FvMYB108, a MYB Gene from Fragaria vesca, Positively Regulates Cold and Salt Tolerance of Arabidopsis
Abstract
1. Introduction
2. Results
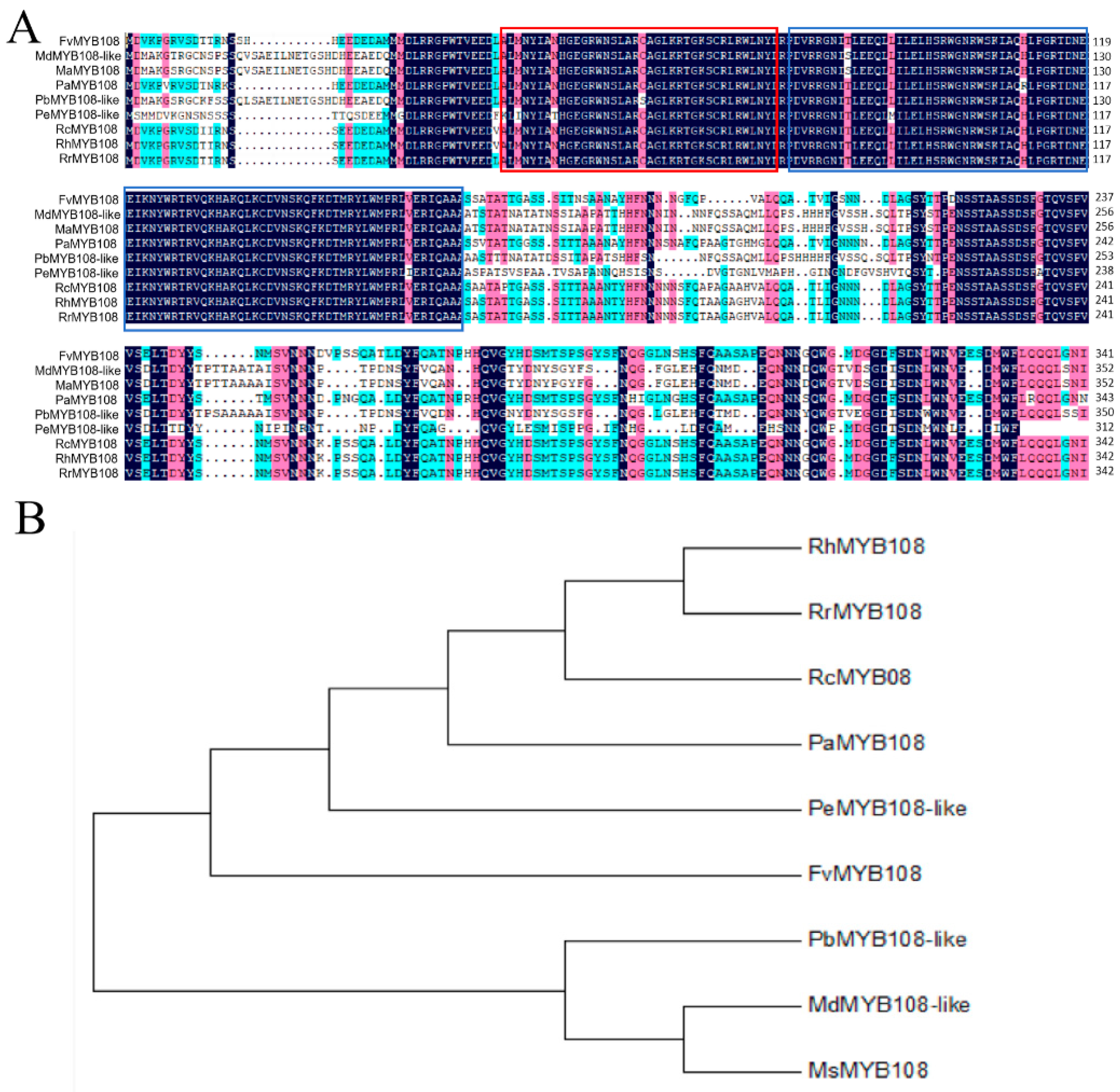
2.1. Cloning and Bioinformatics Analysis of FvMYB108
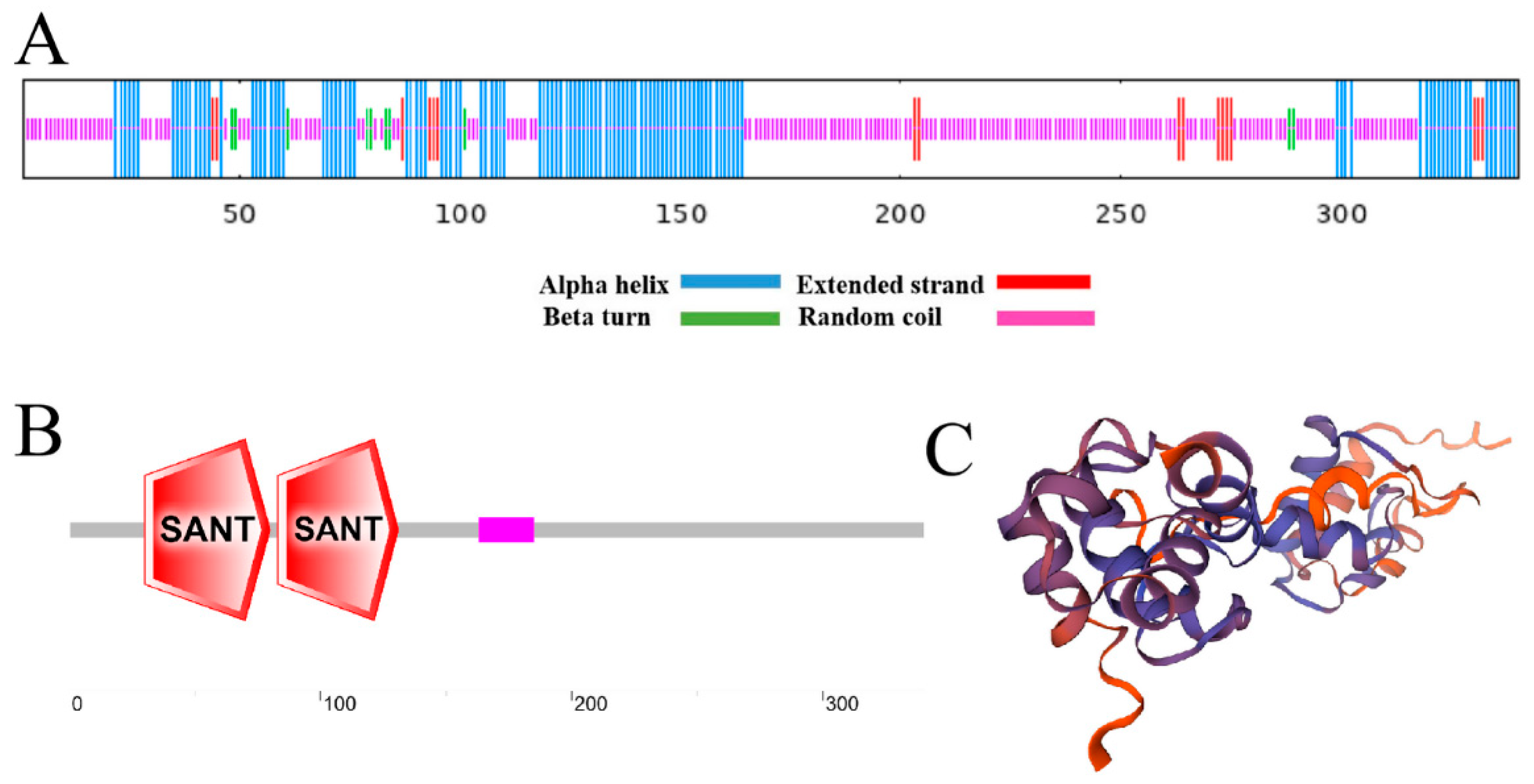
2.2. Structural Prediction of FvMYB108 Protein
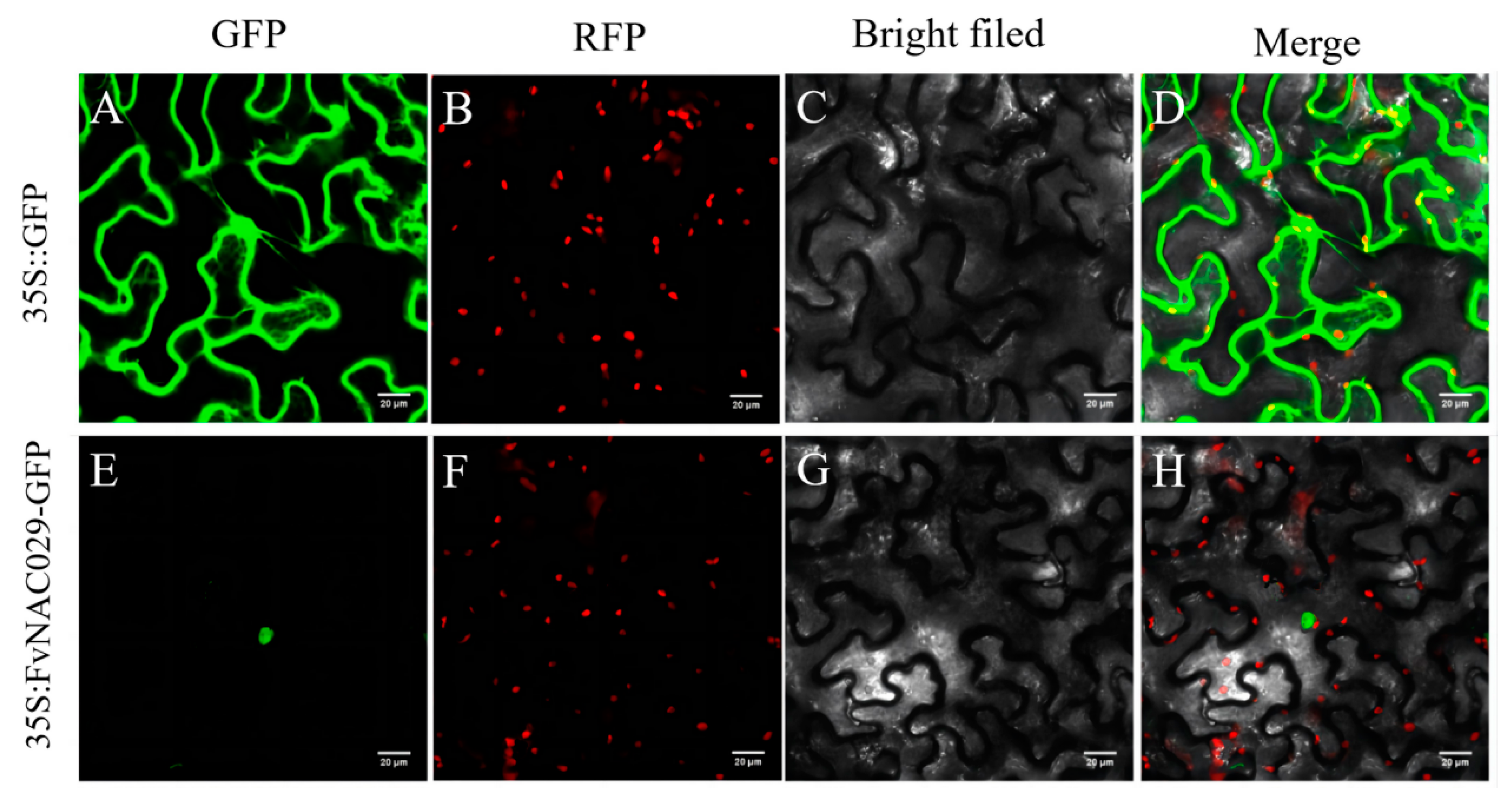
2.3. Subcellular Localization of FvMYB108
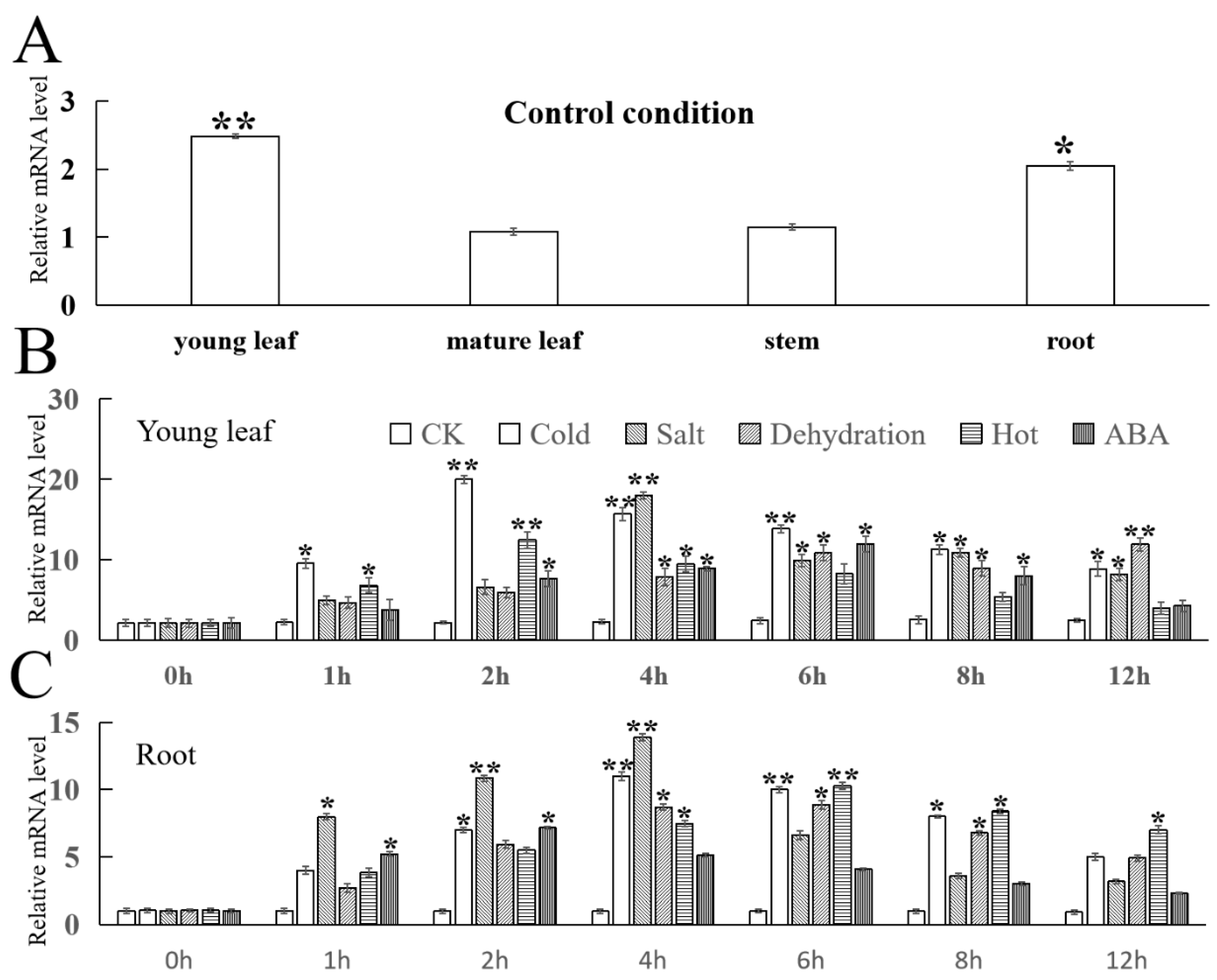
2.4. Expression of FvMYB114 in Strawberry Tissue
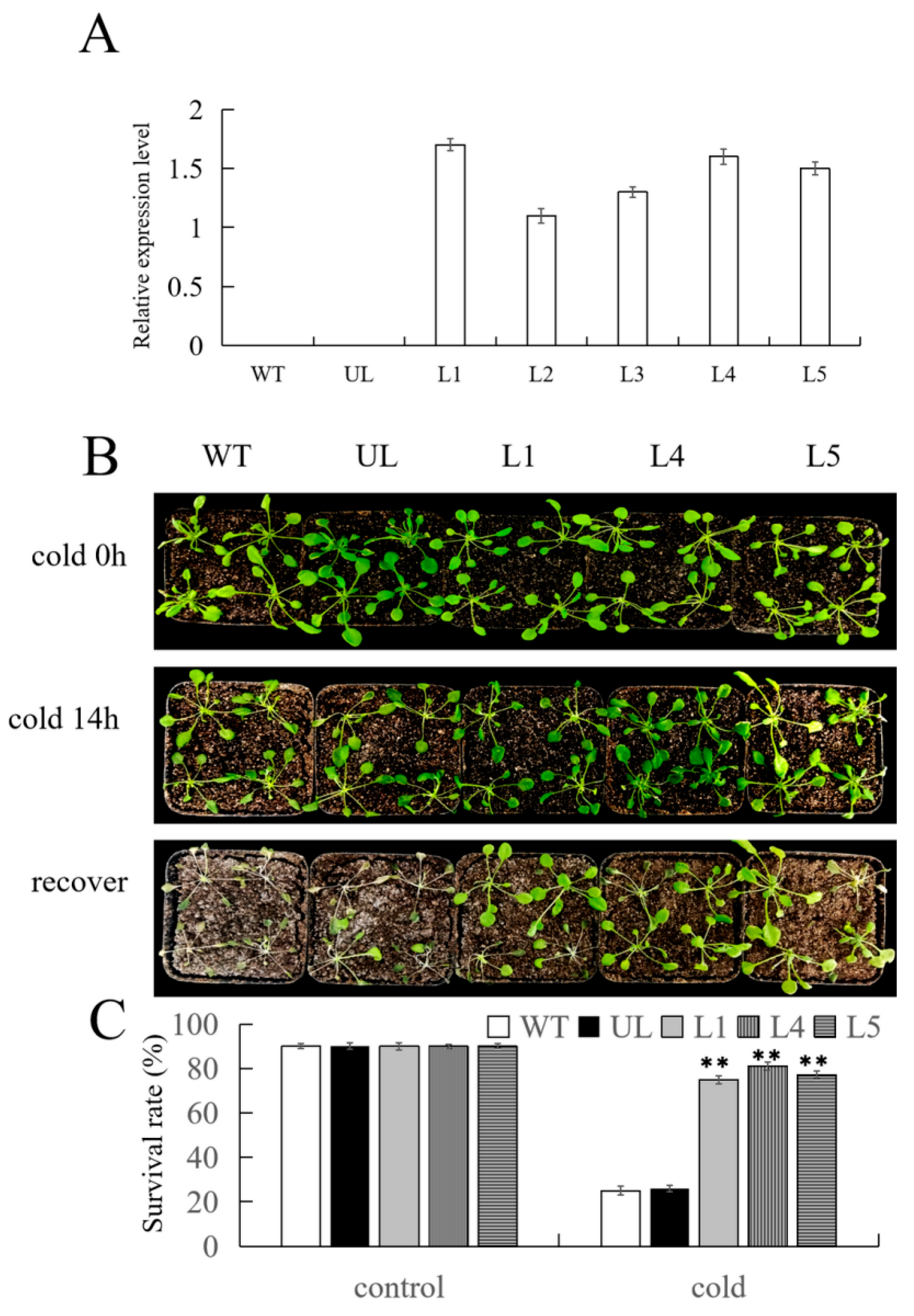
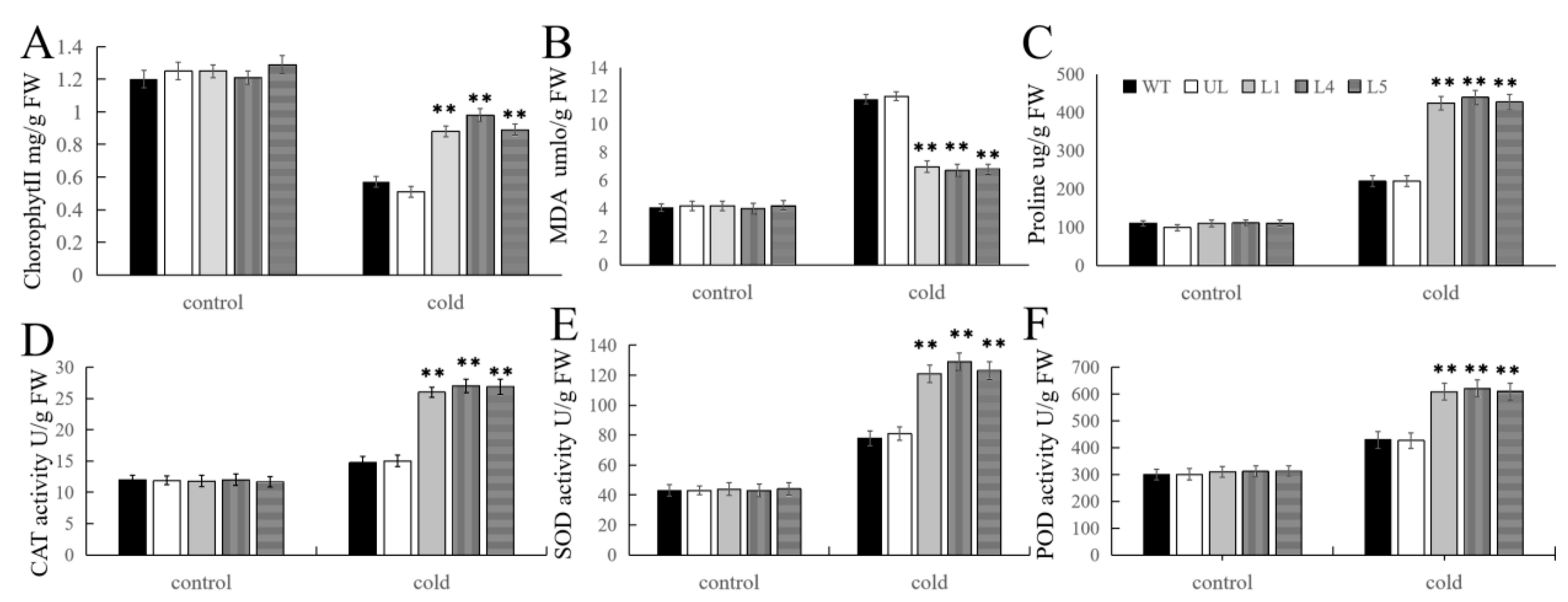
2.5. Overexpression of FvMYB108 Enhances Cold Tolerance in Arabidopsis
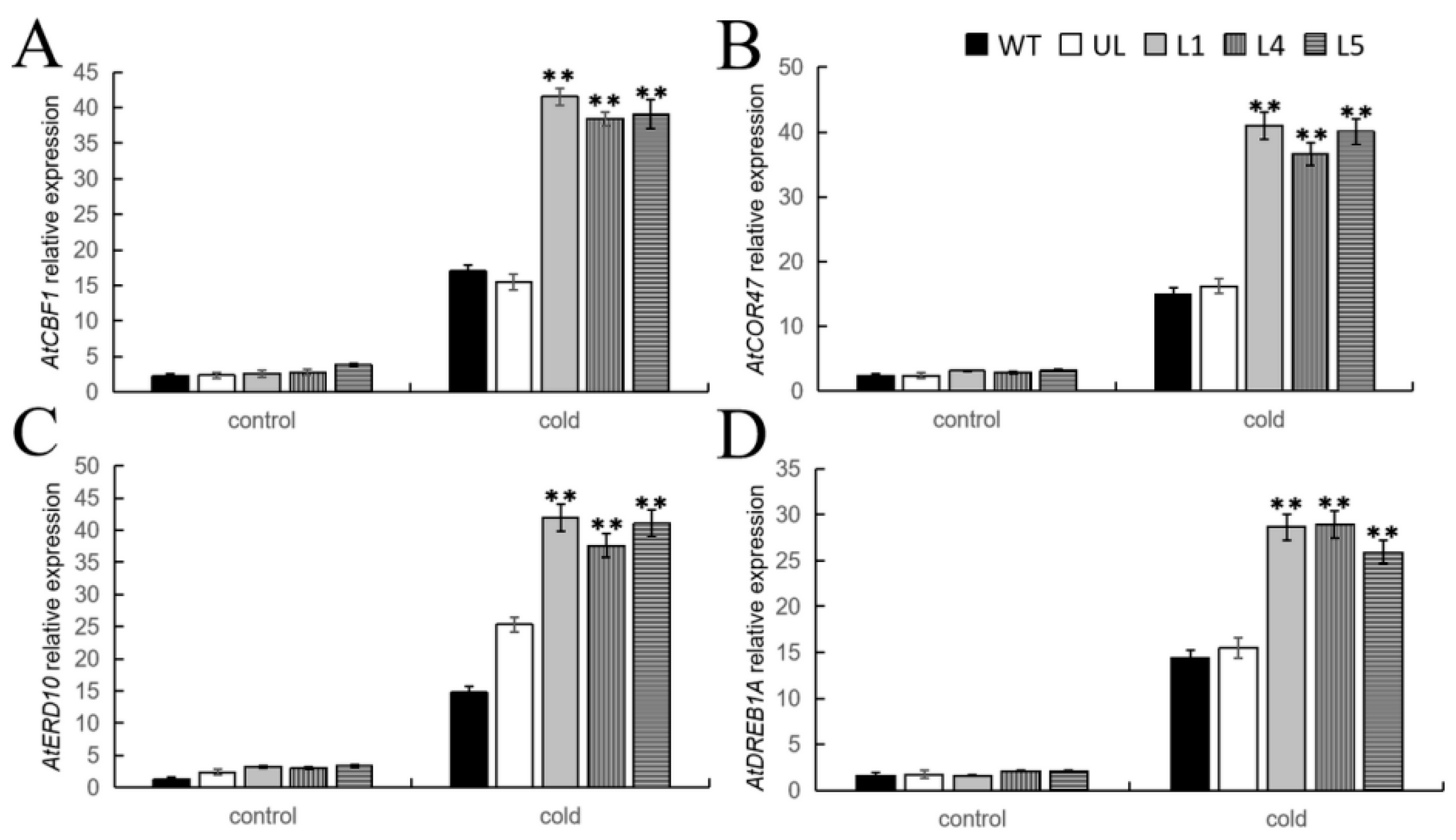
2.6. Overexpression of FvMYB108 Regulates the Expression of Cold Tolerance-Related Genes
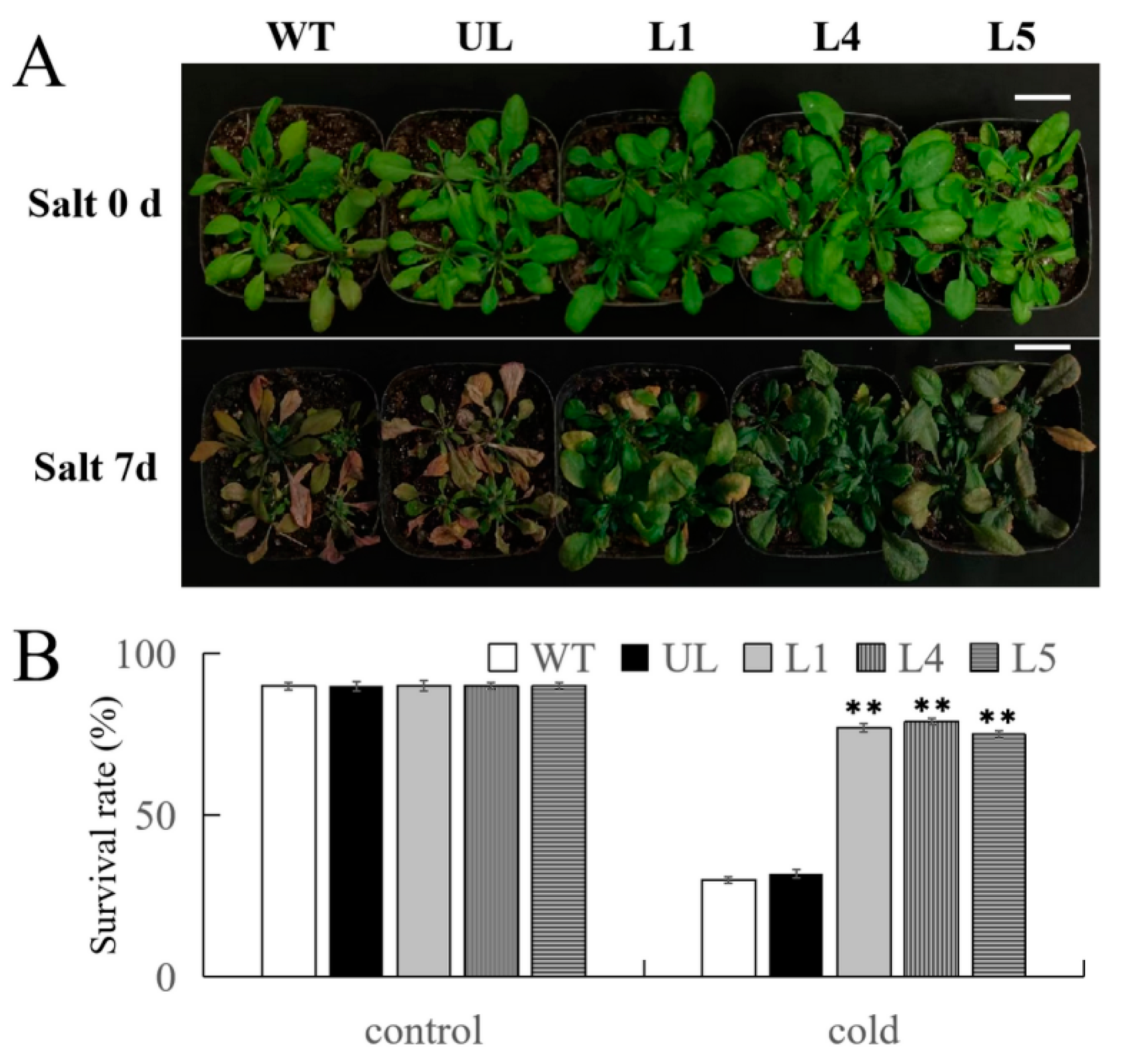
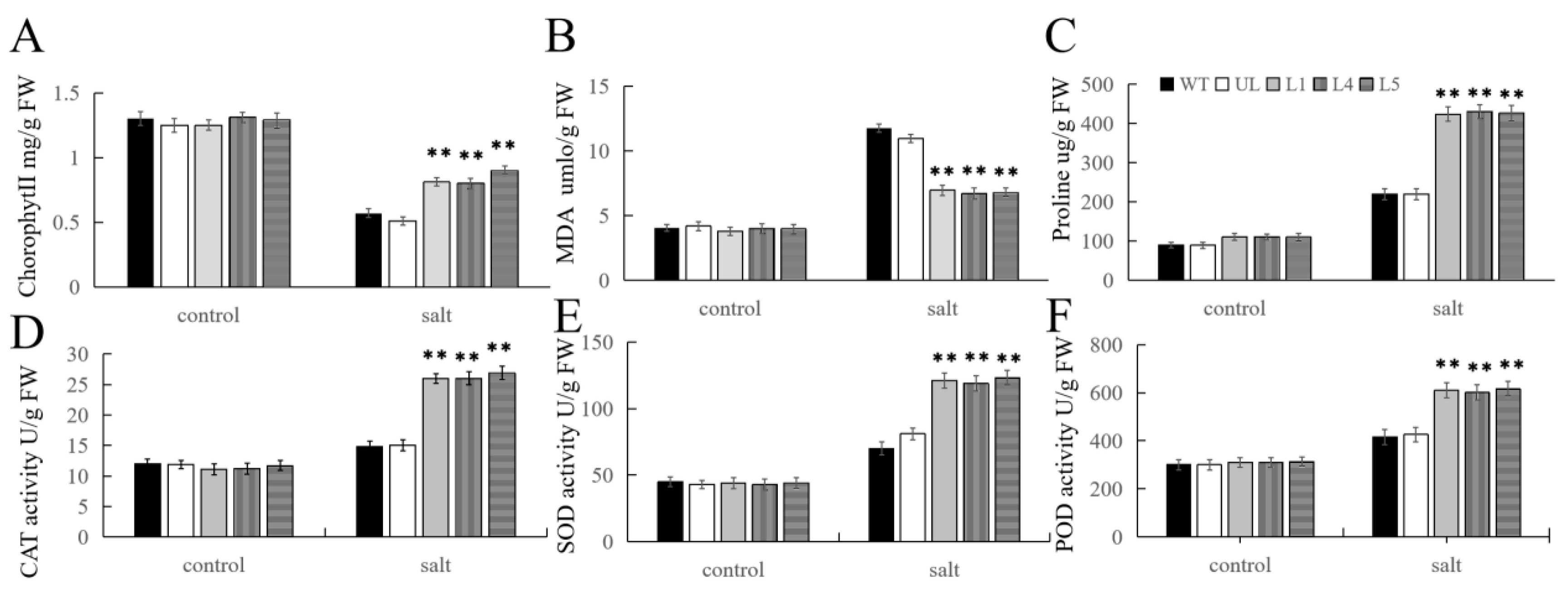
2.7. Overexpression of FvMYB108 Enhances Salt Tolerance in Arabidopsis
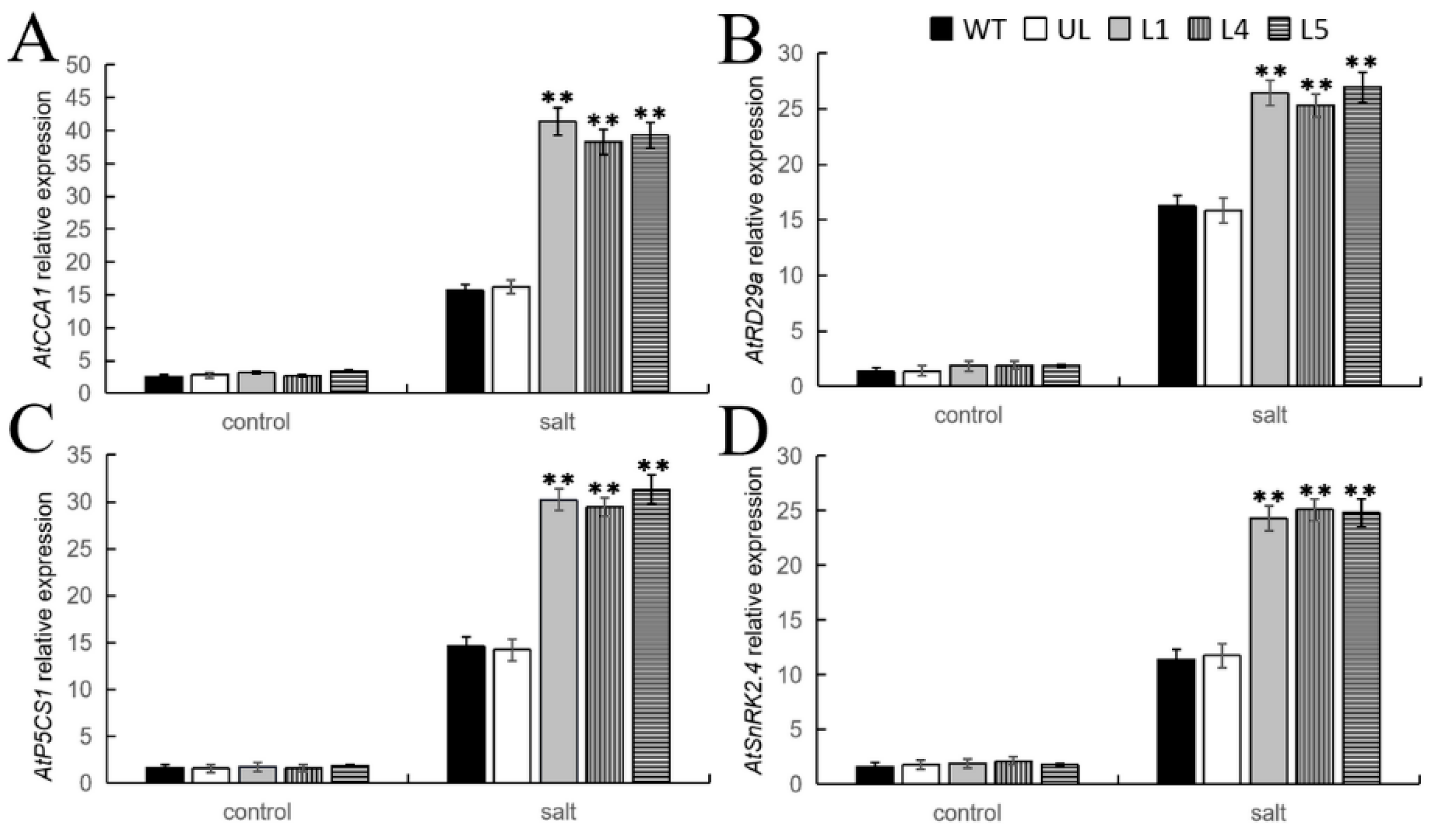
2.8. Overexpression of FvMYB108 Regulates the Expression of Salt Tolerance-Related Genes
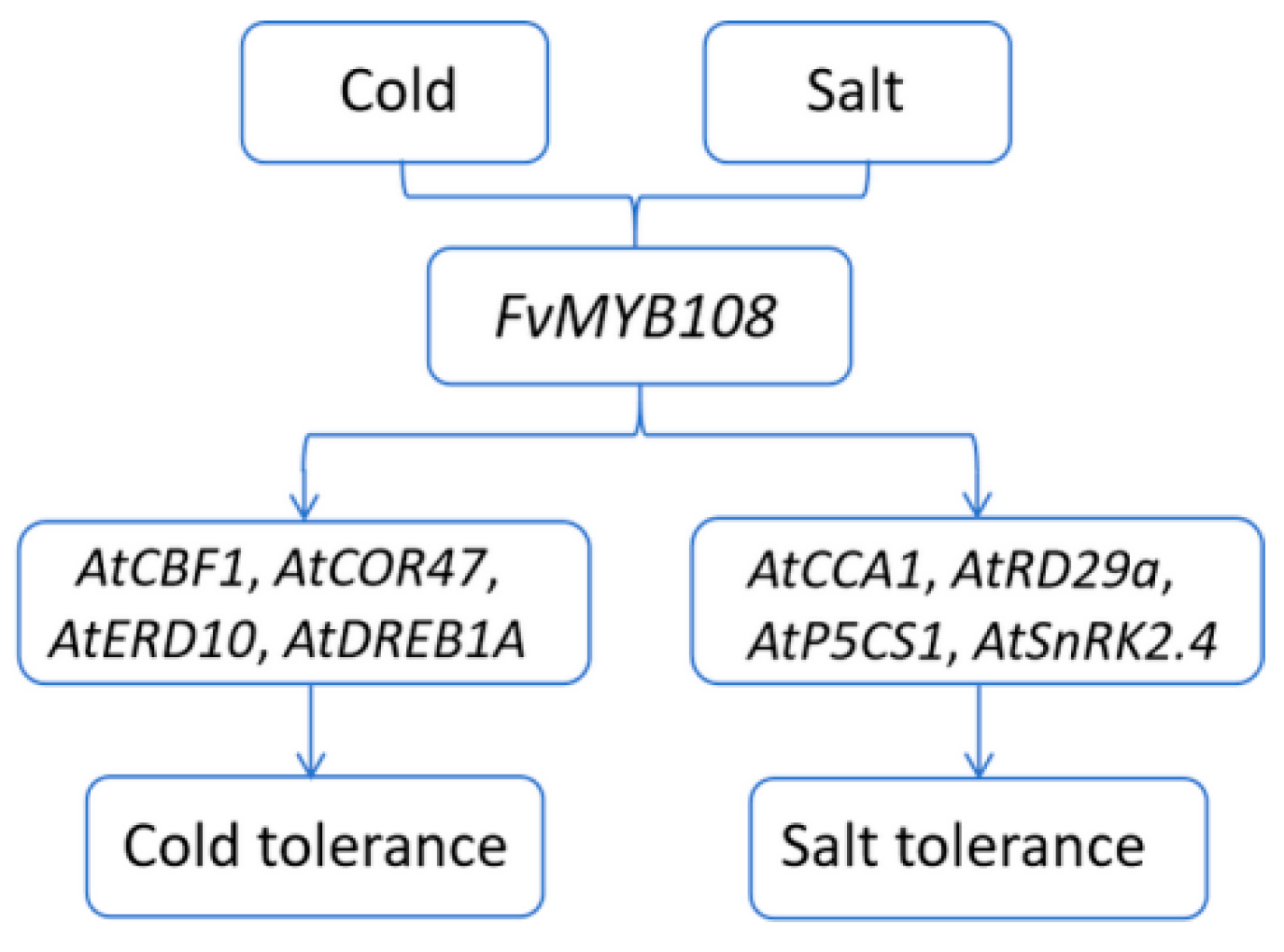
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Treatment
4.2. Cloning and Vector Construction of FvMYB108
4.3. Subcellular Localization of FvMYB108
4.4. Evolutionary and Structural Analyses of FvMYB108
4.5. Fluorescence Quantitative Analysis
4.6. Genetic Transformation of Arabidopsis
4.7. Identification of Cold and Salt Tolerance in Arabidopsis thaliana
4.8. Analysis of Gene Expression Related to Stress Resistance
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, D.; Liang, J.; Xie, H.H.; Wei, X.Y. Norsesquiterpenoids and triterpenoids from strawberry cv. Falandi. Food Chem. 2016, 203, 67–72. [Google Scholar] [CrossRef]
