The Transcriptional Landscape of Immune-Response 3′-UTR Alternative Polyadenylation in Melanoma
Abstract
1. Introduction
2. Results
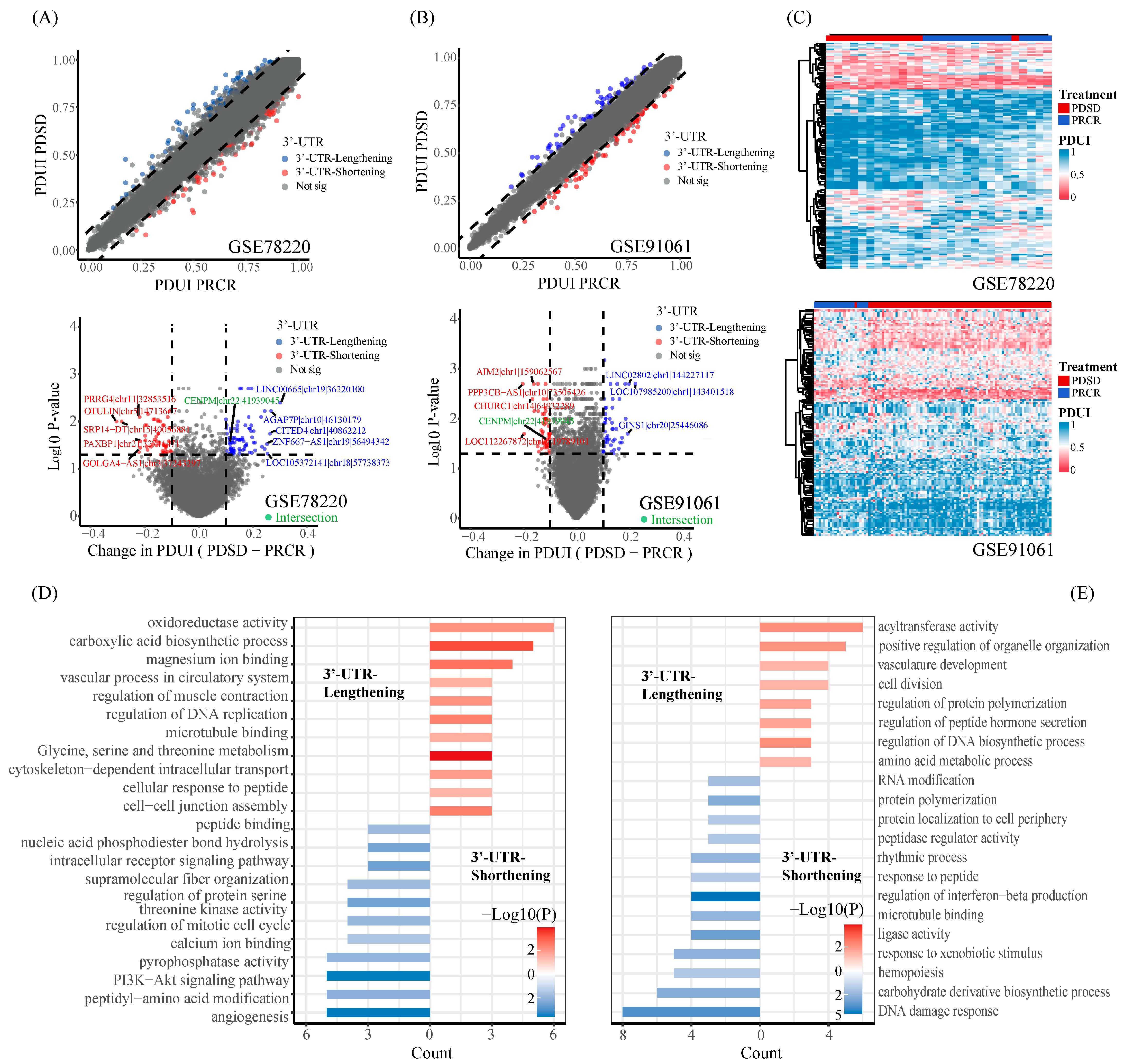
2.1. Landscape of Differential APA Events between Responders and Non-Responders in Melanoma
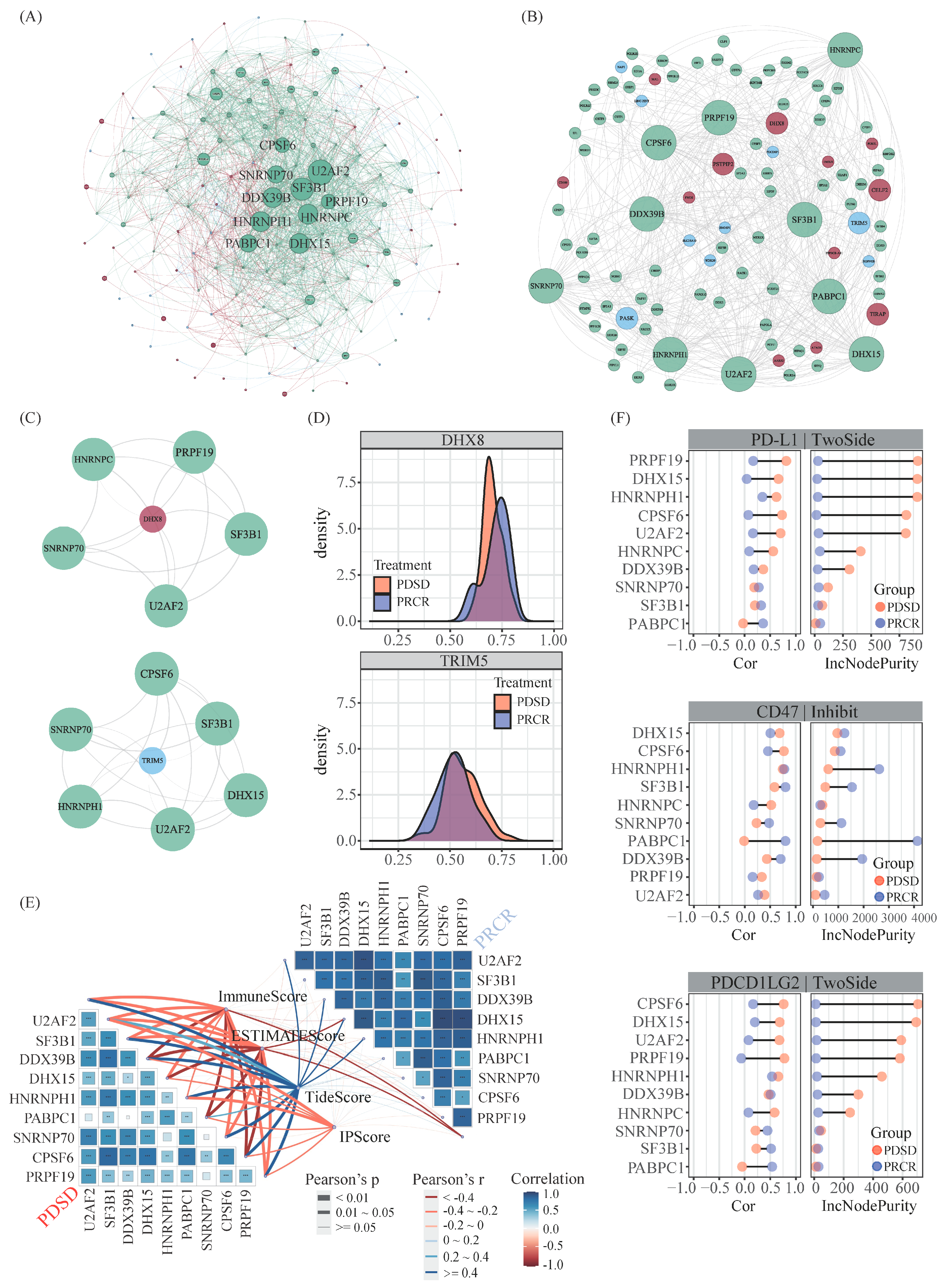
2.2. Master APA Factors Mediating APA Events and Probable Impact on Immunity in Melanoma Patients
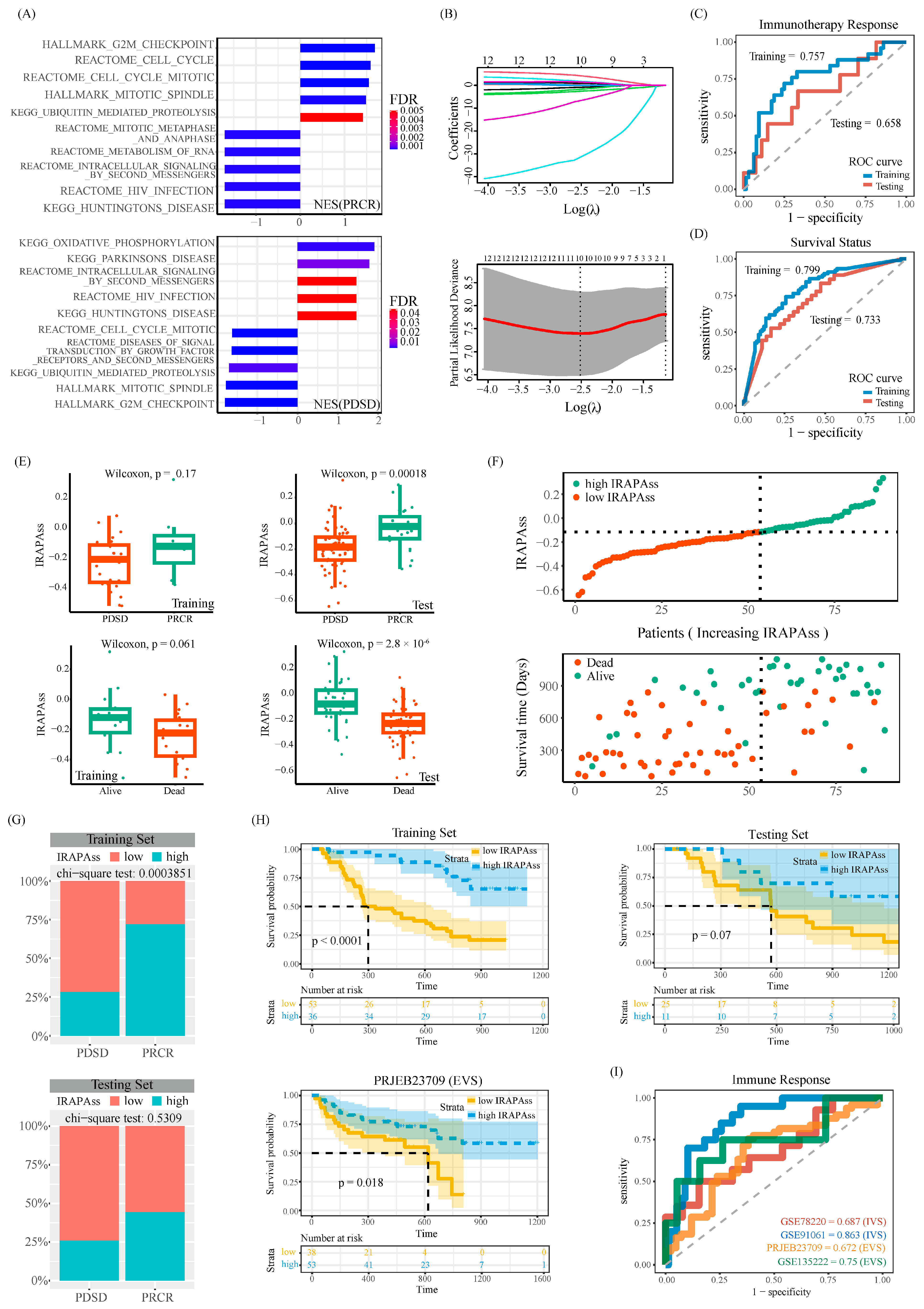
2.3. Establishment of the IRAPAss Model for Predicting the Response to Immunotherapy in Melanoma Patients
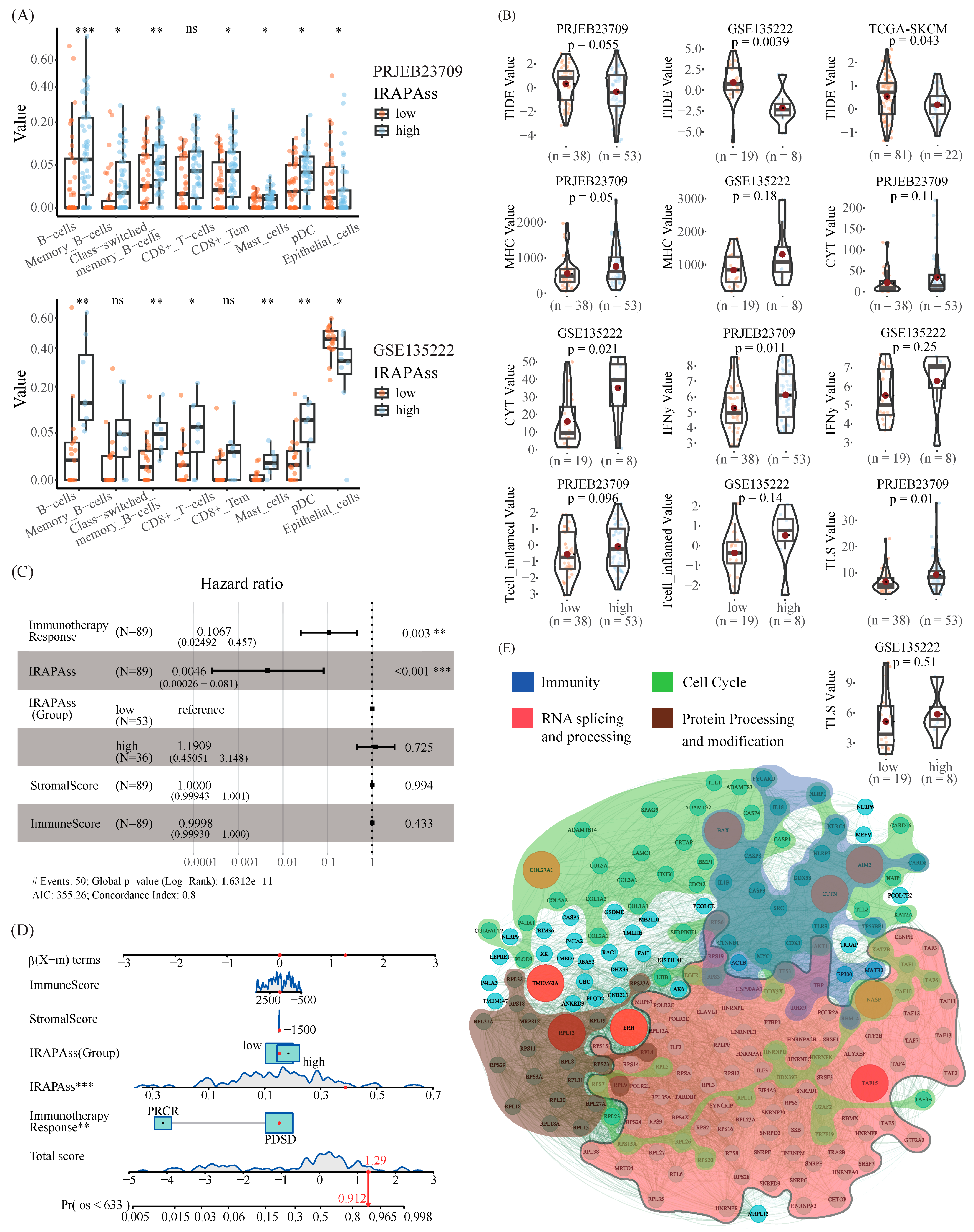
2.4. Correlation Analysis between IRAPAss and TME
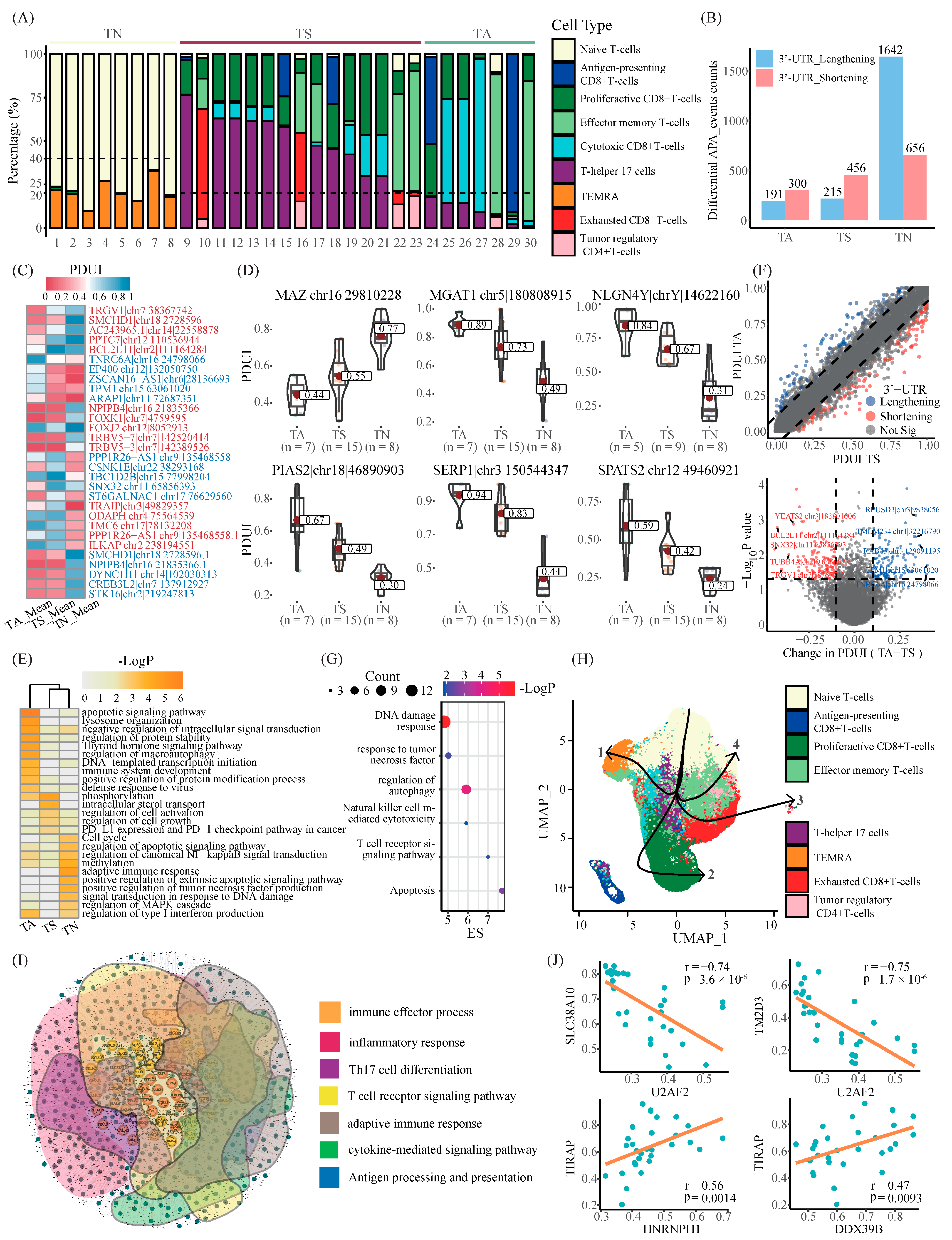
2.5. Master APA Factors Are Prone to Diverging across Distinct T Cell Populations in Melanoma
2.6. Global Discrepancy in 3′-UTR Length across Diverse T Cell Populations May Contribute to Immunoregulation in Melanoma
3. Discussion
4. Materials and Methods
4.1. Data Resources
4.2. Data Pre-Processing
4.3. Differential APA Events Detection and Downstream Analysis
4.4. Computational Assessment of Immune Microenvironment
4.5. Establishment of an Immunotherapy Response-Related APA Scoring System (IRAPAss)
4.6. Braided Target–Proximal Gene PPI Network
4.7. Single-Cell RNA-Seq Analysis
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, Y.; Wang, S.; Liu, S.; Lv, S.; Wang, H.; Li, F. Identification and Verification of Molecular Subtypes with Enhanced Immune Infiltration Based on m6A Regulators in Cutaneous Melanoma. Biomed Res. Int. 2021, 2021, 2769689. [Google Scholar] [CrossRef] [PubMed]
