Neutrophil Extracellular DNA Traps in Response to Infection or Inflammation, and the Roles of Platelet Interactions
Abstract
1. Introduction
1.1. Neutrophil Granules
1.2. Neutrophil Antimicrobial Mechanisms
1.3. Neutrophil Adhesion and Extravasation
2. Neutrophil Extracellular DNA Traps
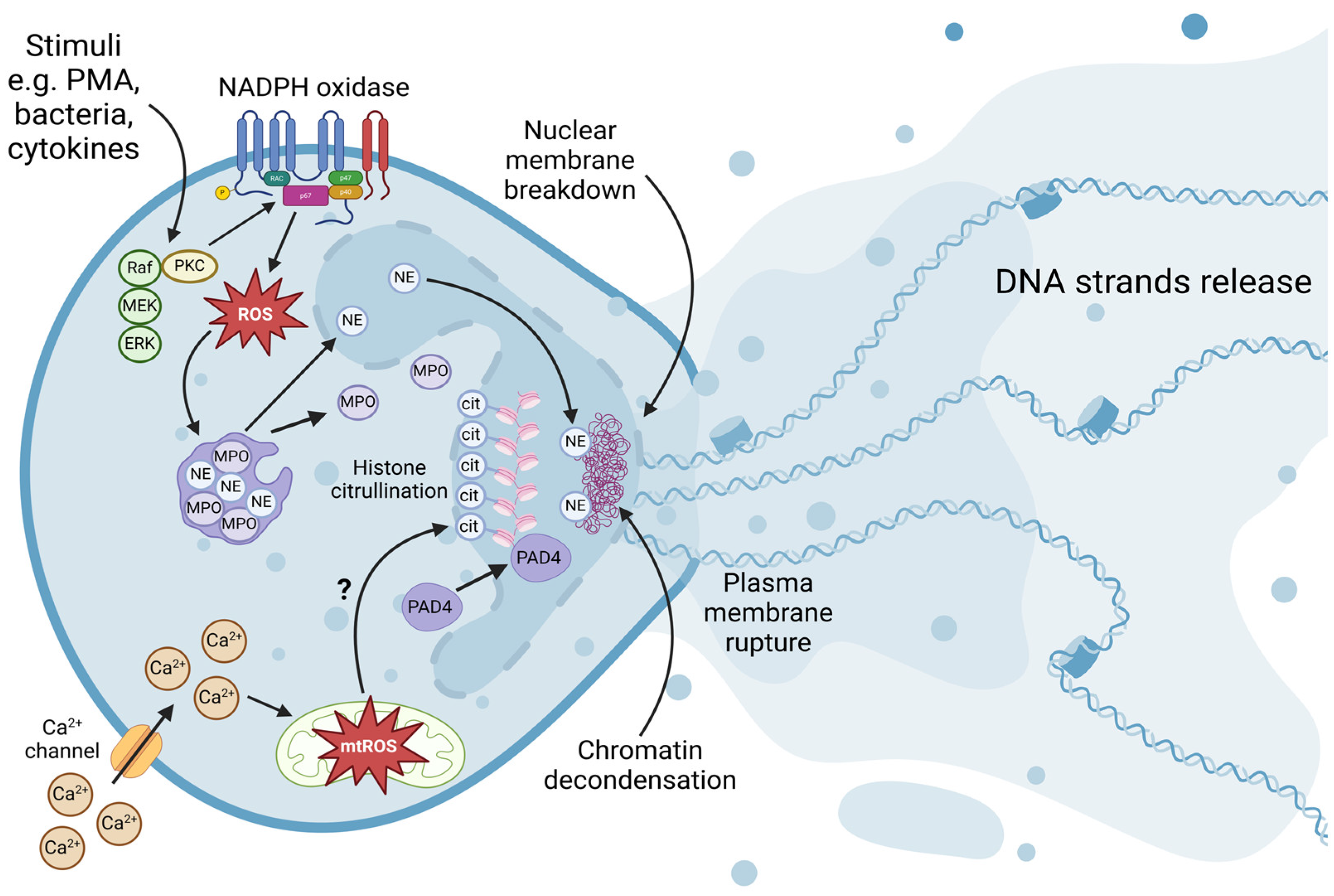
2.1. Formation of NETs
2.1.1. Peptidylarginine Deiminase 4-Mediated Hypercitrullination
2.1.2. Neutrophil Elastase and Myeloperoxidase-Mediated Chromatin Decondensation
2.1.3. Nuclear Envelope Breakdown
2.1.4. Plasma Membrane Rupture
2.1.5. Chromatin Release
2.2. Regulation of NET Formation
2.2.1. NADPH Oxidase-Dependent Pathway
2.2.2. Calcium and Potassium Channel-Dependent Pathway
2.3. NETs and Thrombosis
2.4. NETs and Coagulation
3. NETs and Platelet Functions
3.1. Bacteria Mediated Platelet–Neutrophil Interactions
3.2. Immunothrombosis: Crosstalk between the Hemostatic and Immune Systems
3.2.1. Roles of Neutrophil Toll-like Receptors
3.2.2. Roles of Platelet Toll-like Receptors
3.2.3. Platelet–Neutrophil Interactions
4. Conclusions
Funding
Conflicts of Interest
Abbreviations
References
- Hong, C.-W. Current Understanding in Neutrophil Differentiation and Heterogeneity. Immune Netw. 2017, 17, 298–306. [Google Scholar] [CrossRef]
- Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 2010, 238, 37–46. [Google Scholar] [CrossRef]
- Cowland, J.B.; Borregaard, N. Granulopoiesis and granules of human neutrophils. Immunol. Rev. 2016, 273, 11–28. [Google Scholar] [CrossRef]
- Manz, M.G.; Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 2014, 14, 302–314. [Google Scholar] [CrossRef]
- Adolfsson, J.; Månsson, R.; Buza-Vidas, N.; Hultquist, A.; Liuba, K.; Jensen, C.T.; Bryder, D.; Yang, L.; Borge, O.-J.; Thoren, L.A.; et al. Identification of Flt3+ Lympho-Myeloid Stem Cells Lacking Erythro-Megakaryocytic Potential: A Revised Road Map for Adult Blood Lineage Commitment. Cell 2005, 121, 295–306. [Google Scholar] [CrossRef]
- Görgens, A.; Radtke, S.; Möllmann, M.; Cross, M.; Dürig, J.; Horn, P.A.; Giebel, B. Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. Cell Rep. 2013, 3, 1539–1552. [Google Scholar] [CrossRef]
- Iwasaki, H.; Akashi, K. Myeloid Lineage Commitment from the Hematopoietic Stem Cell. Immunity 2007, 26, 726–740. [Google Scholar] [CrossRef]
- Hu, M.; Krause, D.; Greaves, M.; Sharkis, S.; Dexter, M.; Heyworth, C.; Enver, T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997, 11, 774–785. [Google Scholar] [CrossRef]
- Sionov, R.V.; Fridlender, Z.G.; Granot, Z. The Multifaceted Roles Neutrophils Play in the Tumor Microenvironment. Cancer Microenviron. 2015, 8, 125–158. [Google Scholar] [CrossRef]
- Faurschou, M.; Borregaard, N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 2003, 5, 1317–1327. [Google Scholar] [CrossRef]
- Borregaard, N.; Sørensen, O.E.; Theilgaard-Mönch, K. Neutrophil granules: A library of innate immunity proteins. Trends Immunol. 2007, 28, 340–345. [Google Scholar] [CrossRef]
- Rørvig, S.; Østergaard, O.; Heegaard, N.H.H.; Borregaard, N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: Correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol. 2013, 94, 711–721. [Google Scholar] [CrossRef]
- Lominadze, G.; Powell, D.W.; Luerman, G.C.; Link, A.J.; Ward, R.A.; McLeish, K.R. Proteomic Analysis of Human Neutrophil Granules. Mol. Cell. Proteom. 2005, 4, 1503–1521. [Google Scholar] [CrossRef]
- Vaissiere, C.; Le Cabec, V.; Maridonneau-Parini, I. NADPH oxidase is functionally assembled in specific granules during activation of human neutrophils. J. Leukoc. Biol. 1999, 65, 629–634. [Google Scholar] [CrossRef]
- Jethwaney, D.; Islam, R.; Leidal, K.G.; de Bernabe, D.B.-V.; Campbell, K.P.; Nauseef, W.M.; Gibson, B.W. Proteomic analysis of plasma membrane and secretory vesicles from human neutrophils. Proteome Sci. 2007, 5, 12. [Google Scholar] [CrossRef]
- Uriarte, S.M.; Powell, D.W.; Luerman, G.C.; Merchant, M.L.; Cummins, T.D.; Jog, N.R.; Ward, R.A.; McLeish, K.R. Comparison of Proteins Expressed on Secretory Vesicle Membranes and Plasma Membranes of Human Neutrophils. J. Immunol. 2008, 180, 5575–5581. [Google Scholar] [CrossRef]
- Lollike, K.; Kjeldsen, L.; Sengeløv, H.; Borregaard, N. Lysozyme in human neutrophils and plasma. A parameter of myelopoietic activity. Leukemia 1995, 9, 159–164. [Google Scholar]
- Brieger, K.; Schiavone, S.; Miller, F.J., Jr.; Krause, K.-H. Reactive oxygen species: From health to disease. Swiss Med. Wkly. 2012, 142, w13659. [Google Scholar] [CrossRef]
- Nordenfelt, P.; Tapper, H. Phagosome dynamics during phagocytosis by neutrophils. J. Leukoc. Biol. 2011, 90, 271–284. [Google Scholar] [CrossRef]
- Lacy, P. Mechanisms of Degranulation in Neutrophils. Allergy Asthma Clin. Immunol. 2006, 2, 98–108. [Google Scholar] [CrossRef]
- Sengeløv, H.; Kjeldsen, L.; Borregaard, N. Control of exocytosis in early neutrophil activation. J. Immunol. 1993, 150, 1535–1543. [Google Scholar] [CrossRef]
- Zhang, J.; Alcaide, P.; Liu, L.; Sun, J.; He, A.; Luscinskas, F.W.; Shi, G.-P. Regulation of Endothelial Cell Adhesion Molecule Expression by Mast Cells, Macrophages, and Neutrophils. PLoS ONE 2011, 6, e14525. [Google Scholar] [CrossRef]
- Barthel, S.R.; Gavino, J.D.; Descheny, L.; Dimitroff, C.J. Targeting selectins and selectin ligands in inflammation and cancer. Expert Opin. Ther. Targets 2007, 11, 1473–1491. [Google Scholar] [CrossRef]
- Kolaczkowska, E.; Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 2013, 13, 159–175. [Google Scholar] [CrossRef]
- McEver, R.P. Selectins: Initiators of leucocyte adhesion and signalling at the vascular wall. Cardiovasc. Res. 2015, 107, 331–339. [Google Scholar] [CrossRef]
- Marshall, B.T.; Long, M.; Piper, J.W.; Yago, T.; McEver, R.P.; Zhu, C. Direct observation of catch bonds involving cell-adhesion molecules. Nature 2003, 423, 190–193. [Google Scholar] [CrossRef]
- Yago, T.; Wu, J.; Wey, C.D.; Klopocki, A.G.; Zhu, C.; McEver, R.P. Catch bonds govern adhesion through L-selectin at threshold shear. J. Cell Biol. 2004, 166, 913–923. [Google Scholar] [CrossRef]
- Sundd, P.; Pospieszalska, M.K.; Cheung, L.S.-L.; Konstantopoulos, K.; Ley, K. Biomechanics of leukocyte rolling. Biorheology 2011, 48, 1–35. [Google Scholar] [CrossRef]
- Mócsai, A.; Walzog, B.; Lowell, C.A. Intracellular signalling during neutrophil recruitment. Cardiovasc. Res. 2015, 107, 373–385. [Google Scholar] [CrossRef]
- Ley, K.; Laudanna, C.; Cybulsky, M.I.; Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 2007, 7, 678–689. [Google Scholar] [CrossRef]