- Gleńsk, M.; Dudek, M.K.; Rybacka, A.; Włodarczyk, M.; Fecka, I. Triterpenoids from strawberry Fragaria × ananassa Duch. cultivar Senga Sengana leaves. Ind. Crop. Prod. 2021, 169, 113668. [Google Scholar] [CrossRef]
- Li, W.; Li, P.; Chen, H.H.; Zhong, J.L.; Liang, X.Q.; Wei, Y.F.; Zhang, L.H.; Wang, H.B.; Han, D.G. Overexpression of a Fragaria vesca 1R-MYB Transcription Factor Gene (FvMYB114). Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 5261. [Google Scholar] [CrossRef]
- Perin, E.C.; Messias, R.D.S.; Borowski, T.; Crizel, R.L.; Schott, I.B.; Carvalho, I.R.; Rombaldi, C.V.; Galli, V. ABA-dependent salt and drought stress improve strawberry fruit quality. Food Chem. 2019, 271, 516–526. [Google Scholar] [CrossRef]
- Rajashekar, C.B.; Zhou, H.; Marcum, K.B.; Prakash, O. Glycine betaine accumulation and induction of cold tolerance in strawberry (Fragaria X ananassa Duch.) plants. Plant Sci. 1999, 148, 175–183. [Google Scholar] [CrossRef]
- Kasuga, M.; Miura, S.; Shinozaki, K.; Kazuko, Y.S. A Combination of the Arabidopsis DREB1A Gene and Stress-Inducible rd29A Promoter Improved Drought-and Low-Temperature Stress Tolerance in Tobacco by Gene Transfer. Plant Cell Physiol. 2004, 45, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.B.; Foley, R.C.; Luis, O.S. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Stracke, R.; Holtgrave, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterization of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef]
