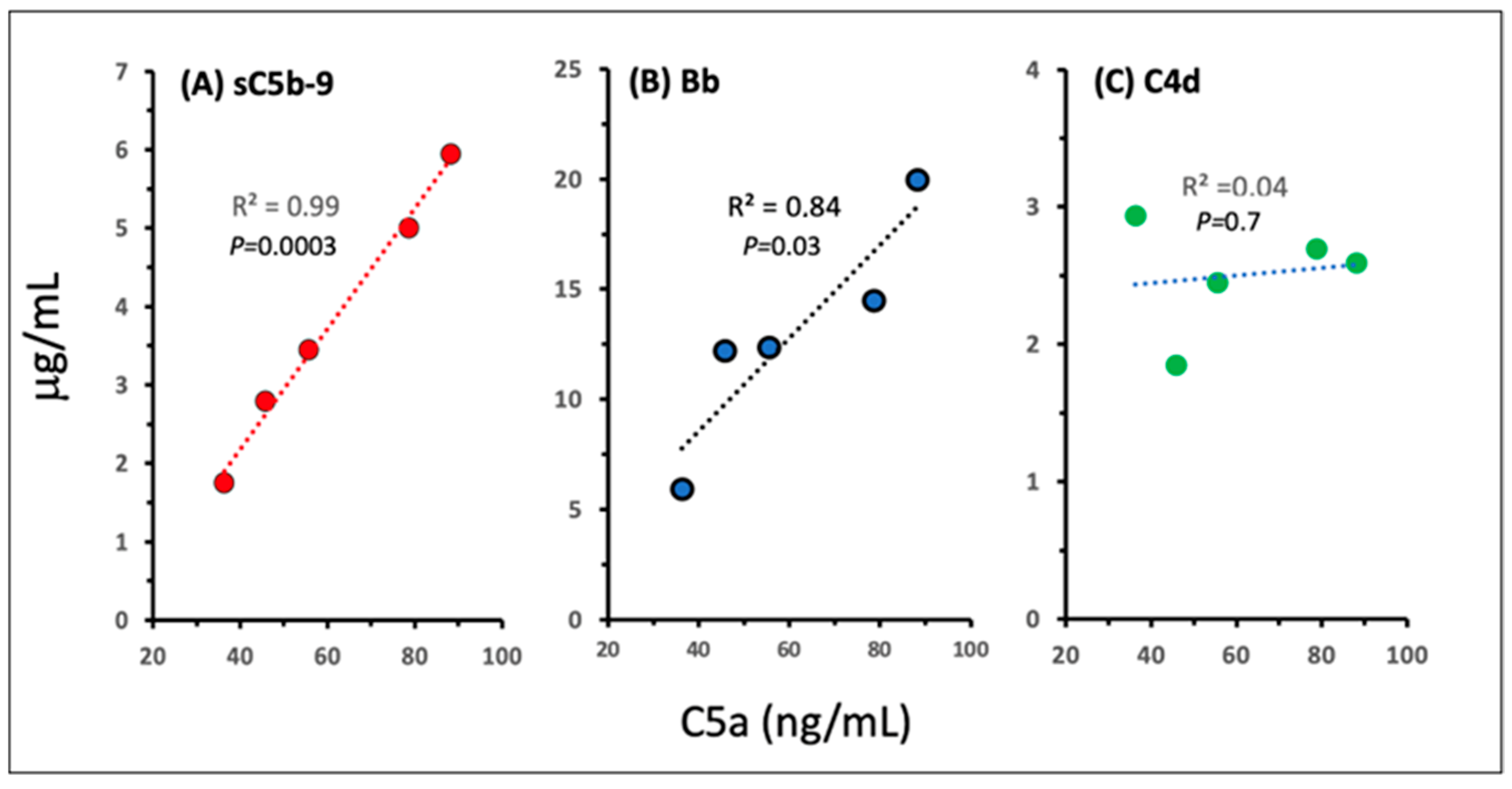
2.3. Pathways and Relative Efficacies of C Activation by Comirnaty and Doxebo
2.4. C Activation by Spikevax
2.5. Lack of C Activation by Naked mRNAs
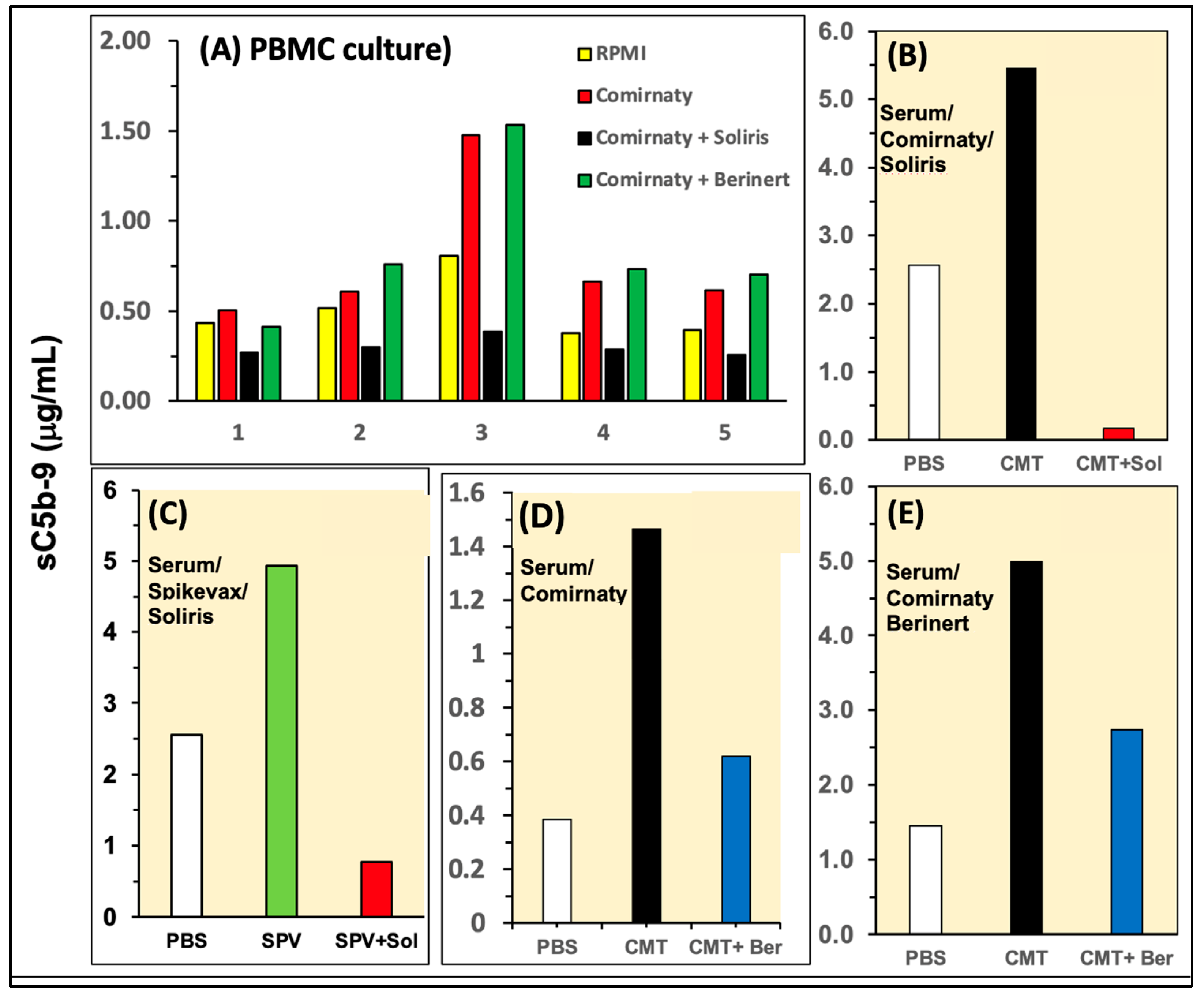
2.6. Effects of C Inhibitors on C Activation in PBMC Culture and 75% Sera
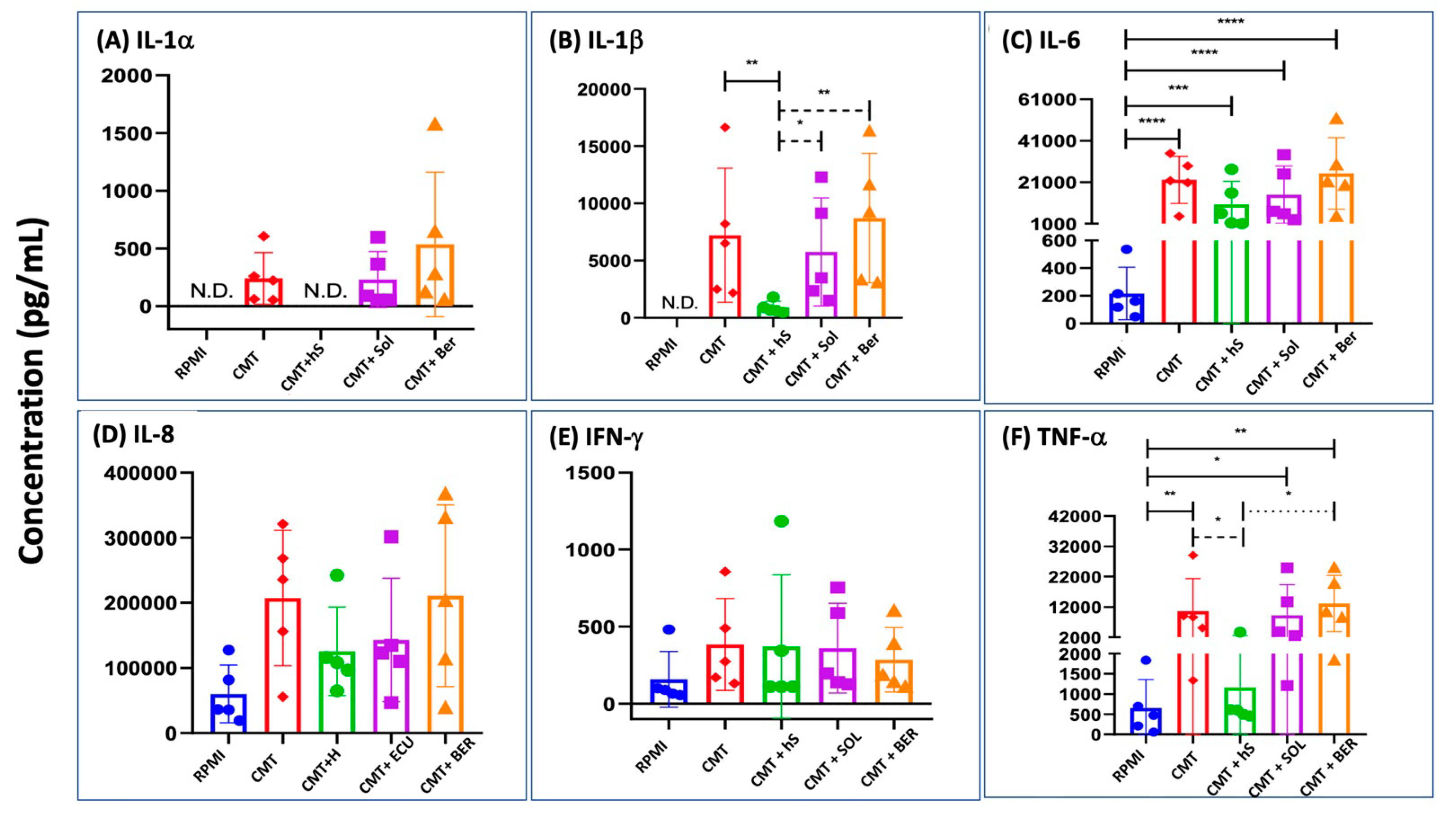
2.7. Cytokine Production in Comirnaty-Exposed, Serum-Substituted PBMC: The Effects of Heat Inactivation and Inhibition of C Activation
3. Discussion
3.1. Complement Activation by Comirnaty: Novel Findings and Mechanism
3.2. Inflammatory Cytokine Production by Comirnaty: Mechanism and Relationship with C Activation
3.3. Broader Implications of In Vitro Data on C Activation and Cytokine Induction by mRNA-LNP Vaccines
3.3.1. The Complement Test as an In Vitro Model for Vaccine-Induced Anaphylaxis
3.3.2. Use of the PBMC-Based Pan-Innate Immune Stimulation Assay for Finding Effective Pharmacological Prevention of mRNA-Vaccine-Induced SAEs
3.3.3. Public Health Implications
4. Materials and Methods
4.1. Materials
4.2. Ethical Permission, Donors, and Blood Collection
4.3. Separation of Serum and PBMC
4.4. Preparation of mRNAs
4.5. PBMC Studies
4.6. Serum Studies
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO: COVID-19 Advice for the Public: Getting Vaccinated. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines/advice (accessed on 18 March 2024).
- CDC: Updated COVID-19 Vaccine Recommendations are Now Available. 2023. Available online: https://www.cdc.gov/ncird/whats-new/covid-vaccine-recommendations-9-12-2023.html (accessed on 18 March 2024).
- Law, B. SO2-D2.1.2 Priority List of COVID-19 Adverse Events of Special Interest: Quarterly Update December 2020. 2020. Available online: https://brightoncollaboration.us/wp-content/uploads/2023/05/SO2_D2.1.2_V1.2_COVID-19_AESI-update_V1.3.pdf (accessed on 18 March 2024).
- Fraiman, J.; Erviti, J.; Jones, M.; Greenland, S.; Whelan, P.; Kaplan, R.M.; Doshi, P. Serious adverse events of special interest following mRNA COVID-19 vaccination in randomized trials in adults. Vaccine 2022, 40, 5798–5805. [Google Scholar] [CrossRef]
- Bozkurt, B. Shedding Light on Mechanisms of Myocarditis with COVID-19 mRNA Vaccines. Circulation 2023, 147, 877–880. [Google Scholar] [CrossRef]
- Patone, M.; Mei, X.W.; Handunnetthi, L.; Dixon, S.; Zaccardi, F.; Shankar-Hari, M.; Watkinson, P.; Khunti, K.; Harnden, A.; Coupland, C.A.C.; et al. Risk of Myocarditis After Sequential Doses of COVID-19 Vaccine and SARS-CoV-2 Infection by Age and Sex. Circulation 2022, 146, 743–754. [Google Scholar] [CrossRef]
- Luo, J.; Hur, K.; Salone, C.; Huang, N.; Burk, M.; Pandey, L.; Thakkar, B.; Donahue, M.; Cunningham, F. Incidence Rates and Clinical Characteristics of Patients With Confirmed Myocarditis or Pericarditis Following COVID-19 mRNA Vaccination: Experience of the Veterans Health Administration Through 9 October 2022. Open Forum Infect. Dis. 2023, 10, ofad268. [Google Scholar] [CrossRef] [PubMed]