- Li, N.; Yang, H.; Wang, M.; Lü, S.; Zhang, Y.; Long, M. Ligand-specific binding forces of LFA-1 and Mac-1 in neutrophil adhesion and crawling. Mol. Biol. Cell 2018, 29, 408–418. [Google Scholar] [CrossRef]
- Hentzen, E.R.; Neelamegham, S.; Kansas, G.S.; Benanti, J.A.; McIntire, L.V.; Smith, C.W.; Simon, S.I. Sequential binding of CD11a/CD18 and CD11b/CD18 defines neutrophil capture and stable adhesion to intercellular adhesion molecule–1. Blood 2000, 95, 911–920. [Google Scholar] [CrossRef]
- Phillipson, M.; Heit, B.; Colarusso, P.; Liu, L.; Ballantyne, C.M.; Kubes, P. Intraluminal crawling of neutrophils to emigration sites: A molecularly distinct process from adhesion in the recruitment cascade. J. Exp. Med. 2006, 203, 2569–2575. [Google Scholar] [CrossRef]
- Huang, M.-T.; Larbi, K.Y.; Scheiermann, C.; Woodfin, A.; Gerwin, N.; Haskard, D.O.; Nourshargh, S. ICAM-2 mediates neutrophil transmigration in vivo: Evidence for stimulus specificity and a role in PECAM-1–independent transmigration. Blood 2006, 107, 4721–4727. [Google Scholar] [CrossRef]
- Sundd, P.; Gutierrez, E.; Koltsova, E.K.; Kuwano, Y.; Fukuda, S.; Pospieszalska, M.K.; Groisman, A.; Ley, K. ‘Slings’ enable neutrophil rolling at high shear. Nature 2012, 488, 399–403. [Google Scholar] [CrossRef]
- Hind, L.E.; Vincent, W.J.; Huttenlocher, A. Leading from the Back: The Role of the Uropod in Neutrophil Polarization and Migration. Dev. Cell 2016, 38, 161–169. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Riel-Mehan, M.; Chen, B.-C.; Lord, S.J.; Goddard, T.D.; E Ferrin, T.; Nicholson-Dykstra, S.M.; Higgs, H.; Johnson, G.T.; Betzig, E.; et al. Actin-based protrusions of migrating neutrophils are intrinsically lamellar and facilitate direction changes. eLife 2017, 6, e26990. [Google Scholar] [CrossRef]
- Smith, L.A.; Aranda-Espinoza, H.; Haun, J.B.; Dembo, M.; Hammer, D.A. Neutrophil Traction Stresses are Concentrated in the Uropod during Migration. Biophys. J. 2007, 92, L58–L60. [Google Scholar] [CrossRef]
- Jannat, R.A.; Dembo, M.; Hammer, D.A. Traction Forces of Neutrophils Migrating on Compliant Substrates. Biophys. J. 2011, 101, 575–584. [Google Scholar] [CrossRef]
- Shaw, S.K.; Bamba, P.S.; Perkins, B.N.; Luscinskas, F.W. Real-Time Imaging of Vascular Endothelial-Cadherin During Leukocyte Transmigration Across Endothelium. J. Immunol. 2001, 167, 2323–2330. [Google Scholar] [CrossRef]
- Van Buul, J.D.; Kanters, E.; Hordijk, P.L. Endothelial Signaling by Ig-Like Cell Adhesion Molecules. Arter. Thromb. Vasc. Biol. 2007, 27, 1870–1876. [Google Scholar] [CrossRef]
- Carman, C.V.; Sage, P.T.; Sciuto, T.E.; de la Fuente, M.A.; Geha, R.S.; Ochs, H.D.; Dvorak, H.F.; Dvorak, A.M.; Springer, T.A. Transcellular Diapedesis Is Initiated by Invasive Podosomes. Immunity 2007, 26, 784–797. [Google Scholar] [CrossRef]
- Filippi, M.-D. Neutrophil transendothelial migration: Updates and new perspectives. Blood 2019, 133, 2149–2158. [Google Scholar] [CrossRef]
- Vestweber, D. How leukocytes cross the vascular endothelium. Nat. Rev. Immunol. 2015, 15, 692–704. [Google Scholar] [CrossRef]
- Woodfin, A.; Voisin, M.-B.; Beyrau, M.; Colom, B.; Caille, D.; Diapouli, F.-M.; Nash, G.B.; Chavakis, T.; Albelda, S.M.; Rainger, G.E.; et al. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 2011, 12, 761–769. [Google Scholar] [CrossRef]
- Martinelli, R.; Zeiger, A.S.; Whitfield, M.; Scuito, T.E.; Dvorak, A.; Van Vliet, K.J.; Greenwood, J.; Carman, C.V. Probing the biomechanical contribution of the endothelium to lymphocyte migration: Diapedesis by the path of least resistance. J. Cell Sci. 2014, 127, 3720–3734. [Google Scholar] [CrossRef]
- Peter, T.S.; Sage, P.T.; Carman, C.V. Settings and mechanisms for trans-cellular diapedesis. Front. Biosci. 2009, 14, 5066–5083. [Google Scholar] [CrossRef]
- Schaefer, A.; Riet, J.T.; Ritz, K.; Hoogenboezem, M.; Anthony, E.C.; Mul, F.P.J.; de Vries, C.J.; Daemen, M.J.; Figdor, C.G.; van Buul, J.D.; et al. Actin-binding proteins differentially regulate endothelial cell stiffness, ICAM-1 function and neutrophil transmigration. J. Cell Sci. 2014, 127, 4470–4482. [Google Scholar] [CrossRef]
- Schimmel, L.; Heemskerk, N.; Van Buul, J.D. Leukocyte transendothelial migration: A local affair. Small GTPases 2016, 8, 1–15. [Google Scholar] [CrossRef]
- Wolburg, H.; Wolburg-Buchholz, K.; Engelhardt, B. Diapedesis of mononuclear cells across cerebral venules during experimental autoimmune encephalomyelitis leaves tight junctions intact. Acta Neuropathol. 2004, 109, 181–190. [Google Scholar] [CrossRef]
- Woodfin, A.; Voisin, M.-B.; Nourshargh, S. Recent developments and complexities in neutrophil transmigration. Curr. Opin. Hematol. 2010, 17, 9–17. [Google Scholar] [CrossRef]
- Yang, L.; Froio, R.M.; Sciuto, T.E.; Dvorak, A.M.; Alon, R.; Luscinskas, F.W. ICAM-1 regulates neutrophil adhesion and transcellular migration of TNF-α-activated vascular endothelium under flow. Blood 2005, 106, 584–592. [Google Scholar] [CrossRef]
- Schauer, C.; Janko, C.; Munoz, L.E.; Zhao, Y.; Kienhöfer, D.; Frey, B.; Lell, M.; Manger, B.; Rech, J.; Naschberger, E.; et al. Aggregated neutrophil extracellular traps limit inflammation by degrading cytokines and chemokines. Nat. Med. 2014, 20, 511–517. [Google Scholar] [CrossRef]
- Urban, C.F.; Reichard, U.; Brinkmann, V.; Zychlinsky, A. Neutrophil extracellular traps capture and kill Candida albicans yeast and hyphal forms. Cell. Microbiol. 2005, 8, 668–676. [Google Scholar] [CrossRef]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef]
- Halverson, T.W.R.; Wilton, M.; Poon, K.K.H.; Petri, B.; Lewenza, S. DNA Is an Antimicrobial Component of Neutrophil Extracellular Traps. PLoS Pathog. 2015, 11, e1004593. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Krumbholz, M.; Schönermarck, U.; Back, W.; Gross, W.L.; Werb, Z.; Gröne, H.-J.; Brinkmann, V.; Jenne, D.E. Netting neutrophils in autoimmune small-vessel vasculitis. Nat. Med. 2009, 15, 623–625. [Google Scholar] [CrossRef]
- Urban, C.F.; Ermert, D.; Schmid, M.; Abu-Abed, U.; Goosmann, C.; Nacken, W.; Brinkmann, V.; Jungblut, P.R.; Zychlinsky, A. Neutrophil Extracellular Traps Contain Calprotectin, a Cytosolic Protein Complex Involved in Host Defense against Candida albicans. PLOS Pathog. 2009, 5, e1000639. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Abed, U.; Goosmann, C.; Hurwitz, R.; Schulze, I.; Wahn, V.; Weinrauch, Y.; Brinkmann, V.; Zychlinsky, A. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 2007, 176, 231–241. [Google Scholar] [CrossRef]