- de Unamuno Bustos, B.; Murria Estal, R.; Perez Simo, G.; Simarro Farinos, J.; Pujol Marco, C.; Navarro Mira, M.; Alegre de Miquel, V.; Ballester Sanchez, R.; Sabater Marco, V.; Llavador Ros, M.; et al. Aberrant DNA methylation is associated with aggressive clinicopathological features and poor survival in cutaneous melanoma. Br. J. Dermatol. 2018, 179, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Frommel, S.C.; Oakes, C.C.; Simon, R.; Grupp, K.; Gerig, C.Y.; Bar, D.; Robinson, M.D.; Baer, C.; Weiss, M.; et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 2015, 47, 22–30. [Google Scholar] [CrossRef]
- Shen, S.; Faouzi, S.; Bastide, A.; Martineau, S.; Malka-Mahieu, H.; Fu, Y.; Sun, X.; Mateus, C.; Routier, E.; Roy, S.; et al. An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat. Commun. 2019, 10, 5713. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Donehower, L.A.; Cooper, T.A.; Neilson, J.R.; Wheeler, D.A.; Wagner, E.J.; Li, W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat. Commun. 2014, 5, 5274. [Google Scholar] [CrossRef]
- Feng, X.; Li, L.; Wagner, E.J.; Li, W. TC3A: The Cancer 3′ UTR Atlas. Nucleic Acids Res. 2018, 46, D1027–D1030. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, Q.; Miao, Y.R.; Yang, J.; Yang, W.; Yu, F.; Wang, D.; Guo, A.Y.; Gong, J. SNP2APA: A database for evaluating effects of genetic variants on alternative polyadenylation in human cancers. Nucleic Acids Res. 2020, 48, D226–D232. [Google Scholar] [CrossRef] [PubMed]