- Mungmunpuntipantip, R.; Wiwanitkit, V. Cardiac inflammation associated with COVID-19 mRNA vaccination and previous myocarditis. Minerva Cardiol. Angiol. 2023, 71, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Schroth, D.; Garg, R.; Bocova, X.; Hansmann, J.; Haass, M.; Yan, A.; Fernando, C.; Chacko, B.; Oikonomou, A.; White, J.; et al. Predictors of persistent symptoms after mRNA SARS-CoV-2 vaccine-related myocarditis (myovacc registry). Front. Cardiovasc. Med. 2023, 10, 1204232. [Google Scholar] [CrossRef]
- Paredes-Vazquez, J.G.; Rubio-Infante, N.; Lopez-de la Garza, H.; Brunck, M.E.G.; Guajardo-Lozano, J.A.; Ramos, M.R.; Vazquez-Garza, E.; Torre-Amione, G.; Garcia-Rivas, G.; Jerjes-Sanchez, C. Soluble factors in COVID-19 mRNA vaccine-induced myocarditis causes cardiomyoblast hypertrophy and cell injury: A case report. Virol. J. 2023, 20, 203. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, T.; Iwabuchi, Y.; Miyazawa, R.; Tonda, K.; Shiga, T.; Strauss, H.W.; Antoniades, C.; Narula, J.; Jinzaki, M. Assessment of Myocardial (18)F-FDG Uptake at PET/CT in Asymptomatic SARS-CoV-2-vaccinated and Nonvaccinated Patients. Radiology 2023, 308, e230743. [Google Scholar] [CrossRef] [PubMed]
- Hviid, A.; Nieminen, T.A.; Pihlström, N.; Gunnes, N.; Dahl, J.; Karlstad, Ø.; Gulseth, H.L.; Sundström, A.; Husby, A.; Hansen, J.V.; et al. Booster vaccination with SARS-CoV-2 mRNA vaccines and myocarditis in adolescents and young adults: A Nordic cohort study. Eur. Heart J. 2024, ehae056. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, J.; Storm, G.; Ljubimova, J.Y.; Castells, M.; Phillips, E.J.; Turjeman, K.; Barenholz, Y.; Crommelin, D.J.A.; Dobrovolskaia, M.A. Applying lessons learned from nanomedicines to understand rare hypersensitivity reactions to mRNA-based SARS-CoV-2 vaccines. Nat. Nanotechnol. 2022, 17, 337–346. [Google Scholar] [CrossRef]
- Dezsi, L.; Meszaros, T.; Kozma, G.; H-Velkei, M.; Olah, C.Z.; Szabo, M.; Patko, Z.; Fulop, T.; Hennies, M.; Szebeni, M.; et al. A naturally hypersensitive porcine model may help understand the mechanism of COVID-19 mRNA vaccine-induced rare (pseudo) allergic reactions: Complement activation as a possible contributing factor. Geroscience 2022, 44, 597–618. [Google Scholar] [CrossRef] [PubMed]
- Kozma, G.T.; Meszaros, T.; Berenyi, P.; Facsko, R.; Patko, Z.; Olah, C.Z.; Nagy, A.; Fulop, T.G.; Glatter, K.A.; Radovits, T.; et al. Role of anti-polyethylene glycol (PEG) antibodies in the allergic reactions to PEG-containing COVID-19 vaccines: Evidence for immunogenicity of PEG. Vaccine 2023, 41, 4561–4570. [Google Scholar] [CrossRef] [PubMed]
- Barta, B.A.; Radovits, T.; Dobos, A.B.; Kozma, G.T.; Mészáros, T.; Berényi, P.; Facskó, R.; Fülöp, T.G.; Merkely, B.; Szebeni, J. The COVID-19 mRNA vaccine Comirnaty induces anaphylactic shock in an anti-PEG hyperimmune large animal model: Role of complement activation in cardiovascular, hematological and inflammatory mediator changes. bioRxiv 2023, bioRxiv:2023.05.19.541479. [Google Scholar] [CrossRef]