- Papayannopoulos, V.; Metzler, K.D.; Hakkim, A.; Zychlinsky, A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. J. Cell Biol. 2010, 191, 677–691. [Google Scholar] [CrossRef]
- Wang, Y.; Li, M.; Stadler, S.; Correll, S.; Li, P.; Wang, D.; Hayama, R.; Leonelli, L.; Han, H.; Grigoryev, S.A.; et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009, 184, 205–213. [Google Scholar] [CrossRef]
- Bicker, K.L.; Thompson, P.R. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers 2013, 99, 155–163. [Google Scholar] [CrossRef]
- Wang, Y.; Wysocka, J.; Sayegh, J.; Lee, Y.-H.; Perlin, J.R.; Leonelli, L.; Sonbuchner, L.S.; McDonald, C.H.; Cook, R.G.; Dou, Y.; et al. Human PAD4 Regulates Histone Arginine Methylation Levels via Demethylimination. Science 2004, 306, 279–283. [Google Scholar] [CrossRef]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone Deimination Antagonizes Arginine Methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef]
- Iwasaki, W.; Miya, Y.; Horikoshi, N.; Osakabe, A.; Taguchi, H.; Tachiwana, H.; Shibata, T.; Kagawa, W.; Kurumizaka, H. Contribution of histone N-terminal tails to the structure and stability of nucleosomes. FEBS Open Bio 2013, 3, 363–369. [Google Scholar] [CrossRef]
- Pepenella, S.; Murphy, K.J.; Hayes, J.J. Intra- and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma 2013, 123, 3–13. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Neeli, I.; Khan, S.N.; Radic, M. Histone Deimination As a Response to Inflammatory Stimuli in Neutrophils. J. Immunol. 2008, 180, 1895–1902. [Google Scholar] [CrossRef]
- Li, P.; Li, M.; Lindberg, M.R.; Kennett, M.J.; Xiong, N.; Wang, Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J. Exp. Med. 2010, 207, 1853–1862. [Google Scholar] [CrossRef]
- György, B.; Tóth, E.; Tarcsa, E.; Falus, A.; Buzás, E.I. Citrullination: A posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 2006, 38, 1662–1677. [Google Scholar] [CrossRef]
- Sørensen, O.E.; Borregaard, N. Neutrophil extracellular traps—The dark side of neutrophils. J. Clin. Investig. 2016, 126, 1612–1620. [Google Scholar] [CrossRef]
- Takahashi, H.; Nukiwa, T.; Yoshimura, K.; Quick, C.D.; States, D.J.; Holmes, M.D.; Whang-Peng, J.; Knutsen, T.; Crystal, R.G. Structure of the human neutrophil elastase gene. J. Biol. Chem. 1988, 263, 14739–14747. [Google Scholar] [CrossRef]
- Belaaouaj, A.; McCarthy, R.; Baumann, M.; Gao, Z.; Ley, T.J.; Abraham, S.N.; Shapiro, S.D. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 1998, 4, 615–618. [Google Scholar] [CrossRef]
- Furtmüller, P.G.; Obinger, C.; Hsuanyu, Y.; Dunford, H.B. Mechanism of reaction of myeloperoxidase with hydrogen peroxide and chloride ion. JBIC J. Biol. Inorg. Chem. 2000, 267, 5858–5864. [Google Scholar] [CrossRef]
- Hirche, T.O.; Gaut, J.P.; Heinecke, J.W.; Belaaouaj, A. Myeloperoxidase Plays Critical Roles in Killing Klebsiella pneumoniae and Inactivating Neutrophil Elastase: Effects on Host Defense. J. Immunol. 2005, 174, 1557–1565. [Google Scholar] [CrossRef]
- Harshman, S.W.; Young, N.L.; Parthun, M.R.; Freitas, M.A. H1 histones: Current perspectives and challenges. Nucleic Acids Res. 2013, 41, 9593–9609. [Google Scholar] [CrossRef]
- Routh, A.; Sandin, S.; Rhodes, D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. USA 2008, 105, 8872–8877. [Google Scholar] [CrossRef]
- Metzler, K.D.; Goosmann, C.; Lubojemska, A.; Zychlinsky, A.; Papayannopoulos, V. A Myeloperoxidase-Containing Complex Regulates Neutrophil Elastase Release and Actin Dynamics during NETosis. Cell Rep. 2014, 8, 883–896. [Google Scholar] [CrossRef]
- Roos, D. Chronic granulomatous disease. Br. Med. Bull. 2016, 118, 50–63. [Google Scholar] [CrossRef]
- De Bont, C.M.; Koopman, W.J.; Boelens, W.C.; Pruijn, G.J. Stimulus-dependent chromatin dynamics, citrullination, calcium signalling and ROS production during NET formation. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2018, 1865, 1621–1629. [Google Scholar] [CrossRef]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef]
- Dobrzynska, A.; Gonzalo, S.; Shanahan, C.; Askjaer, P. The nuclear lamina in health and disease. Nucleus 2016, 7, 233–248. [Google Scholar] [CrossRef]
- Liu, B.; Wang, J.; Chan, K.M.; Tjia, W.M.; Deng, W.; Guan, X.; Huang, J.-D.; Li, K.M.; Chau, P.Y.; Chen, D.J.; et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005, 11, 780–785. [Google Scholar] [CrossRef]
- Nikolova, V.; Leimena, C.; McMahon, A.C.; Tan, J.C.; Chandar, S.; Jogia, D.; Kesteven, S.H.; Michalicek, J.; Otway, R.; Verheyen, F.; et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C–deficient mice. J. Clin. Investig. 2004, 113, 357–369. [Google Scholar] [CrossRef]
- Swift, J.; Ivanovska, I.L.; Buxboim, A.; Harada, T.; Dingal, P.C.D.P.; Pinter, J.; Pajerowski, J.D.; Spinler, K.R.; Shin, J.-W.; Tewari, M.; et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 2013, 341, 1240104. [Google Scholar] [CrossRef]
- Neubert, E.; Meyer, D.; Rocca, F.; Günay, G.; Kwaczala-Tessmann, A.; Grandke, J.; Senger-Sander, S.; Geisler, C.; Egner, A.; Schön, M.P.; et al. Chromatin swelling drives neutrophil extracellular trap release. Nat. Commun. 2018, 9, 3767. [Google Scholar] [CrossRef]
- Thiam, H.R.; Wong, S.L.; Qiu, R.; Kittisopikul, M.; Vahabikashi, A.; Goldman, A.E.; Goldman, R.D.; Wagner, D.D.; Waterman, C.M. NETosis proceeds by cytoskeleton and endomembrane disassembly and PAD4-mediated chromatin decondensation and nuclear envelope rupture. Proc. Natl. Acad. Sci. USA 2020, 117, 7326–7337. [Google Scholar] [CrossRef]
- Li, Y.; Li, M.; Weigel, B.; Mall, M.; Werth, V.P.; Liu, M. Nuclear envelope rupture and NET formation is driven by PKCα-mediated lamin B disassembly. Embo Rep. 2020, 21, e48779. [Google Scholar] [CrossRef]
- Mall, M.; Walter, T.; Gorjánácz, M.; Davidson, I.F.; Ly-Hartig, T.B.N.; Ellenberg, J.; Mattaj, I.W. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J. Cell Biol. 2012, 198, 981–990. [Google Scholar] [CrossRef]
- Amulic, B.; Knackstedt, S.L.; Abu Abed, U.; Deigendesch, N.; Harbort, C.J.; Caffrey, B.E.; Brinkmann, V.; Heppner, F.L.; Hinds, P.W.; Zychlinsky, A. Cell-Cycle Proteins Control Production of Neutrophil Extracellular Traps. Dev. Cell 2017, 43, 449–462.e5. [Google Scholar] [CrossRef]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef]
- Heald, R.; McKeon, F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 1990, 61, 579–589. [Google Scholar] [CrossRef]
- Neeli, I.; Dwivedi, N.; Khan, S.; Radic, M. Regulation of Extracellular Chromatin Release from Neutrophils. J. Innate Immun. 2009, 1, 194–201. [Google Scholar] [CrossRef]