- Albittar, A.A.; Alhalabi, O.; Glitza Oliva, I.C. Immunotherapy for Melanoma. Adv. Exp. Med. Biol. 2020, 1244, 51–68. [Google Scholar] [CrossRef]
- Jia, X.; Yuan, S.; Wang, Y.; Fu, Y.; Ge, Y.; Ge, Y.; Lan, X.; Feng, Y.; Qiu, F.; Li, P.; et al. The role of alternative polyadenylation in the antiviral innate immune response. Nat. Commun. 2017, 8, 14605. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, L.; Qiu, Q.; Zhou, Q.; Ding, J.; Lu, Y.; Liu, P. Alternative polyadenylation: Methods, mechanism, function, and role in cancer. J. Exp. Clin. Cancer Res. 2021, 40, 51. [Google Scholar] [CrossRef]
- Zhang, J.; Dang, F.; Ren, J.; Wei, W. Biochemical Aspects of PD-L1 Regulation in Cancer Immunotherapy. Trends Biochem. Sci. 2018, 43, 1014–1032. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Li, C.; Jiang, T. Systemic Analysis of the Prognosis-Associated Alternative Polyadenylation Events in Breast Cancer. Front. Genet. 2020, 11, 590770. [Google Scholar] [CrossRef]
- Lianoglou, S.; Garg, V.; Yang, J.L.; Leslie, C.S.; Mayr, C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013, 27, 2380–2396. [Google Scholar] [CrossRef]
- Davis, A.G.; Shima, T.; Wang, R.; Zheng, D.; Tian, B.; Zhang, D.-E. FIP1L1 Regulates Alternative Polyadenylation of Leukemia-Associated Genes in Acute Myeloid Leukemia. Blood 2018, 132, 3882. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, J.-X.; Luo, J.-H.; Wu, S.; Yuan, G.-J.; Ma, N.-F.; Feng, Y.; Cai, M.