- Guo, M.; Liu, X.; Chen, X.; Li, Q. Insights into new-onset autoimmune diseases after COVID-19 vaccination. Autoimmun. Rev. 2023, 22, 103340. [Google Scholar] [CrossRef]
- Root-Bernstein, R. COVID-19 coagulopathies: Human blood proteins mimic SARS-CoV-2 virus, vaccine proteins and bacterial co-infections inducing autoimmunity: Combinations of bacteria and SARS-CoV-2 synergize to induce autoantibodies targeting cardiolipin, cardiolipin-binding proteins, platelet factor 4, prothrombin, and coagulation factors. Bioessays 2021, 43, e2100158. [Google Scholar] [CrossRef]
- Brambilla, M.; Canzano, P.; Valle, P.D.; Becchetti, A.; Conti, M.; Alberti, M.; Galotta, A.; Biondi, M.L.; Lonati, P.A.; Veglia, F.; et al. Head-to-head comparison of four COVID-19 vaccines on platelet activation, coagulation and inflammation. The TREASURE study. Thromb. Res. 2023, 223, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, S.R.; Sogaard, O.S.; Tolstrup, M.; Staerke, N.B.; Lundgren, J.; Ostergaard, L.; Hvas, A.M. Inflammation and Platelet Activation After COVID-19 Vaccines—Possible Mechanisms Behind Vaccine-Induced Immune Thrombocytopenia and Thrombosis. Front. Immunol. 2021, 12, 779453. [Google Scholar] [CrossRef]
- Lorente, E. Idiopathic Ipsilateral External Jugular Vein Thrombophlebitis After Coronavirus Disease (COVID-19) Vaccination. AJR Am. J. Roentgenol. 2021, 217, 767. [Google Scholar] [CrossRef]
- Lopatynsky-Reyes, E.Z.; Acosta-Lazo, H.; Ulloa-Gutierrez, R.; Avila-Aguero, M.L.; Chacon-Cruz, E. BCG Scar Local Skin Inflammation as a Novel Reaction Following mRNA COVID-19 Vaccines in Two International Healthcare Workers. Cureus 2021, 13, e14453. [Google Scholar] [CrossRef]
- Ben-Fredj, N.; Chahed, F.; Ben-Fadhel, N.; Mansour, K.; Ben-Romdhane, H.; Mabrouk, R.S.E.; Chadli, Z.; Ghedira, D.; Belhadjali, H.; Chaabane, A.; et al. Case series of chronic spontaneous urticaria following COVID-19 vaccines: An unusual skin manifestation. Eur. J. Clin. Pharmacol. 2022, 78, 1959–1964. [Google Scholar] [CrossRef]
- Grieco, T.; Ambrosio, L.; Trovato, F.; Vitiello, M.; Demofonte, I.; Fanto, M.; Paolino, G.; Pellacani, G. Effects of Vaccination against COVID-19 in Chronic Spontaneous and Inducible Urticaria (CSU/CIU) Patients: A Monocentric Study. J. Clin. Med. 2022, 11, 1822. [Google Scholar] [CrossRef] [PubMed]
- Magen, E.; Yakov, A.; Green, I.; Israel, A.; Vinker, S.; Merzon, E. Chronic spontaneous urticaria after BNT162b2 mRNA (Pfizer-BioNTech) vaccination against SARS-CoV-2. Allergy Asthma Proc. 2022, 43, 30–36. [Google Scholar] [CrossRef]
- Ang, T.; Tong, J.Y.; Patel, S.; Khong, J.J.; Selva, D. Orbital inflammation following COVID-19 vaccination: A case series and literature review. Int. Ophthalmol. 2023, 43, 3391–3401. [Google Scholar] [CrossRef]
- Li, S.; Ho, M.; Mak, A.; Lai, F.; Brelen, M.; Chong, K.; Young, A. Intraocular inflammation following COVID-19 vaccination: The clinical presentations. Int. Ophthalmol. 2023, 43, 2971–2981. [Google Scholar] [CrossRef] [PubMed]