- Shi, J.; Gao, W.; Shao, F. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem. Sci. 2017, 42, 245–254. [Google Scholar] [CrossRef]
- Chen, K.W.; Monteleone, M.; Boucher, D.; Sollberger, G.; Ramnath, D.; Condon, N.D.; von Pein, J.B.; Broz, P.; Sweet, M.J.; Schroder, K. Noncanonical inflammasome signaling elicits gasdermin D–dependent neutrophil extracellular traps. Sci. Immunol. 2018, 3, eaar6676. [Google Scholar] [CrossRef]
- Mukherjee, S.; Karmakar, S.; Babu, S.P.S. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz. J. Infect. Dis. 2016, 20, 193–204. [Google Scholar] [CrossRef]
- Sollberger, G.; Choidas, A.; Burn, G.L.; Habenberger, P.; Di Lucrezia, R.; Kordes, S.; Menninger, S.; Eickhoff, J.; Nussbaumer, P.; Klebl, B.; et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol. 2018, 3, eaar6689. [Google Scholar] [CrossRef]
- Kenny, E.F.; Herzig, A.; Krüger, R.; Muth, A.; Mondal, S.; Thompson, P.R.; Brinkmann, V.; von Bernuth, H.; Zychlinsky, A. Diverse stimuli engage different neutrophil extracellular trap pathways. eLife 2017, 6, e24437. [Google Scholar] [CrossRef]
- McDonald, B.; Urrutia, R.; Yipp, B.G.; Jenne, C.N.; Kubes, P. Intravascular Neutrophil Extracellular Traps Capture Bacteria from the Bloodstream during Sepsis. Cell Host Microbe 2012, 12, 324–333. [Google Scholar] [CrossRef]
- Mori, Y.; Yamaguchi, M.; Terao, Y.; Hamada, S.; Ooshima, T.; Kawabata, S. α-Enolase of Streptococcus pneumoniae Induces Formation of Neutrophil Extracellular Traps. J. Biol. Chem. 2012, 287, 10472–10481. [Google Scholar] [CrossRef]
- Thanabalasuriar, A.; Scott, B.N.V.; Peiseler, M.; Willson, M.E.; Zeng, Z.; Warrener, P.; Keller, A.E.; Surewaard, B.G.J.; Dozier, E.A.; Korhonen, J.T.; et al. Neutrophil Extracellular Traps Confine Pseudomonas aeruginosa Ocular Biofilms and Restrict Brain Invasion. Cell Host Microbe 2019, 25, 526–536.e4. [Google Scholar] [CrossRef]
- Demers, M.; Krause, D.S.; Schatzberg, D.; Martinod, K.; Voorhees, J.R.; Fuchs, T.A.; Scadden, D.T.; Wagner, D.D. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc. Natl. Acad. Sci. USA 2012, 109, 13076–13081. [Google Scholar] [CrossRef]
- Keshari, R.S.; Jyoti, A.; Dubey, M.; Kothari, N.; Kohli, M.; Bogra, J.; Barthwal, M.K.; Dikshit, M. Cytokines Induced Neutrophil Extracellular Traps Formation: Implication for the Inflammatory Disease Condition. PLoS ONE 2012, 7, e48111. [Google Scholar] [CrossRef]
- Xiao, Y.; Cong, M.; Li, J.; He, D.; Wu, Q.; Tian, P.; Wang, Y.; Yang, S.; Liang, C.; Liang, Y.; et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell 2021, 39, 423–437.e7. [Google Scholar] [CrossRef]
- Martinelli, S.; Urosevic, M.; Daryadel, A.; Oberholzer, P.A.; Baumann, C.; Fey, M.F.; Dummer, R.; Simon, H.-U.; Yousefi, S. Induction of Genes Mediating Interferon-dependent Extracellular Trap Formation during Neutrophil Differentiation. J. Biol. Chem. 2004, 279, 44123–44132. [Google Scholar] [CrossRef]
- Delgado-Rizo, V.; Martínez-Guzmán, M.A.; Iñiguez-Gutierrez, L.; García-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef]
- Marin-Esteban, V.; Turbica, I.; Dufour, G.; Semiramoth, N.; Gleizes, A.; Gorges, R.; Beau, I.; Servin, A.L.; Lievin-Le Moal, V.; Sandré, C.; et al. Afa/Dr Diffusely Adhering Escherichia coli Strain C1845 Induces Neutrophil Extracellular Traps That Kill Bacteria and Damage Human Enterocyte-Like Cells. Infect. Immun. 2012, 80, 1891–1899. [Google Scholar] [CrossRef]
- Young, R.L.; Malcolm, K.C.; Kret, J.E.; Caceres, S.M.; Poch, K.R.; Nichols, D.P.; Taylor-Cousar, J.L.; Saavedra, M.T.; Randell, S.H.; Vasil, M.L.; et al. Neutrophil Extracellular Trap (NET)-Mediated Killing of Pseudomonas aeruginosa: Evidence of Acquired Resistance within the CF Airway, Independent of CFTR. PLoS ONE 2011, 6, e23637. [Google Scholar] [CrossRef]
- Seper, A.; Hosseinzadeh, A.; Gorkiewicz, G.; Lichtenegger, S.; Roier, S.; Leitner, D.R.; Röhm, M.; Grutsch, A.; Reidl, J.; Urban, C.F.; et al. Vibrio cholerae Evades Neutrophil Extracellular Traps by the Activity of Two Extracellular Nucleases. PLoS Pathog. 2013, 9, e1003614. [Google Scholar] [CrossRef]
- Möllerherm, H.; Neumann, A.; Schilcher, K.; Blodkamp, S.; E Zeitouni, N.; Dersch, P.; Lüthje, P.; Naim, H.Y.; Zinkernagel, A.S.; von Köckritz-Blickwede, M. Yersinia enterocolitica-mediated degradation of neutrophil extracellular traps (NETs). FEMS Microbiol. Lett. 2015, 362, fnv192. [Google Scholar] [CrossRef][Green Version]
- Pilsczek, F.H.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A Novel Mechanism of Rapid Nuclear Neutrophil Extracellular Trap Formation in Response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef]
- Kothary, V.; Doster, R.S.; Rogers, L.M.; Kirk, L.A.; Boyd, K.L.; Romano-Keeler, J.; Haley, K.P.; Manning, S.D.; Aronoff, D.M.; Gaddy, J.A. Group B Streptococcus Induces Neutrophil Recruitment to Gestational Tissues and Elaboration of Extracellular Traps and Nutritional Immunity. Front. Cell. Infect. Microbiol. 2017, 7, 19. [Google Scholar] [CrossRef]
- Hirschfeld, J.; White, P.C.; Milward, M.R.; Cooper, P.R.; Chapple, I.L.C. Modulation of Neutrophil Extracellular Trap and Reactive Oxygen Species Release by Periodontal Bacteria. Infect. Immun. 2017, 85, e00297-17. [Google Scholar] [CrossRef]
- Jung, C.-J.; Yeh, C.-Y.; Hsu, R.-B.; Lee, C.-M.; Shun, C.-T.; Chia, J.-S. Endocarditis Pathogen Promotes Vegetation Formation by Inducing Intravascular Neutrophil Extracellular Traps Through Activated Platelets. Circulation 2015, 131, 571–581. [Google Scholar] [CrossRef]
- De Buhr, N.; Neumann, A.; Jerjomiceva, N.; von Köckritz-Blickwede, M.; Baums, C.G. Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 2014, 160, 385–395. [Google Scholar] [CrossRef]
- Beiter, K.; Wartha, F.; Albiger, B.; Normark, S.; Zychlinsky, A.; Henriques-Normark, B. An Endonuclease Allows Streptococcus pneumoniae to Escape from Neutrophil Extracellular Traps. Curr. Biol. 2006, 16, 401–407. [Google Scholar] [CrossRef]
- Cole, J.N.; Pence, M.A.; von Köckritz-Blickwede, M.; Hollands, A.; Gallo, R.L.; Walker, M.J.; Nizet, V. M Protein and Hyaluronic Acid Capsule Are Essential for In Vivo Selection of covRS Mutations Characteristic of Invasive Serotype M1T1 Group A Streptococcus. mBio 2010, 1, e00191-10. [Google Scholar] [CrossRef]
- Berends, E.T.; Horswill, A.R.; Haste, N.M.; Monestier, M.; Nizet, V.; von Köckritz-Blickwede, M. Nuclease Expression by Staphylococcus aureus Facilitates Escape from Neutrophil Extracellular Traps. J. Innate Immun. 2010, 2, 576–586. [Google Scholar] [CrossRef]