-Y.; Chen, R.-X.; Lu, J.; et al. CSTF2-Induced Shortening of the RAC1 3′UTR Promotes the Pathogenesis of Urothelial Carcinoma of the Bladder. Cancer Res. 2018, 78, 5848–5862. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Liu, C.; Li, J.; Yu, T.; Dong, J.; Chen, B.; Du, Y.; Tang, X.; Xi, Y. Construction of Two Alternative Polyadenylation Signatures to Predict the Prognosis of Sarcoma Patients. Front. Cell Dev. Biol. 2021, 9, 595331. [Google Scholar] [CrossRef] [PubMed]
- Hage, A.; Bharaj, P.; van Tol, S.; Giraldo, M.I.; Gonzalez-Orozco, M.; Valerdi, K.M.; Warren, A.N.; Aguilera-Aguirre, L.; Xie, X.; Widen, S.G.; et al. The RNA helicase DHX16 recognizes specific viral RNA to trigger RIG-I-dependent innate antiviral immunity. Cell Rep. 2022, 38, 110434. [Google Scholar] [CrossRef] [PubMed]
- Masamha, C.P.; Xia, Z.; Yang, J.; Albrecht, T.R.; Li, M.; Shyu, A.B.; Li, W.; Wagner, E.J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature 2014, 510, 412–416. [Google Scholar] [CrossRef]
- Xiang, Y.; Ye, Y.; Lou, Y.; Yang, Y.; Cai, C.; Zhang, Z.; Mills, T.; Chen, N.Y.; Kim, Y.; Muge Ozguc, F.; et al. Comprehensive Characterization of Alternative Polyadenylation in Human Cancer. J. Natl. Cancer Inst. 2018, 110, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Zingone, A.; Sinha, S.; Ante, M.; Nguyen, C.; Daujotyte, D.; Bowman, E.D.; Sinha, N.; Mitchell, K.A.; Chen, Q.; Yan, C.; et al. A comprehensive map of alternative polyadenylation in African American and European American lung cancer patients. Nat. Commun. 