- Yasaka, Y.; Hasegawa, E.; Keino, H.; Usui, Y.; Maruyama, K.; Yamamoto, Y.; Kaburaki, T.; Iwata, D.; Takeuchi, M.; Kusuhara, S.; et al. A multicenter study of ocular inflammation after COVID-19 vaccination. Jpn. J. Ophthalmol. 2023, 67, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Le Vu, S.; Bertrand, M.; Botton, J.; Jabagi, M.J.; Drouin, J.; Semenzato, L.; Weill, A.; Dray-Spira, R.; Zureik, M. Risk of Guillain-Barre Syndrome Following COVID-19 Vaccines: A Nationwide Self-Controlled Case Series Study. Neurology 2023, 101, e2094–e2102. [Google Scholar] [CrossRef]
- Lee, H.; Kwon, D.; Park, S.; Park, S.R.; Chung, D.; Ha, J. Temporal association between the age-specific incidence of Guillain-Barre syndrome and SARS-CoV-2 vaccination in Republic of Korea: A nationwide time-series correlation study. Osong Public Health Res. Perspect. 2023, 14, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Reddy, Y.M.; Murthy, J.M.; Osman, S.; Jaiswal, S.K.; Gattu, A.K.; Pidaparthi, L.; Boorgu, S.K.; Chavan, R.; Ramakrishnan, B.; Yeduguri, S.R. Guillain-Barre syndrome associated with SARS-CoV-2 vaccination: How is it different? a systematic review and individual participant data meta-analysis. Clin. Exp. Vaccine Res. 2023, 12, 143–155. [Google Scholar] [CrossRef]
- Algahtani, H.A.; Shirah, B.H.; Albeladi, Y.K.; Albeladi, R.K. Guillain-Barre Syndrome Following the BNT162b2 mRNA COVID-19 Vaccine. Acta Neurol. Taiwan. 2023, 32, 82–85. [Google Scholar]
- Ogunjimi, O.B.; Tsalamandris, G.; Paladini, A.; Varrassi, G.; Zis, P. Guillain-Barre Syndrome Induced by Vaccination Against COVID-19: A Systematic Review and Meta-Analysis. Cureus 2023, 15, e37578. [Google Scholar] [CrossRef]
- Ha, J.; Park, S.; Kang, H.; Kyung, T.; Kim, N.; Kim, D.K.; Kim, H.; Bae, K.; Song, M.C.; Lee, K.J.; et al. Real-world data on the incidence and risk of Guillain-Barre syndrome following SARS-CoV-2 vaccination: A prospective surveillance study. Sci. Rep. 2023, 13, 3773. [Google Scholar] [CrossRef]
- Abara, W.E.; Gee, J.; Marquez, P.; Woo, J.; Myers, T.R.; DeSantis, A.; Baumblatt, J.A.G.; Woo, E.J.; Thompson, D.; Nair, N.; et al. Reports of Guillain-Barre Syndrome After COVID-19 Vaccination in the United States. JAMA Netw. Open 2023, 6, e2253845. [Google Scholar] [CrossRef]
- Walter, A.; Kraemer, M. A neurologist’s rhombencephalitis after comirnaty vaccination. A change of perspective. Neurol. Res. Pract. 2021, 3, 56. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, R.; Askari, N. A review of neurological side effects of COVID-19 vaccination. Eur. J. Med. Res. 2023, 28, 102. [Google Scholar] [CrossRef] [PubMed]
- Alosaimi, B.; Mubarak, A.; Hamed, M.E.; Almutairi, A.Z.; Alrashed, A.A.; AlJuryyan, A.; Enani, M.; Alenzi, F.Q.; Alturaiki, W. Complement Anaphylatoxins and Inflammatory Cytokines as Prognostic Markers for COVID-19 Severity and In-Hospital Mortality. Front. Immunol. 2021, 12, 668725. [Google Scholar] [CrossRef] [PubMed]