- Juneau, R.A.; Pang, B.; Weimer, K.E.D.; Armbruster, C.E.; Swords, W.E. Nontypeable Haemophilus influenzae Initiates Formation of Neutrophil Extracellular Traps. Infect. Immun. 2011, 79, 431–438. [Google Scholar] [CrossRef]
- El-Benna, J.; Hurtado-Nedelec, M.; Marzaioli, V.; Marie, J.; Gougerot-Pocidalo, M.; Dang, P.M. Priming of the neutrophil respiratory burst: Role in host defense and inflammation. Immunol. Rev. 2016, 273, 180–193. [Google Scholar] [CrossRef]
- Belambri, S.A.; Rolas, L.; Raad, H.; Hurtado-Nedelec, M.; Dang, P.M.; El-Benna, J. NADPH oxidase activation in neutrophils: Role of the phosphorylation of its subunits. Eur. J. Clin. Investig. 2018, 48, e12951. [Google Scholar] [CrossRef]
- Nguyen, G.T.; Green, E.R.; Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH Oxidase Activation and Bacterial Resistance. Front. Cell. Infect. Microbiol. 2017, 7, 373. [Google Scholar] [CrossRef]
- Poli, V.; Zanoni, I. Neutrophil intrinsic and extrinsic regulation of NETosis in health and disease. Trends Microbiol. 2022, 31, 280–293. [Google Scholar] [CrossRef]
- Hakkim, A.; Fuchs, T.A.; Martinez, N.E.; Hess, S.; Prinz, H.; Zychlinsky, A.; Waldmann, H. Activation of the Raf-MEK-ERK pathway is required for neutrophil extracellular trap formation. Nat. Chem. Biol. 2011, 7, 75–77. [Google Scholar] [CrossRef]
- Fonseca, Z.; Díaz-Godínez, C.; Mora, N.; Alemán, O.R.; Uribe-Querol, E.; Carrero, J.C.; Rosales, C. Entamoeba histolytica Induce Signaling via Raf/MEK/ERK for Neutrophil Extracellular Trap (NET) Formation. Front. Cell. Infect. Microbiol. 2018, 8, 226. [Google Scholar] [CrossRef]
- Parker, H.; Albrett, A.M.; Kettle, A.J.; Winterbourn, C.C. Myeloperoxidase associated with neutrophil extracellular traps is active and mediates bacterial killing in the presence of hydrogen peroxide. J. Leukoc. Biol. 2011, 91, 369–376. [Google Scholar] [CrossRef]
- Björnsdottir, H.; Welin, A.; Michaëlsson, E.; Osla, V.; Berg, S.; Christenson, K.; Sundqvist, M.; Dahlgren, C.; Karlsson, A.; Bylund, J. Neutrophil NET formation is regulated from the inside by myeloperoxidase-processed reactive oxygen species. Free. Radic. Biol. Med. 2015, 89, 1024–1035. [Google Scholar] [CrossRef]
- Bylund, J.; Björnsdottir, H.; Sundqvist, M.; Karlsson, A.; Dahlgren, C. Measurement of respiratory burst products, released or retained, during activation of professional phagocytes. Methods Mol. Biol. 2014, 1124, 321–338. [Google Scholar] [CrossRef]
- Ravindran, M.; Khan, M.A.; Palaniyar, N. Neutrophil Extracellular Trap Formation: Physiology, Pathology, and Pharmacology. Biomolecules 2019, 9, 365. [Google Scholar] [CrossRef]
- Douda, D.N.; Khan, M.A.; Grasemann, H.; Palaniyar, N. SK3 channel and mitochondrial ROS mediate NADPH oxidase-independent NETosis induced by calcium influx. Proc. Natl. Acad. Sci. USA 2015, 112, 2817–2822. [Google Scholar] [CrossRef]
- Fay, A.J.; Qian, X.; Jan, Y.N.; Jan, L.Y. SK channels mediate NADPH oxidase-independent reactive oxygen species production and apoptosis in granulocytes. Proc. Natl. Acad. Sci. USA 2006, 103, 17548–17553. [Google Scholar] [CrossRef]
- Engelmann, B.; Massberg, S. Thrombosis as an intravascular effector of innate immunity. Nat. Rev. Immunol. 2013, 13, 34–45. [Google Scholar] [CrossRef]
- Thålin, C.; Hisada, Y.; Lundström, S.; Mackman, N.; Wallén, H. Neutrophil Extracellular Traps: Villains and Targets in Arterial, Venous, and Cancer-Associated Thrombosis. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1724–1738. [Google Scholar] [CrossRef]
- Noubouossie, D.F.; Whelihan, M.F.; Yu, Y.-B.; Sparkenbaugh, E.; Pawlinski, R.; Monroe, D.M.; Key, N.S. In vitro activation of coagulation by human neutrophil DNA and histone proteins but not neutrophil extracellular traps. Blood 2017, 129, 1021–1029. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, L.; Braun, O.; Westman, J.; Madhi, R.; Herwald, H.; Mörgelin, M.; Thorlacius, H. Neutrophil extracellular trap-microparticle complexes enhance thrombin generation via the intrinsic pathway of coagulation in mice. Sci. Rep. 2018, 8, 4020. [Google Scholar] [CrossRef]
- Hisada, Y.; Grover, S.P.; Maqsood, A.; Houston, R.; Ay, C.; Noubouossie, D.F.; Cooley, B.C.; Wallén, H.; Key, N.S.; Thålin, C.; et al. Neutrophils and neutrophil extracellular traps enhance venous thrombosis in mice bearing human pancreatic tumors. Haematologica 2019, 105, 218–225. [Google Scholar] [CrossRef]
- Kumar, S.V.; Kulkarni, O.P.; Mulay, S.R.; Darisipudi, M.N.; Romoli, S.; Thomasova, D.; Scherbaum, C.R.; Hohenstein, B.; Hugo, C.; Müller, S.; et al. Neutrophil Extracellular Trap-Related Extracellular Histones Cause Vascular Necrosis in Severe GN. J. Am. Soc. Nephrol. 2015, 26, 2399–2413. [Google Scholar] [CrossRef]
- Brill, A.; Fuchs, T.A.; Savchenko, A.S.; Thomas, G.M.; Martinod, K.; DE Meyer, S.F.; Bhandari, A.A.; Wagner, D.D. Neutrophil extracellular traps promote deep vein thrombosis in mice. J. Thromb. Haemost. 2011, 10, 136–144. [Google Scholar] [CrossRef]
- Ammollo, C.T.; Semeraro, F.; Xu, J.; Esmon, N.L.; Esmon, C.T. Extracellular histones increase plasma thrombin generation by impairing thrombomodulin-dependent protein C activation. J. Thromb. Haemost. 2011, 9, 1795–1803. [Google Scholar] [CrossRef]
- Von Brühl, M.L.; Stark, K.; Steinhart, A.; Chandraratne, S.; Konrad, I.; Lorenz, M.; Khandoga, A.; Tirniceriu, A.; Coletti, R.; Köllnberger, M.; et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J. Exp. Med. 2012, 209, 819–835. [Google Scholar] [CrossRef]
- Massberg, S.; Grahl, L.; von Bruehl, M.-L.; Manukyan, D.; Pfeiler, S.; Goosmann, C.; Brinkmann, V.; Lorenz, M.; Bidzhekov, K.; Khandagale, A.B.; et al. Reciprocal coupling of coagulation and innate immunity via neutrophil serine proteases. Nat. Med. 2010, 16, 887–896. [Google Scholar] [CrossRef]
- Perdomo, J.; Leung, H.H.L.; Ahmadi, Z.; Yan, F.; Chong, J.J.H.; Passam, F.H.; Chong, B.H. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat. Commun. 2019, 10, 1322. [Google Scholar] [CrossRef]
- Yalavarthi, S.; Gould, T.J.; Rao, A.N.; Mazza, L.F.; Morris, A.E.; Núñez-Álvarez, C.; Hernández-Ramírez, D.; Bockenstedt, P.L.; Liaw, P.C.; Cabral, A.R.; et al. Release of Neutrophil Extracellular Traps by Neutrophils Stimulated With Antiphospholipid Antibodies: A Newly Identified Mechanism of Thrombosis in the Antiphospholipid Syndrome. Arthritis Rheumatol. 2015, 67, 2990–3003. [Google Scholar] [CrossRef]
- De Groot, P.G.; Meijers, J.C.M. β2-Glycoprotein I: Evolution, structure and function. J. Thromb. Haemost. 2011, 9, 1275–1284. [Google Scholar] [CrossRef]