2021, 12, 5605. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Ji, P.; Niu, X.; Zhao, X.; Chen, Y.; Liu, W.; Liu, Y.; Fan, W.; Sun, Y.; Miao, C. CstF64-Induced Shortening of the BID 3′ UTR Promotes Esophageal Squamous Cell Carcinoma Progression by Disrupting ceRNA Cross-talk with ZFP36L2Shortening of the BID 3′ UTR Promotes Esophageal Cancer. Cancer Res. 2021, 81, 5638–5651. [Google Scholar] [CrossRef]
- Wang, G.; Xie, Z.; Su, J.; Chen, M.; Du, Y.; Gao, Q.; Zhang, G.; Zhang, H.; Chen, X.; Liu, H.; et al. Characterization of Immune-Related Alternative Polyadenylation Events in Cancer Immunotherapy. Cancer Res. 2022, 82, 3474–3485. [Google Scholar] [CrossRef] [PubMed]
- Deen, D.; Butter, F.; Daniels, D.E.; Ferrer-Vicens, I.; Ferguson, D.C.; Holland, M.L.; Samara, V.; Sloane-Stanley, J.A.; Ayyub, H.; Mann, M. Identification of the transcription factor MAZ as a regulator of erythropoiesis. Blood Adv. 2021, 5, 3002–3015. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Southard, R.C.; Allred, C.D.; Talbert, D.R.; Wilson, M.E.; Kilgore, M.W. MAZ drives tumor-specific expression of PPAR gamma 1 in breast cancer cells. Breast Cancer Res. Treat. 2008, 111, 103–111. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mayr, C. What Are 3′ UTRs Doing? Cold Spring Harb. Perspect. Biol. 2019, 11, a034728. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Ye, Y.; Zhang, Z.; Han, L. Maximizing the Utility of Cancer Transcriptomic Data. Trends Cancer 2018, 4, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Yousaf, N.; Larkin, J. Melanoma epidemiology, biology and prognosis. EJC Suppl. 