- Li, W.H.; Zhong, J.L.; Zhang, L.H.; Wang, Y.; Song, P.H.; Liu, W.D.; Li, X.G.; Han, D.G. Overexpression of a Fragaria vesca MYB Transcription Factor Gene (FvMYB82) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 10538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Zhang, Y.; Fan, C.J.; Wei, Y.H.; Meng, J.X.; Li, Z.; Zhong, C.L. Genome-wide analysis of MYB transcription factors and their responses to salt stress in Casuarina equisetifolia. BMC Plant Biol. 2021, 21, 328. [Google Scholar] [CrossRef]
- Chen, B.; Niu, F.F.; Liu, W.Z.; Yang, B.; Zhang, J.X.; Ma, J.Y.; Cheng, H.; Han, F.; Jiang, Y.Q. Identification, cloning and characterization of R2R3-MYB gene family in canola (Brassica napus L.) identify a novel member modulating ROS accumulation and hypersensitive-like cell death. DNA Res. 2016, 23, 101–114. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.-Y.; Wang, S.-Y.; Li, X.-F.; Xu, H.; Zhang, H.; Zhu, Y.; Wang, L.-C. Progress of MYB transcription factors involved in plant abiotic stress response and plant hormone response. Zhejiang J. Agric. 2020, 32, 1317–1328. [Google Scholar]
- Cathie, M.; Javier, P.A. MYB transcription factors in plants. Trends Genet. 1997, 13, 67–73. [Google Scholar]
- Yang, K.Y.; Dong, Q.L.; Wu, J.H.; Li, H.; Luan, H.A.; Jia, P.; Zhang, X.M.; Guo, S.P.; Yang, M.S.; Qi, G.H. Genome-wide analysis of the R2R3-MYB transcription factor gene family expressed in Juglans regia under abiotic and biotic stresses. Ind. Crop. Prod. 2023, 198, 116709. [Google Scholar] [CrossRef]
- Li, B.Z.; Fan, R.N.; Guo, S.Y.; Wang, P.T.; Zhu, X.H.; Fan, Y.T.; Chen, Y.X.; He, K.Y.; Kumar, A.; Shi, J.P.; et al. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 2019, 166, 103807. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. The MYB96-HHP module integrates cold and abscisic acid signaling to activate the CBF-COR pathway in Arabidopsis. Plant J. 2015, 82, 962–977. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- He, Y.; Wei, L.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 1511–1522. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Rao, Y.; Wang, B.; Tian, R.; Tan, Y.; Peng, T. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii. J. Plant Physiol. 2023, 281, 153925. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.Y.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold and dehyderation tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Xie, Y.P.; Chen, P.X.; Yan, Y.; Bao, C.; Li, X.W.; Wang, L.P.; Shen, X.X.; Li, H.Y.; Liu, X.F.; Niu, C.; et al. An atypical R2R3 MYB transcription factor increases coldhardiness by CBF-dependent and CBF-independent pathways inapple. New Phytol. 2017, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yu, S.; Shi, G.F.; Yan, L.; Lv, R.T.; Ma, Z.; Wang, L. Comparative analysis of R2R3-MYB transcription factors in the flowers of Iris Ieavigata identifies a novel gene regulating tobacco cold tolerance. Plant Biol. 2022, 24, 1066–1075. [Google Scholar] [CrossRef]
- Han, J.X.; Li, X.G.; Li, W.H.; Yang, Q.; Li, Z.H.; Cheng, Z.; Lv, L.; Zhang, L.H.; Han, D.G. Isolation and preliminary functional analysis of FvICE1, involved in cold and drought tolerance in Fragaria vesca through overexpression and CRISPR/Cas9 technologies. Plant Physiol. Biochem. 2023, 196, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Q.; Xie, Q.; Yan, J.H.; Chen, J.W.; Chen, Q.X. Genome-wide identification and characterization of the abiotic-stress-responsive lipoxygenase gene family in diploid woodland strawberry (Fragaria vesca). J. Integr. Agric. 2022, 21, 1982–1996. [Google Scholar] [CrossRef]