- Bruni, F.; Charitos, P.; Lampart, M.; Moser, S.; Siegemund, M.; Bingisser, R.; Osswald, S.; Bassetti, S.; Twerenbold, R.; Trendelenburg, M.; et al. Complement and endothelial cell activation in COVID-19 patients compared to controls with suspected SARS-CoV-2 infection: A prospective cohort study. Front. Immunol. 2022, 13, 941742. [Google Scholar] [CrossRef] [PubMed]
- Hurler, L.; Szilagyi, A.; Mescia, F.; Bergamaschi, L.; Mezo, B.; Sinkovits, G.; Reti, M.; Muller, V.; Ivanyi, Z.; Gal, J.; et al. Complement lectin pathway activation is associated with COVID-19 disease severity, independent of MBL2 genotype subgroups. Front. Immunol. 2023, 14, 1162171. [Google Scholar] [CrossRef] [PubMed]
- Meroni, P.L.; Croci, S.; Lonati, P.A.; Pregnolato, F.; Spaggiari, L.; Besutti, G.; Bonacini, M.; Ferrigno, I.; Rossi, A.; Hetland, G.; et al. Complement activation predicts negative outcomes in COVID-19: The experience from Northen Italian patients. Autoimmun. Rev. 2023, 22, 103232. [Google Scholar] [CrossRef] [PubMed]
- Pires, B.G.; Calado, R.T. Hyper-inflammation and complement in COVID-19. Am. J. Hematol. 2023, 98 (Suppl. S4), S74–S81. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, T.; De Wit, Y.; Scharz, N.; van Mierlo, G.; Angelillo-Scherrer, A.; Brodard, J.; Schefold, J.; Hirzel, C.; Jongerius, I.; Zeerleder, S. Immunothrombosis and complement activation contribute to disease severity and adverse outcome in COVID-19. J. Innate Immun. 2023, 15, 850–864. [Google Scholar] [CrossRef]
- Tsai, C.L.; Lai, C.C.; Chen, C.Y.; Lee, H.S. The efficacy and safety of complement C5a inhibitors for patients with severe COVID-19: A systematic review and meta-analysis. Expert. Rev. Anti Infect. Ther. 2023, 21, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, Y.; Holland, L.Z. COVID-19 microthrombosis: Unusually large VWF multimers are a platform for activation of the alternative complement pathway under cytokine storm. Int. J. Hematol. 2022, 115, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Laurence, J.; Mulvey, J.J.; Seshadri, M.; Racanelli, A.; Harp, J.; Schenck, E.J.; Zappetti, D.; Horn, E.M.; Magro, C.M. Anti-complement C5 therapy with eculizumab in three cases of critical COVID-19. Clin. Immunol. 2020, 219, 108555. [Google Scholar] [CrossRef] [PubMed]
- Annane, D.; Heming, N.; Grimaldi-Bensouda, L.; Fremeaux-Bacchi, V.; Vigan, M.; Roux, A.L.; Marchal, A.; Michelon, H.; Rottman, M.; Moine, P.; et al. Eculizumab as an emergency treatment for adult patients with severe COVID-19 in the intensive care unit: A proof-of-concept study. EClinicalMedicine 2020, 28, 100590. [Google Scholar] [CrossRef] [PubMed]
- Mastellos, D.C.; Pires da Silva, B.G.P.; Fonseca, B.A.L.; Fonseca, N.P.; Auxiliadora-Martins, M.; Mastaglio, S.; Ruggeri, A.; Sironi, M.; Radermacher, P.; Chrysanthopoulou, A.; et al. Complement C3 vs C5 inhibition in severe COVID-19: Early clinical findings reveal differential biological efficacy. Clin. Immunol. 2020, 220, 108598. [Google Scholar] [CrossRef] [PubMed]
- Fodil, S.; Annane, D. Complement Inhibition and COVID-19: The Story so Far. ImmunoTargets Ther. 2021, 10, 273–284. [Google Scholar] [CrossRef]