2013, 11, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Liu, X.; Zeng, X.; Liu, Y.; Zhang, C.; Zhang, Q.; Xu, J. Comprehensive Analysis of Cuproptosis-Related Genes in Immune Infiltration and Prognosis in Melanoma. Front. Pharmacol. 2022, 13, 930041. [Google Scholar] [CrossRef]
- Rothberg, B.E.G.; Bracken, M.B.; Rimm, D.L. Tissue Biomarkers for Prognosis in Cutaneous Melanoma: A Systematic Review and Meta-analysis. JNCI J. Natl. Cancer Inst. 2009, 101, 452–474. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, Z.; Qiu, N.; Ling, L.; Jia, X.; Song, Y.; Li, H.; Li, J.; Lyu, H.; Liu, H.; et al. Disruption of FOXO3a-miRNA feedback inhibition of IGF2/IGF-1R/IRS1 signaling confers Herceptin resistance in HER2-positive breast cancer. Nat. Commun. 2021, 12, 2699. [Google Scholar] [CrossRef]
- Aksentijevich, I.; Zhou, Q. NF-κB Pathway in Autoinflammatory Diseases: Dysregulation of Protein Modifications by Ubiquitin Defines a New Category of Autoinflammatory Diseases. Front. Immunol. 2017, 8, 399. [Google Scholar] [CrossRef]
- Zou, W.; Restifo, N.P. T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 2010, 10, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gao, F.H. Th17 Cells Paradoxical Roles in Melanoma and Potential Application in Immunotherapy. Front. Immunol. 2019, 10, 187. [Google Scholar] [CrossRef] [PubMed]