- Wei, W.; Hu, Y.; Han, Y.T.; Zhang, K.; Zhao, F.L.; Feng, J.Y. The WRKY transcription factors in the diploid woodland strawberry Fragaria vesca: Identification and expression analysis under biotic and abiotic stresses. Plant Physiol. Biochem. 2016, 105, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.X.; Wei, X.X.; Lan, H.Y. CgMYB1, an R2R3-MYB transcription factor, can alleviate abiotic stress in an annual halophyte Chenopodium glaucum. Plant Physiol. Biochem. 2023, 196, 484–496. [Google Scholar] [CrossRef]
- Wen, X.; Geng, F.; Cheng, Y.; Wang, J.Q. Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 166, 777–788. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Liang, X.Q.; Luo, G.J.; Li, W.H.; Yao, A.Q.; Liu, W.D.; Xie, L.P.; Han, M.N.; Li, X.G.; Han, D.G. Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, Increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol Bioch. 2022, 192, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Moseki, B.; Musil, C.F. Characterisation of low temperature stress effects on photosynthetic performance of maize cultivars using chlorophyll fluorescence. S. Afr. J. Bot. 2004, 70, 730–733. [Google Scholar] [CrossRef][Green Version]
- Aydın, A. The Growth, Leaf Antioxidant Enzymes and Amino Acid Content of Tomato as Affected by Grafting on Wild Tomato Rootstocks 1 (S. Pimpinellifolium and S. Habrochaites) Under Salt Stress. Sci. Hortic. 2024, 325, 112679. [Google Scholar] [CrossRef]
- Min, Y.F.; Yu, D.; Yang, J.H.; Zhao, W.D.; Zhang, L.H.; Bai, Y.; Guo, C.H. Bioinformatics and expression analysis of proline metabolism-related gene families in alfalfa under saline-alkali stress. Plant Physiol. Biochem. 2023, 205, 108182. [Google Scholar] [CrossRef] [PubMed]