- Shi, T.; Giannakopoulos, B.; Yan, X.; Yu, P.; Berndt, M.C.; Andrews, R.K.; Rivera, J.; Iverson, G.M.; Cockerill, K.A.; Linnik, M.D.; et al. Anti–β2-glycoprotein I antibodies in complex with β2-glycoprotein I can activate platelets in a dysregulated manner via glycoprotein Ib-IX-V. Arthritis Rheum. 2006, 54, 2558–2567. [Google Scholar] [CrossRef]
- Zha, C.; Zhang, W.; Gao, F.; Xu, J.; Jia, R.; Cai, J.; Liu, Y. Anti-β2GPI/β2GPI induces neutrophil extracellular traps formation to promote thrombogenesis via the TLR4/MyD88/MAPKs axis activation. Neuropharmacology 2018, 138, 140–150. [Google Scholar] [CrossRef]
- Healy, L.D.; Puy, C.; Itakura, A.; Chu, T.; Robinson, D.K.; Bylund, A.; Phillips, K.G.; Gardiner, E.E.; McCarty, O.J. Colocalization of neutrophils, extracellular DNA and coagulation factors during NETosis: Development and utility of an immunofluorescence-based microscopy platform. J. Immunol. Methods 2016, 435, 77–84. [Google Scholar] [CrossRef]
- Yang, C.; Sun, W.; Cui, W.; Li, X.; Yao, J.; Jia, X.; Li, C.; Wu, H.; Hu, Z.; Zou, X. Procoagulant role of neutrophil extracellular traps in patients with gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 14075–14086. [Google Scholar]
- McDonald, B.; Davis, R.P.; Kim, S.-J.; Tse, M.; Esmon, C.T.; Kolaczkowska, E.; Jenne, C.N. Platelets and neutrophil extracellular traps collaborate to promote intravascular coagulation during sepsis in mice. Blood 2017, 129, 1357–1367. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D., Jr.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885. [Google Scholar] [CrossRef]
- Evangelista, V.; Pamuklar, Z.; Piccoli, A.; Manarini, S.; Dell’Elba, G.; Pecce, R.; Martelli, N.; Federico, L.; Rojas, M.; Berton, G.; et al. Src family kinases mediate neutrophil adhesion to adherent platelets. Blood 2006, 109, 2461–2469. [Google Scholar] [CrossRef]
- Blair, P.; Rex, S.; Vitseva, O.; Beaulieu, L.; Tanriverdi, K.; Chakrabarti, S.; Hayashi, C.; Genco, C.A.; Iafrati, M.; Freedman, J.E.; et al. Stimulation of Toll-Like Receptor 2 in Human Platelets Induces a Thromboinflammatory Response Through Activation of Phosphoinositide 3-Kinase. Circ. Res. 2009, 104, 346–354. [Google Scholar] [CrossRef]
- Assinger, A.; Laky, M.; Schabbauer, G.; Hirschl, A.M.; Buchberger, E.; Binder, B.R.; Volf, I. Efficient phagocytosis of periodontopathogens by neutrophils requires plasma factors, platelets and TLR2. J. Thromb. Haemost. 2011, 9, 799–809. [Google Scholar] [CrossRef]
- Ståhl, A.-L.; Svensson, M.; Mörgelin, M.; Svanborg, C.; Tarr, P.I.; Mooney, J.C.; Watkins, S.L.; Johnson, R.; Karpman, D. Lipopolysaccharide from enterohemorrhagic Escherichia coli binds to platelets through TLR4 and CD62 and is detected on circulating platelets in patients with hemolytic uremic syndrome. Blood 2006, 108, 167–176. [Google Scholar] [CrossRef]
- Grässle, S.; Huck, V.; Pappelbaum, K.I.; Gorzelanny, C.; Aponte-Santamaría, C.; Baldauf, C.; Gräter, F.; Schneppenheim, R.; Obser, T.; Schneider, S.W.; et al. von Willebrand Factor Directly Interacts With DNA From Neutrophil Extracellular Traps. Arter. Thromb. Vasc. Biol. 2014, 34, 1382–1389. [Google Scholar] [CrossRef]
- Gould, T.J.; Vu, T.T.; Swystun, L.L.; Dwivedi, D.J.; Mai, S.H.; Weitz, J.I.; Liaw, P.C. Neutrophil extracellular traps promote thrombin generation through platelet-dependent and platelet-independent mechanisms. Arter. Thromb. Vasc. Biol. 2014, 34, 1977–1984. [Google Scholar] [CrossRef]
- Bäck, J.; Lang, M.H.; Elgue, G.; Kalbitz, M.; Sanchez, J.; Ekdahl, K.N.; Nilsson, B. Distinctive regulation of contact activation by antithrombin and C1-inhibitor on activated platelets and material surfaces. Biomaterials 2009, 30, 6573–6580. [Google Scholar] [CrossRef]
- Hoeksema, M.; van Eijk, M.; Haagsman, H.P.; Hartshorn, K.L. Histones as mediators of host defense, inflammation and thrombosis. Futur. Microbiol. 2016, 11, 441–453. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, X.; Pelayo, R.; Monestier, M.; Ammollo, C.T.; Semeraro, F.; Taylor, F.B.; Esmon, N.L.; Lupu, F.; Esmon, C.T. Extracellular histones are major mediators of death in sepsis. Nat. Med. 2009, 15, 1318–1321. [Google Scholar] [CrossRef]
- Fuchs, T.A.; Bhandari, A.A.; Wagner, D.D. Histones induce rapid and profound thrombocytopenia in mice. Blood 2011, 118, 3708–3714. [Google Scholar] [CrossRef]
- Lam, F.W.; Vijayan, K.V.; Rumbaut, R.E. Platelets and their interactions with other immune cells. Compr. Physiol. 2015, 5, 1265–1280. [Google Scholar] [CrossRef]
- Dowling, J.K.; Mansell, A. Toll-like receptors: The swiss army knife of immunity and vaccine development. Clin. Transl. Immunol. 2016, 5, e85. [Google Scholar] [CrossRef]
- Semeraro, F.; Ammollo, C.T.; Morrissey, J.H.; Dale, G.L.; Friese, P.; Esmon, N.L.; Esmon, C.T. Extracellular histones promote thrombin generation through platelet-dependent mechanisms: Involvement of platelet TLR2 and TLR4. Blood 2011, 118, 1952–1961. [Google Scholar] [CrossRef]
- Reasor, D.A.; Mehrabadi, M.; Ku, D.N.; Aidun, C.K. Determination of Critical Parameters in Platelet Margination. Ann. Biomed. Eng. 2012, 41, 238–249. [Google Scholar] [CrossRef]
- Stalker, T.J.; Traxler, E.A.; Wu, J.; Wannemacher, K.M.; Cermignano, S.L.; Voronov, R.; Diamond, S.L.; Brass, L.F. Hierarchical organization in the hemostatic response and its relationship to the platelet-signaling network. Blood 2013, 121, 1875–1885. [Google Scholar] [CrossRef]
- Qiu, Y.; Brown, A.C.; Myers, D.R.; Sakurai, Y.; Mannino, R.G.; Tran, R.; Ahn, B.; Hardy, E.T.; Kee, M.F.; Kumar, S.; et al. Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc. Natl. Acad. Sci. USA 2014, 111, 14430–14435. [Google Scholar] [CrossRef]
- Semple, J.W.; Italiano, J.E.; Freedman, J. Platelets and the immune continuum. Nat. Rev. Immunol. 2011, 11, 264–274. [Google Scholar] [CrossRef]
- Parimon, T.; Li, Z.; Bolz, D.D.; McIndoo, E.R.; Bayer, C.R.; Stevens, D.L.; Bryant, A.E. Staphylococcus aureus α-Hemolysin Promotes Platelet-Neutrophil Aggregate Formation. J. Infect. Dis. 2013, 208, 761–770. [Google Scholar] [CrossRef]
- Hurley, S.M.; Kahn, F.; Nordenfelt, P.; Mörgelin, M.; Sørensen, O.E.; Shannon, O. Platelet-Dependent Neutrophil Function Is Dysregulated by M Protein from Streptococcus pyogenes. Infect. Immun. 2015, 83, 3515–3525. [Google Scholar] [CrossRef]
- Hurley, S.M.; Lutay, N.; Holmqvist, B.; Shannon, O. The Dynamics of Platelet Activation during the Progression of Streptococcal Sepsis. PLoS ONE 2016, 11, e0163531. [Google Scholar] [CrossRef]