- Perez-Lorenzo, R.; Gill, K.Z.; Shen, C.-H.; Zhao, F.X.; Zheng, B.; Schulze, H.-J.; Silvers, D.N.; Brunner, G.; Horst, B.A. A tumor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. J. Investig. Dermatol. 2014, 134, 1359–1368. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, L.; Huang, K.L.; Gao, Y.; Cui, Y.; Wang, G.; Elrod, N.D.; Li, Y.; Chen, Y.E.; Ji, P.; Peng, F.; et al. An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat. Genet. 2021, 53, 994–1005. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z. Complex heatmap visualization. iMeta 2022, 1, e43. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Hankey, W.; Wagner, E.J.; Li, W.; Wang, Q. Alternative polyadenylation of mRNA and its role in cancer. Genes Dis. 2021, 8, 61–72. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martinez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Ye, Z.; Shen, R.; Yu, G.; Wu, J.; Xiong, Y.; Zhou, R.; Qiu, W.; Huang, N.; Sun, L.; et al. IOBR: Multi-Omics Immuno-Oncology Biological Research to Decode Tumor Microenvironment and Signatures. Front. Immunol. 2021, 12, 687975. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. arXiv 2016, arXiv:1603.04467. [Google Scholar]
- Fernandez-Torras, A.; Duran-Frigola, M.; Aloy, P. Encircling the regions of the pharmacogenomic landscape that determine drug response. Genome Med. 2019, 11, 17. [Google Scholar] [CrossRef]