- Peymaei, M.; Sarabi, V.; Hashempour, H. Improvement of the yield and essential oil of fennel (Foeniculum vulgare Mill.) using external proline, uniconazole and methyl jasmonate under drought stress conditions. Sci. Hort. 2024, 323, 112488. [Google Scholar] [CrossRef]
- Xu, W.B.; Miao, Y.M.; Kong, J.; Lindsey, K.; Zhang, X.L.; Min, L. ROS signaling and its involvement in abiotic stress with emphasis on heat stress-driven anther sterility in plants. Crop Environ. 2023, in press. [Google Scholar] [CrossRef]
- Huang, H.N.; Zheng, X.; Yang, S.Y.; Chen, Y.G. More than sulfidation: Roles of biogenic sulfide in attenuating the impacts of CuO nanoparticle on antibiotic resistance genes during sludge anaerobic digestion. Water Res. 2019, 158, 1–10. [Google Scholar] [CrossRef]
- Xu, Z.C.; Xin, T.Y.; Bartels, D.; Li, Y.; Gu, W.; Yao, H.; Liu, S.; Yu, H.Y.; Pu, X.D.; Zhou, J.G.; et al. Genome Analysis of the Ancient Tracheophyte Selaginella tamariscina Reveals Evolutionary Features Relevant to the Acquisition of Desiccation Tolerance. Mol. Plant. 2018, 11, 983–994. [Google Scholar] [CrossRef]
- Salih, H.; Bai, W.W.; Liang, Y.Q.; Yang, R.R.; Zhao, M.Q.; Muhammd, S.M.; Zhang, D.Y.; Li, X.H. ROS scavenging enzyme-encoding genes play important roles in the desert moss Syntrichia caninervis response to extreme cold and desiccation stresses. Int. J. Biol. Macromol. 2024, 254, 127778. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Zhu, L.; Song, A.P.; Wang, H.B.; Chen, S.M.; Jiang, J.F.; Chen, F.D. Chrysanthemum (Chrysanthemum morifolium) CmICE2 conferred freezing tolerance in Arabidopsis. Plant Physiol. Biochem. 2020, 146, 31–41. [Google Scholar] [CrossRef]