- Lim, E.H.T.; van Amstel, R.B.E.; de Boer, V.V.; van Vught, L.A.; de Bruin, S.; Brouwer, M.C.; Vlaar, A.P.J.; van de Beek, D. Complement activation in COVID-19 and targeted therapeutic options: A scoping review. Blood Rev. 2023, 57, 100995. [Google Scholar] [CrossRef]
- Estape Senti, M.; de Jongh, C.A.; Dijkxhoorn, K.; Verhoef, J.J.F.; Szebeni, J.; Storm, G.; Hack, C.E.; Schiffelers, R.M.; Fens, M.H.; Boross, P. Anti-PEG antibodies compromise the integrity of PEGylated lipid-based nanoparticles via complement. J. Control. Release 2022, 341, 475–486. [Google Scholar] [CrossRef]
- Alameh, M.G.; Tombacz, I.; Bettini, E.; Lederer, K.; Sittplangkoon, C.; Wilmore, J.R.; Gaudette, B.T.; Soliman, O.Y.; Pine, M.; Hicks, P.; et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity 2021, 54, 2877–2892.e2877. [Google Scholar] [CrossRef]
- Tahtinen, S.; Tong, A.J.; Himmels, P.; Oh, J.; Paler-Martinez, A.; Kim, L.; Wichner, S.; Oei, Y.; McCarron, M.J.; Freund, E.C.; et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2022, 23, 532–542. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Afonin, K.A. Use of human peripheral blood mononuclear cells to define immunological properties of nucleic acid nanoparticles. Nat. Protoc. 2020, 15, 3678–3698. [Google Scholar] [CrossRef]
- Kozma, G.T.; Meszaros, T.; Bakos, T.; Hennies, M.; Bencze, D.; Uzonyi, B.; Gyorffy, B.; Cedrone, E.; Dobrovolskaia, M.A.; Jozsi, M.; et al. Mini-Factor H Modulates Complement-Dependent IL-6 and IL-10 Release in an Immune Cell Culture (PBMC) Model: Potential Benefits Against Cytokine Storm. Front. Immunol. 2021, 12, 642860. [Google Scholar] [CrossRef]
- Szebeni, J.; Kiss, B.; Bozo, T.; Turjeman, K.; Levi-Kalisman, Y.; Barenholz, Y.; Kellermayer, M. Insights into the Structure of Comirnaty Covid-19 Vaccine: A Theory on Soft, Partially Bilayer-Covered Nanoparticles with Hydrogen Bond-Stabilized mRNA-Lipid Complexes. ACS Nano 2023, 17, 13147–13157. [Google Scholar] [CrossRef]
- Szebeni, J.; Bedocs, P.; Urbanics, R.; Bunger, R.; Rosivall, L.; Toth, M.; Barenholz, Y. Prevention of infusion reactions to PEGylated liposomal doxorubicin via tachyphylaxis induction by placebo vesicles: A porcine model. J. Control. Release 2012, 160, 382–387. [Google Scholar] [CrossRef]
- Harder, M.J.; Kuhn, N.; Schrezenmeier, H.; Hochsmann, B.; von Zabern, I.; Weinstock, C.; Simmet, T.; Ricklin, D.; Lambris, J.D.; Skerra, A.; et al. Incomplete inhibition by eculizumab: Mechanistic evidence for residual C5 activity during strong complement activation. Blood 2017, 129, 970–980. [Google Scholar] [CrossRef]
- Preissner, K.P.; Podack, E.R.; Muller-Eberhard, H.J. SC5b-7, SC5b-8 and SC5b-9 complexes of complement: Ultrastructure and localization of the S-protein (vitronectin) within the macromolecules. Eur. J. Immunol. 1989, 19, 69–75. [Google Scholar] [CrossRef]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. Author Correction: A new coronavirus associated with human respiratory disease in China. Nature 2020, 580, E7. [Google Scholar] [CrossRef]