- Kong, J.; Lee, H.; Kim, D.; Han, S.K.; Ha, D.; Shin, K.; Kim, S. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat. Commun. 2020, 11, 5485. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, A.; Swart, P.; Chult, D.S. Exploring Network Structure, Dynamics, and Function Using NetworkX; Los Alamos National Lab. (LANL): Los Alamos, NM, USA, 2008. [Google Scholar]
- Street, K.; Risso, D.; Fletcher, R.B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Iasonos, A.; Schrag, D.; Raj, G.V.; Panageas, K.S. How to build and interpret a nomogram for cancer prognosis. J. Clin. Oncol. 2008, 26, 1364–1370. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Wu, Y.; Chen, X.; Qiu, J.; Huang, C. The Transcriptional Landscape of Immune-Response 3′-UTR Alternative Polyadenylation in Melanoma. Int. J. Mol. Sci. 2024, 25, 3041. https://doi.org/10.3390/ijms25053041
Yang X, Wu Y, Chen X, Qiu J, Huang C. The Transcriptional Landscape of Immune-Response 3′-UTR Alternative Polyadenylation in Melanoma. International Journal of Molecular Sciences. 2024; 25(5):3041. https://doi.org/10.3390/ijms25053041
Chicago/Turabian StyleYang, Xiao, Yingyi Wu, Xingyu Chen, Jiayue Qiu, and Chen Huang. 2024. "The Transcriptional Landscape of Immune-Response 3′-UTR Alternative Polyadenylation in Melanoma" International Journal of Molecular Sciences 25, no. 5: 3041. https://doi.org/10.3390/ijms25053041
APA StyleYang, X., Wu, Y., Chen, X., Qiu, J., & Huang, C. (2024). The Transcriptional Landscape of Immune-Response 3′-UTR Alternative Polyadenylation in Melanoma. International Journal of Molecular Sciences, 25(5), 3041. https://doi.org/10.3390/ijms25053041