- Han, J.X.; Li, X.G.; Li, W.H.; Yao, A.Q.; Niu, C.G.; Hou, R.N.; Liu, W.D.; Wang, Y.; Zhang, L.H.; Han, D.G. Overexpression of Malus baccata WRKY40 (MbWRKY40) enhances stress tolerance in Arabidopsis subjected to cold and drought. Plant Stress. 2023, 10, 100209. [Google Scholar] [CrossRef]
- Han, D.G.; Han, J.X.; Yang, G.H.; Wang, S.; Xu, T.L.; Li, W.H. An ERF transcription factor gene from Malus baccata (L.) borkh, MbERF11, affects cold and salt stress tolerance in Arabidopsis. Forests 2020, 11, 514. [Google Scholar] [CrossRef]
- Jiang, K.Y.; Zhou, M.B. Cloning and functional characterization of PjPORB, a memberof the POR gene family in Pseudosasa japonica cv. Akebonosuji. Plant Growth Regul. 2016, 79, 95–106. [Google Scholar] [CrossRef]
- Wang, M.; Dai, W.; Du, J.; Ming, R.H.; Bachar, D.; Liu, J.H. ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 2019, 17, 1316–1332. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Senuma, M.; Dobashi, S.; Bando, Y.; Ko, S.; Shiota, H. Overexpression of eelgrass Rare Cold Inducible 2 (RCI2) maintains chlorophyll content in Arabidopsis subjected to high salinity and dehydration. Plant Stress. 2022, 6, 100116. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Gitelson, A.; Lang, M. Non-Destructive Determination of Chlorophyll Content of Leaves of a Green and an Aurea Mutant of Tobacco by Reflectance Measurements. J. Plant Physiol. 1996, 148, 483–493. [Google Scholar] [CrossRef]