- Szebeni, J.; Simberg, D.; Gonzalez-Fernandez, A.; Barenholz, Y.; Dobrovolskaia, M.A. Roadmap and strategy for overcoming infusion reactions to nanomedicines. Nat. Nanotechnol. 2018, 13, 1100–1108. [Google Scholar] [CrossRef] [PubMed]
- Kozma, G.T.; Meszaros, T.; Vashegyi, I.; Fulop, T.; Orfi, E.; Dezsi, L.; Rosivall, L.; Bavli, Y.; Urbanics, R.; Mollnes, T.E.; et al. Pseudo-anaphylaxis to Polyethylene Glycol (PEG)-Coated Liposomes: Roles of Anti-PEG IgM and Complement Activation in a Porcine Model of Human Infusion Reactions. ACS Nano 2019, 13, 9315–9324. [Google Scholar] [CrossRef] [PubMed]
- Rissanou, A.N.; Ouranidis, A.; Karatasos, K. Complexation of single stranded RNA with an ionizable lipid: An all-atom molecular dynamics simulation study. Soft Matter 2020, 16, 6993–7005. [Google Scholar] [CrossRef] [PubMed]
- Plank, C.; Mechtler, K.; Szoka, F.C.; Wagner, E. Activation of the Complement System by Synthetic DNA Complexes: A Potential Barrier for Intravenous Gene Delivery. Hum. Gene Ther. 1996, 7, 1437–1446. [Google Scholar] [CrossRef]
- Chonn, A.; Cullis, P.R.; Devine, D.V. The role of surface charge in the activation of the classical and alternative pathways of complement by liposomes. J. Immunol. 1991, 146, 4234–4241. [Google Scholar] [CrossRef]
- Szebeni, J.; Alving, C.R.; Savay, S.; Barenholz, Y.; Priev, A.; Danino, D.; Talmon, Y. Formation of complement-activating particles in aqueous solutions of Taxol: Possible role in hypersensitivity reactions. Intern. Immunopharm. 2001, 1, 721–735. [Google Scholar] [CrossRef]
- Zhu, X.L.; Pacheco, N.D.; Dick, E.J.; Rollwagen, F.M. Differentially increased IL-6 mRNA expression in liver and spleen following injection of liposome-encapsulated haemoglobin. Cytokine 1999, 11, 696–703. [Google Scholar] [CrossRef]
- O’Barr, S.; Cooper, N.R. The C5a complement activation peptide increases IL-1beta and IL-6 release from amyloid-beta primed human monocytes: Implications for Alzheimer’s disease. J. Neuroimmunol. 2000, 109, 87–94. [Google Scholar] [CrossRef]
- Yang, J.; Wise, L.; Fukuchi, K.I. TLR4 Cross-Talk With NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol. 2020, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Kokeny, G.; Bakos, T.; Barta, B.A.; Nagy, G.V.; Meszaros, T.; Kozma, G.T.; Szabo, A.; Szebeni, J.; Merkely, B.; Radovits, T. Zymosan Particle-Induced Hemodynamic, Cytokine and Blood Cell Changes in Pigs: An Innate Immune Stimulation Model with Relevance to Cytokine Storm Syndrome and Severe COVID-19. Int. J. Mol. Sci. 2023, 24, 1138. [Google Scholar] [CrossRef]
- Pfizer; Biontech. COMIRNATY® (COVID-19 Vaccine, mRNA) Suspension for Injection, for Intramuscular Use 2023–2024 Formula. 2023. Available online: https://labeling.pfizer.com/ShowLabeling.aspx?id=19542 (accessed on 18 March 2024).
- Prescribing Information, DOXIL® (Doxorubicin Hydrochloride Liposome Injection) 2022. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2022/050718Orig1s060lbl.pdf (accessed on 18 March 2024).