- Chen, W.A.; Fletcher, H.M.; Payne, K.J.; Aka, S.; Thornburg, M.B.; Gheorghe, J.D.; Safi, S.B.; Shavlik, D.; Oyoyo, U.; Boskovic, D.S. Platelet and neutrophil responses to Porphyromonas gingivalis in human whole blood. Mol. Oral Microbiol. 2021, 36, 202–213. [Google Scholar] [CrossRef]
- Amison, R.T.; O’shaughnessy, B.G.; Arnold, S.; Cleary, S.J.; Nandi, M.; Pitchford, S.C.; Bragonzi, A.; Page, C.P. Platelet Depletion Impairs Host Defense to Pulmonary Infection with Pseudomonas aeruginosa in Mice. Am. J. Respir. Cell Mol. Biol. 2018, 58, 331–340. [Google Scholar] [CrossRef]
- Gros, A.; Syvannarath, V.; Lamrani, L.; Ollivier, V.; Loyau, S.; Goerge, T.; Nieswandt, B.; Jandrot-Perrus, M.; Ho-Tin-Noé, B. Single platelets seal neutrophil-induced vascular breaches via GPVI during immune-complex–mediated inflammation in mice. Blood 2015, 126, 1017–1026. [Google Scholar] [CrossRef]
- A Abdulla, A.; Awla, D.; Hartman, H.; Rahman, M.; Jeppsson, B.; Regnér, S.; Thorlacius, H. Role of platelets in experimental acute pancreatitis. Br. J. Surg. 2010, 98, 93–103. [Google Scholar] [CrossRef]
- Hidalgo, A.; Chang, J.; Jang, J.-E.; Peired, A.J.; Chiang, E.Y.; Frenette, P.S. Heterotypic interactions enabled by polarized neutrophil microdomains mediate thromboinflammatory injury. Nat. Med. 2009, 15, 384–391. [Google Scholar] [CrossRef]
- Polanowska-Grabowska, R.; Wallace, K.; Field, J.J.; Chen, L.; Marshall, M.A.; Figler, R.; Gear, A.R.; Linden, J. P-Selectin–Mediated Platelet-Neutrophil Aggregate Formation Activates Neutrophils in Mouse and Human Sickle Cell Disease. Arter. Thromb. Vasc. Biol. 2010, 30, 2392–2399. [Google Scholar] [CrossRef]
- De Stoppelaar, S.F.; Veer, C.v.; Claushuis, T.A.M.; Albersen, B.J.A.; Roelofs, J.J.T.H.; van der Poll, T. Thrombocytopenia impairs host defense in gram-negative pneumonia–derived sepsis in mice. Blood 2014, 124, 3781–3790. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Konkel, J.; Sarmadi, M.; Eskan, M.A.; Wild, T.; Dutzan, N.; Abusleme, L.; Zenobia, C.; Hosur, K.B.; Abe, T.; et al. Defective Neutrophil Recruitment in Leukocyte Adhesion Deficiency Type I Disease Causes Local IL-17–Driven Inflammatory Bone Loss. Sci. Transl. Med. 2014, 6, 229ra40. [Google Scholar] [CrossRef]
- Ambruso, D.R.; Knall, C.; Abell, A.N.; Panepinto, J.; Kurkchubasche, A.; Thurman, G.; Gonzalez-Aller, C.; Hiester, A.; Deboer, M.; Harbeck, R.J.; et al. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc. Natl. Acad. Sci. USA 2000, 97, 4654–4659. [Google Scholar] [CrossRef]
- Zuchtriegel, G.; Uhl, B.; Puhr-Westerheide, D.; Pörnbacher, M.; Lauber, K.; Krombach, F.; Reichel, C.A. Platelets Guide Leukocytes to Their Sites of Extravasation. PLoS Biol. 2016, 14, e1002459. [Google Scholar] [CrossRef]
- Castellon, X.; Bogdanova, V. Chronic Inflammatory Diseases and Endothelial Dysfunction. Aging Dis. 2016, 7, 81–89. [Google Scholar] [CrossRef]
- Sreeramkumar, V.; Adrover, J.M.; Ballesteros, I.; Cuartero, M.I.; Rossaint, J.; Bilbao, I.; Nácher, M.; Pitaval, C.; Radovanovic, I.; Fukui, Y.; et al. Neutrophils scan for activated platelets to initiate inflammation. Science 2014, 346, 1234–1238. [Google Scholar] [CrossRef]
- Simon, D.I.; Chen, Z.; Xu, H.; Li, C.Q.; Dong, J.-F.; McIntire, L.V.; Ballantyne, C.M.; Zhang, L.; Furman, M.I.; Berndt, M.C.; et al. Platelet Glycoprotein Ibα Is a Counterreceptor for the Leukocyte Integrin Mac-1 (CD11b/CD18). J. Exp. Med. 2000, 192, 193–204. [Google Scholar] [CrossRef]
- Li, J.; Kim, K.; Barazia, A.; Tseng, A.; Cho, J. Platelet–neutrophil interactions under thromboinflammatory conditions. Cell. Mol. Life Sci. 2015, 72, 2627–2643. [Google Scholar] [CrossRef]
- Prince, L.R.; Whyte, M.K.; Sabroe, I.; Parker, L.C. The role of TLRs in neutrophil activation. Curr. Opin. Pharmacol. 2011, 11, 397–403. [Google Scholar] [CrossRef]
- Dudley, D.D.; Chaudhuri, J.; Bassing, C.H.; Alt, F.W. Mechanism and control of V (D) J recombina-tion versus class switch recombination: Similarities and differences. Adv. Immunol. 2005, 86, 43–112. [Google Scholar] [CrossRef]
- Takeuchi, O.; Akira, S. Pattern Recognition Receptors and Inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef]
- Hayashi, F.; Means, T.K.; Luster, A.D. Toll-like receptors stimulate human neutrophil function. Blood 2003, 102, 2660–2669. [Google Scholar] [CrossRef]
- Koupenova, M.; Mick, E.; Mikhalev, E.; Benjamin, E.J.; Tanriverdi, K.; Freedman, J.E. Sex Differences in Platelet Toll-Like Receptors and Their Association With Cardiovascular Risk Factors. Arter. Thromb. Vasc. Biol. 2015, 35, 1030–1037. [Google Scholar] [CrossRef]
- Medzhitov, R.; Janeway, C.A., Jr. Decoding the Patterns of Self and Nonself by the Innate Immune System. Science 2002, 296, 298–300. [Google Scholar] [CrossRef]
- Parker, L.C.; Whyte, M.K.B.; Dower, S.K.; Sabroe, I. The expression and roles of Toll-like receptors in the biology of the human neutrophil. J. Leukoc. Biol. 2005, 77, 886–892. [Google Scholar] [CrossRef]
- Sabroe, I.; Prince, L.R.; Jones, E.C.; Horsburgh, M.J.; Foster, S.J.; Vogel, S.N.; Dower, S.K.; Whyte, M.K.B. Selective Roles for Toll-Like Receptor (TLR)2 and TLR4 in the Regulation of Neutrophil Activation and Life Span. J. Immunol. 2003, 170, 5268–5275. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-Like Receptor Signaling Pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Alvarez-Arellano, L.; Camorlinga-Ponce, M.; Maldonado-Bernal, C.; Torres, J. Activation of human neutrophils with Helicobacter pylori and the role of Toll-like receptors 2 and 4 in the response. FEMS Immunol. Med. Microbiol. 2007, 51, 473–479. [Google Scholar] [CrossRef]
- Huang, X.; Barrett, R.P.; McClellan, S.A.; Hazlett, L.D. Silencing Toll-like Receptor-9 in Pseudomonas aeruginosa Keratitis. Investig. Opthalmol. Vis. Sci. 2005, 46, 4209–4216. [Google Scholar] [CrossRef]
- Hemmi, H.; Takeuchi, O.; Kawai, T.; Kaisho, T.; Sato, S.; Sanjo, H.; Matsumoto, M.; Hoshino, K.; Wagner, H.; Takeda, K.; et al. A Toll-like receptor recognizes bacterial DNA. Nature 2000, 408, 740–745. [Google Scholar] [CrossRef]
- Andrews, K.; Abdelsamed, H.; Yi, A.-K.; Miller, M.A.; Fitzpatrick, E.A. TLR2 Regulates Neutrophil Recruitment and Cytokine Production with Minor Contributions from TLR9 during Hypersensitivity Pneumonitis. PLoS ONE 2013, 8, e73143. [Google Scholar] [CrossRef]
- Echchannaoui, H.; Frei, K.; Schnell, C.; Leib, S.L.; Zimmerli, W.; Landmann, R. Toll-Like Receptor 2–Deficient Mice Are Highly Susceptible to Streptococcus pneumoniae Meningitis because of Reduced Bacterial Clearing and Enhanced Inflammation. J. Infect. Dis. 2002, 186, 798–806. [Google Scholar] [CrossRef]