- Ding, Z.S.; Zhou, B.Y.; Sun, X.F.; Zhao, M. High Light Tolerance is Enhanced by Overexpressed PEPC in Rice Under Drought Stress. Acta Agron. Sin. 2012, 38, 285–292. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Wan, H.; Tang, J.; Ni, Z. The sea-island cotton GbTCP4 transcription factor positively regulates drought and salt stress responses. Plant Sci. 2022, 322, 111329. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.J.; Wei, G.; Zhang, Z.Q.; Dai, X.Z.; Zou, X.X. Effects of low temperature and low irradiance on the physiological characteristics and related gene expression of different pepper species. Photosynthetica 2015, 53, 85–94. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, P.; Yang, R.; Jiao, K.; Guo, B.; Zhang, L.; Li, Y.; Zhang, K.; Zhou, S.; Wu, X.; Li, X. FvMYB108, a MYB Gene from Fragaria vesca, Positively Regulates Cold and Salt Tolerance of Arabidopsis. Int. J. Mol. Sci. 2024, 25, 3405. https://doi.org/10.3390/ijms25063405
Song P, Yang R, Jiao K, Guo B, Zhang L, Li Y, Zhang K, Zhou S, Wu X, Li X. FvMYB108, a MYB Gene from Fragaria vesca, Positively Regulates Cold and Salt Tolerance of Arabidopsis. International Journal of Molecular Sciences. 2024; 25(6):3405. https://doi.org/10.3390/ijms25063405
Chicago/Turabian StyleSong, Penghui, Ruihua Yang, Kuibao Jiao, Baitao Guo, Lei Zhang, Yuze Li, Kun Zhang, Shuang Zhou, Xinjuan Wu, and Xingguo Li. 2024. "FvMYB108, a MYB Gene from Fragaria vesca, Positively Regulates Cold and Salt Tolerance of Arabidopsis" International Journal of Molecular Sciences 25, no. 6: 3405. https://doi.org/10.3390/ijms25063405
APA StyleSong, P., Yang, R., Jiao, K., Guo, B., Zhang, L., Li, Y., Zhang, K., Zhou, S., Wu, X., & Li, X. (2024). FvMYB108, a MYB Gene from Fragaria vesca, Positively Regulates Cold and Salt Tolerance of Arabidopsis. International Journal of Molecular Sciences, 25(6), 3405. https://doi.org/10.3390/ijms25063405