- Ikeda, Z.; Kamei, T.; Sasaki, Y.; Reynolds, M.; Sakai, N.; Yoshikawa, M.; Tawada, M.; Morishita, N.; Dougan, D.R.; Chen, C.H.; et al. Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway. J. Med. Chem. 2023, 66, 6354–6371. [Google Scholar] [CrossRef]
- Girmenia, C.; Barcellini, W.; Bianchi, P.; Di Bona, E.; Iori, A.P.; Notaro, R.; Sica, S.; Zanella, A.; De Vivo, A.; Barosi, G.; et al. Management of infection in PNH patients treated with eculizumab or other complement inhibitors: Unmet clinical needs. Blood Rev. 2023, 58, 101013. [Google Scholar] [CrossRef] [PubMed]
- Wooden, B.; Tarragon, B.; Navarro-Torres, M.; Bomback, A.S. Complement inhibitors for kidney disease. Nephrol. Dial. Transplant. 2023, 38 (Suppl. S2), ii29–ii39. [Google Scholar] [CrossRef] [PubMed]
- Goury, A.; Mourvillier, B. Treatment of severe COVID-19: A role for JAK and complement inhibitors? Lancet Respir. Med. 2023, 11, 1036–1037. [Google Scholar] [CrossRef]
- Schubart, A.; Flohr, S.; Junt, T.; Eder, J. Low-molecular weight inhibitors of the alternative complement pathway. Immunol. Rev. 2023, 313, 339–357. [Google Scholar] [CrossRef] [PubMed]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.J.; Pardi, N. mRNA Vaccines in the COVID-19 Pandemic and Beyond. Annu. Rev. Med. 2022, 73, 17–39. [Google Scholar] [CrossRef] [PubMed]
- Kariko, K.; Whitehead, K.; van der Meel, R. What does the success of mRNA vaccines tell us about the future of biological therapeutics? Cell Syst. 2021, 12, 757–758. [Google Scholar] [CrossRef]
- Bevers, S.; Kooijmans, S.A.A.; Van de Velde, E.; Evers, M.J.W.; Seghers, S.; Gitz-Francois, J.; van Kronenburg, N.C.H.; Fens, M.; Mastrobattista, E.; Hassler, L.; et al. mRNA-LNP vaccines tuned for systemic immunization induce strong antitumor immunity by engaging splenic immune cells. Mol. Ther. 2022, 30, 3078–3094. [Google Scholar] [CrossRef]

| Symptoms | Cardiovascular | Coagulation | Enteral | Immune | Neural | Respiratory | Skin |
| ST elevation/AMI | cerebral venous sinus thrombosis | hepatitis | anaphylaxis | encephalitis/ myelitis/ encephalomyelitis | ARDS | acute urticaria | |
| tachy- or bradycardia, arrhythmias | disseminated intravascular coagulation | cholecystitis | hypersensitivity reactions | Bell’s palsy | pulmonary embolism | chronic urticaria | |
| vascular inflammation (Kawasaki disease) | immune thrombocytopenia | colitis | lymphadenopathy (Kawasaki disease) | Guillain–Barré syndrome | stridor, hoarseness | skin graphia, dermatographia | |
| myocarditis/ pericarditis | pulmonary embolisms | enteritis | autoimmune glomerulonephritis | narcolepsy/ catalepsy | dyspnea | dermatographic urticaria | |
| hypo/ hypertension | stroke (hemorrhagic/ ischemic) | diarrhea | autoimmune rheumatic disease | seizures/ convulsions/ epilepsy | coughing | rash | |
| stroke (hemorrhagic/ ischemic) | thrombosis with thrombocytopenia (VITT) | appendicitis | autoimmune hepatitis | transverse myelitis | ocular/orbital inflammation | ||
| arteriosclerosis | venous thrombo-embolism | CARPA | delirium | ||||
| chest/back pain | amenorrhea/ dysmenorrhea/ oligomenorrhea | akathisa (psychomotor restlessness) | |||||
| other forms of cardiac injury | thrombocytopenic purpura | multiple sclerosis | |||||
| lip, tongue, face edema | intracerebral hemorrhage | ||||||
| death | fibrous white clots † |
| Test Agent | sTCC (sC5b-9, μg/mL) * | CAI | SCA (TCC/CAI) *** | Zym % # | CMT/Dox ## | |
|---|---|---|---|---|---|---|
| Mean | SEM (n = 5) | (mg/mL) ** | ||||
| Comirnaty | 2.3 | 0.6 | 0.5 | 4.6 | 0.9 | 4.6 |
| Doxebo | 3.1 | 1.1 | 3.2 | 1.0 | 0.2 | |
| Zymosan | 148.4 | 22.9 | 0.30 | 494.7 | 100.0 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakos, T.; Mészáros, T.; Kozma, G.T.; Berényi, P.; Facskó, R.; Farkas, H.; Dézsi, L.; Heirman, C.; de Koker, S.; Schiffelers, R.; et al. mRNA-LNP COVID-19 Vaccine Lipids Induce Complement Activation and Production of Proinflammatory Cytokines: Mechanisms, Effects of Complement Inhibitors, and Relevance to Adverse Reactions. Int. J. Mol. Sci. 2024, 25, 3595. https://doi.org/10.3390/ijms25073595
Bakos T, Mészáros T, Kozma GT, Berényi P, Facskó R, Farkas H, Dézsi L, Heirman C, de Koker S, Schiffelers R, et al. mRNA-LNP COVID-19 Vaccine Lipids Induce Complement Activation and Production of Proinflammatory Cytokines: Mechanisms, Effects of Complement Inhibitors, and Relevance to Adverse Reactions. International Journal of Molecular Sciences. 2024; 25(7):3595. https://doi.org/10.3390/ijms25073595
Chicago/Turabian StyleBakos, Tamás, Tamás Mészáros, Gergely Tibor Kozma, Petra Berényi, Réka Facskó, Henriette Farkas, László Dézsi, Carlo Heirman, Stefaan de Koker, Raymond Schiffelers, and et al. 2024. "mRNA-LNP COVID-19 Vaccine Lipids Induce Complement Activation and Production of Proinflammatory Cytokines: Mechanisms, Effects of Complement Inhibitors, and Relevance to Adverse Reactions" International Journal of Molecular Sciences 25, no. 7: 3595. https://doi.org/10.3390/ijms25073595
APA StyleBakos, T., Mészáros, T., Kozma, G. T., Berényi, P., Facskó, R., Farkas, H., Dézsi, L., Heirman, C., de Koker, S., Schiffelers, R., Glatter, K. A., Radovits, T., Szénási, G., & Szebeni, J. (2024). mRNA-LNP COVID-19 Vaccine Lipids Induce Complement Activation and Production of Proinflammatory Cytokines: Mechanisms, Effects of Complement Inhibitors, and Relevance to Adverse Reactions. International Journal of Molecular Sciences, 25(7), 3595. https://doi.org/10.3390/ijms25073595