- Letiembre, M.; Echchannaoui, H.; Bachmann, P.; Ferracin, F.; Nieto, C.; Espinosa, M.; Landmann, R. Toll-Like Receptor 2 Deficiency Delays Pneumococcal Phagocytosis and Impairs Oxidative Killing by Granulocytes. Infect. Immun. 2005, 73, 8397–8401. [Google Scholar] [CrossRef]
- Cognasse, F.; Nguyen, K.A.; Damien, P.; McNicol, A.; Pozzetto, B.; Hamzeh-Cognasse, H.; Garraud, O. The inflammatory role of platelets via their TLRs and Siglec receptors. Front. Immunol. 2015, 6, 83. [Google Scholar] [CrossRef]
- Guidetti, G.F.; Canobbio, I.; Torti, M. PI3K/Akt in platelet integrin signaling and implications in thrombosis. Adv. Biol. Regul. 2015, 59, 36–52. [Google Scholar] [CrossRef]
- Keane, C.; Tilley, D.; Cunningham, A.; Smolenski, A.; Kadioglu, A.; Cox, D.; Jenkinson, H.F.; Kerrigan, S.W. Invasive Streptococcus pneumoniae trigger platelet activation via Toll-like receptor 2. J. Thromb. Haemost. 2010, 8, 2757–2765. [Google Scholar] [CrossRef]
- Li, Z.; Delaney, M.K.; O’Brien, K.A.; Du, X. Signaling During Platelet Adhesion and Activation. Arter. Thromb. Vasc. Biol. 2010, 30, 2341–2349. [Google Scholar] [CrossRef]
- Assinger, A.; Laky, M.; Badrnya, S.; Esfandeyari, A.; Volf, I. Periodontopathogens induce expression of CD40L on human platelets via TLR2 and TLR4. Thromb. Res. 2012, 130, e73–e78. [Google Scholar] [CrossRef]
- Vowinkel, T.; Anthoni, C.; Wood, K.C.; Stokes, K.Y.; Russell, J.; Gray, L.; Bharwani, S.; Senninger, N.; Alexander, J.S.; Krieglstein, C.F.; et al. CD40–CD40 Ligand Mediates the Recruitment of Leukocytes and Platelets in the Inflamed Murine Colon. Gastroenterology 2007, 132, 955–965. [Google Scholar] [CrossRef]
- Aloui, C.; Prigent, A.; Sut, C.; Tariket, S.; Hamzeh-Cognasse, H.; Pozzetto, B.; Richard, Y.; Cognasse, F.; Laradi, S.; Garraud, O. The Signaling Role of CD40 Ligand in Platelet Biology and in Platelet Component Transfusion. Int. J. Mol. Sci. 2014, 15, 22342–22364. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Varghese, S.; Vitseva, O.; Tanriverdi, K.; Freedman, J.E. CD40 Ligand Influences Platelet Release of Reactive Oxygen Intermediates. Arter. Thromb. Vasc. Biol. 2005, 25, 2428–2434. [Google Scholar] [CrossRef]
- Jin, R.; Yu, S.; Song, Z.; Zhu, X.; Wang, C.; Yan, J.; Wu, F.; Nanda, A.; Granger, D.N.; Li, G. Soluble CD40 Ligand Stimulates CD40-Dependent Activation of the β2 Integrin Mac-1 and Protein Kinase C Zeda (PKCζ) in Neutrophils: Implications for Neutrophil-Platelet Interactions and Neutrophil Oxidative Burst. PLoS ONE 2013, 8, e64631. [Google Scholar] [CrossRef]
- Von Hundelshausen, P.; Weber, C. Platelets as Immune Cells: Bridging inflammation and cardiovascular disease. Circ. Res. 2007, 100, 27–40. [Google Scholar] [CrossRef]
- Ehlers, R.; Ustinov, V.; Chen, Z.; Zhang, X.; Rao, R.; Luscinskas, F.W.; Lopez, J.; Plow, E.; Simon, D.I. Targeting Platelet–Leukocyte Interactions. J. Exp. Med. 2003, 198, 1077–1088. [Google Scholar] [CrossRef]
- Sumagin, R.; Prizant, H.; Lomakina, E.; Waugh, R.E.; Sarelius, I.H. LFA-1 and Mac-1 Define Characteristically Different Intralumenal Crawling and Emigration Patterns for Monocytes and Neutrophils In Situ. J. Immunol. 2010, 185, 7057–7066. [Google Scholar] [CrossRef]
- Clark, S.R.; Ma, A.C.; Tavener, S.A.; McDonald, B.; Goodarzi, Z.; Kelly, M.M.; Patel, K.D.; Chakrabarti, S.; McAvoy, E.; Sinclair, G.D.; et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 2007, 13, 463–469. [Google Scholar] [CrossRef]
- Carestia, A.; Kaufman, T.; Rivadeneyra, L.; Landoni, V.I.; Pozner, R.G.; Negrotto, S.; D’atri, L.P.; Gómez, R.M.; Schattner, M. Mediators and molecular pathways involved in the regulation of neutrophil extracellular trap formation mediated by activated platelets. J. Leukoc. Biol. 2015, 99, 153–162. [Google Scholar] [CrossRef]
| Classification | Bacterial Species | References |
|---|---|---|
| Gram-negative bacteria | Escherichia coli | [108] |
| Klebsiella pneumoniae | [60] | |
| Pseudomonas aeruginosa | [109] | |
| Salmonella typhimurium | [55] | |
| Shigella flexneri | [69] | |
| Vibrio cholerae | [110] | |
| Yersinia enterocolitica | [111] | |
| Gram-positive bacteria | Staphylococcus aureus | [112] |
| Streptococcus agalactiae | [113] | |
| Streptococcus gordonii | [114] | |
| Streptococcus mutans | [115] | |
| Streptococcus pneumoniae | [101] | |
| Streptococcus pyogenes | [81] | |
| Streptococcus suis | [116] |
| Ligand | Platelet Receptor | Description | References |
|---|---|---|---|
| ADP | P2Y1 P2Y12 | Weak activator of platelets that can induce P-selectin expression, which facilitates platelet–neutrophil interactions | [152] |
| Collagen | GPVI Integrin α2β1 | Can trigger platelet activation and mediate formation of NETs | [151,153] |
| Fibrinogen | Integrin αIIbβ3 | Promote platelet aggregation and interact with extracellular chromatin | [148] |
| LPS | TLR4 | Can trigger platelet or neutrophil activation, mediate platelet–neutrophil interactions, and induce NET release | [100,154,155] |
| Pam3CSK4 | TLR2 | Can induce platelet activation, promote platelet aggregation, and mediate NET formation | [96,153,154] |
| Thrombin | PAR1 PAR4 | Strong activator of platelets that can induce P-selectin expression, which facilitates platelet–neutrophil interactions | [150] |
| vWF | GPIbα Integrin αIIbβ3 | Mediates platelet adhesion to endothelium, induce platelet aggregation, and promote platelet adhesion to extracellular DNA | [151,156] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.A.; Boskovic, D.S. Neutrophil Extracellular DNA Traps in Response to Infection or Inflammation, and the Roles of Platelet Interactions. Int. J. Mol. Sci. 2024, 25, 3025. https://doi.org/10.3390/ijms25053025
Chen WA, Boskovic DS. Neutrophil Extracellular DNA Traps in Response to Infection or Inflammation, and the Roles of Platelet Interactions. International Journal of Molecular Sciences. 2024; 25(5):3025. https://doi.org/10.3390/ijms25053025
Chicago/Turabian StyleChen, William A., and Danilo S. Boskovic. 2024. "Neutrophil Extracellular DNA Traps in Response to Infection or Inflammation, and the Roles of Platelet Interactions" International Journal of Molecular Sciences 25, no. 5: 3025. https://doi.org/10.3390/ijms25053025
APA StyleChen, W. A., & Boskovic, D. S. (2024). Neutrophil Extracellular DNA Traps in Response to Infection or Inflammation, and the Roles of Platelet Interactions. International Journal of Molecular Sciences, 25(5), 3025. https://doi.org/10.3390/ijms25053025





