Pulmonary Effects of Traumatic Brain Injury in Mice: A Gene Set Enrichment Analysis
Abstract
1. Introduction
2. Results
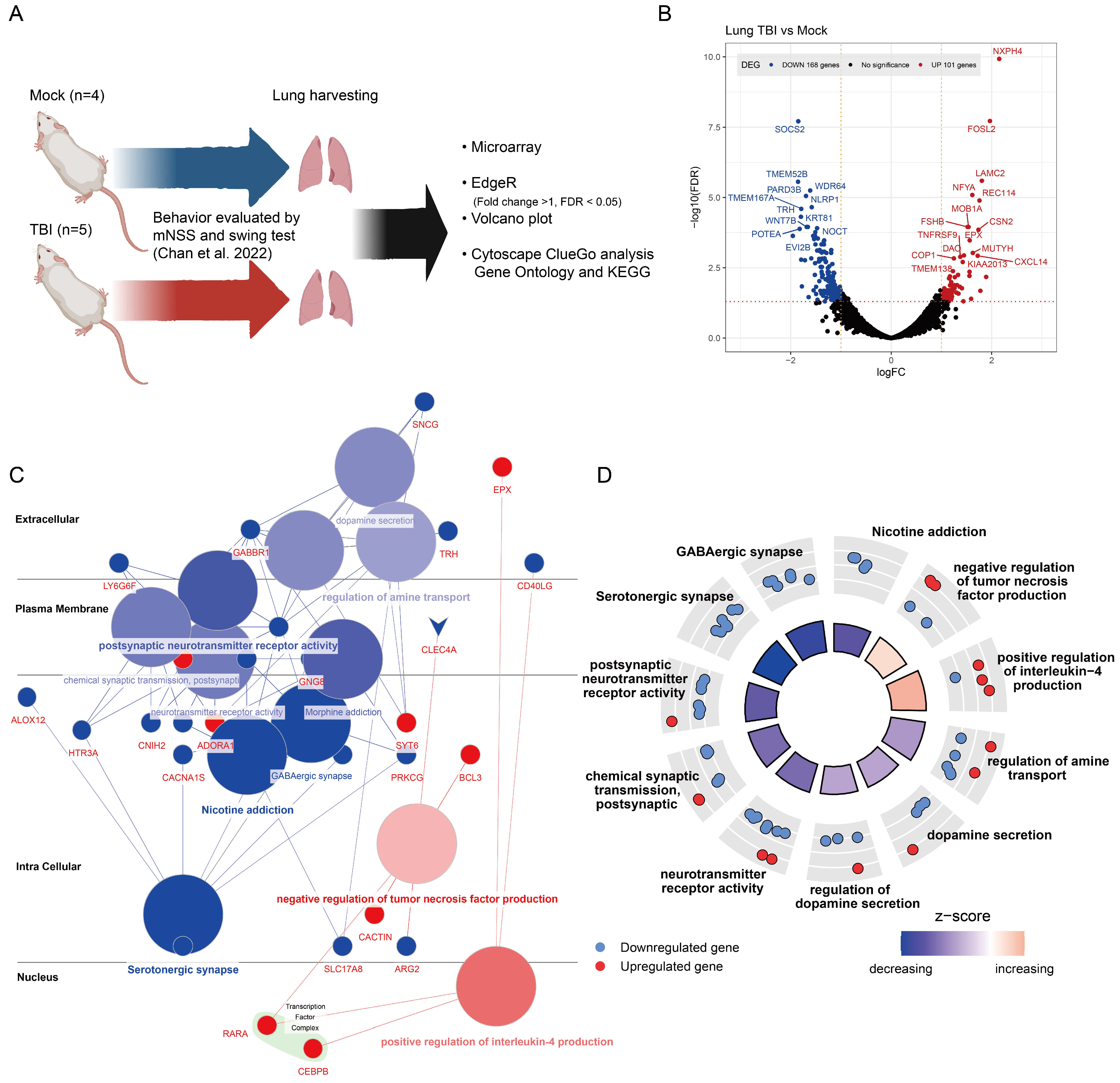
2.1. Gene Expression Alterations in TBI-Affected Lung Tissues
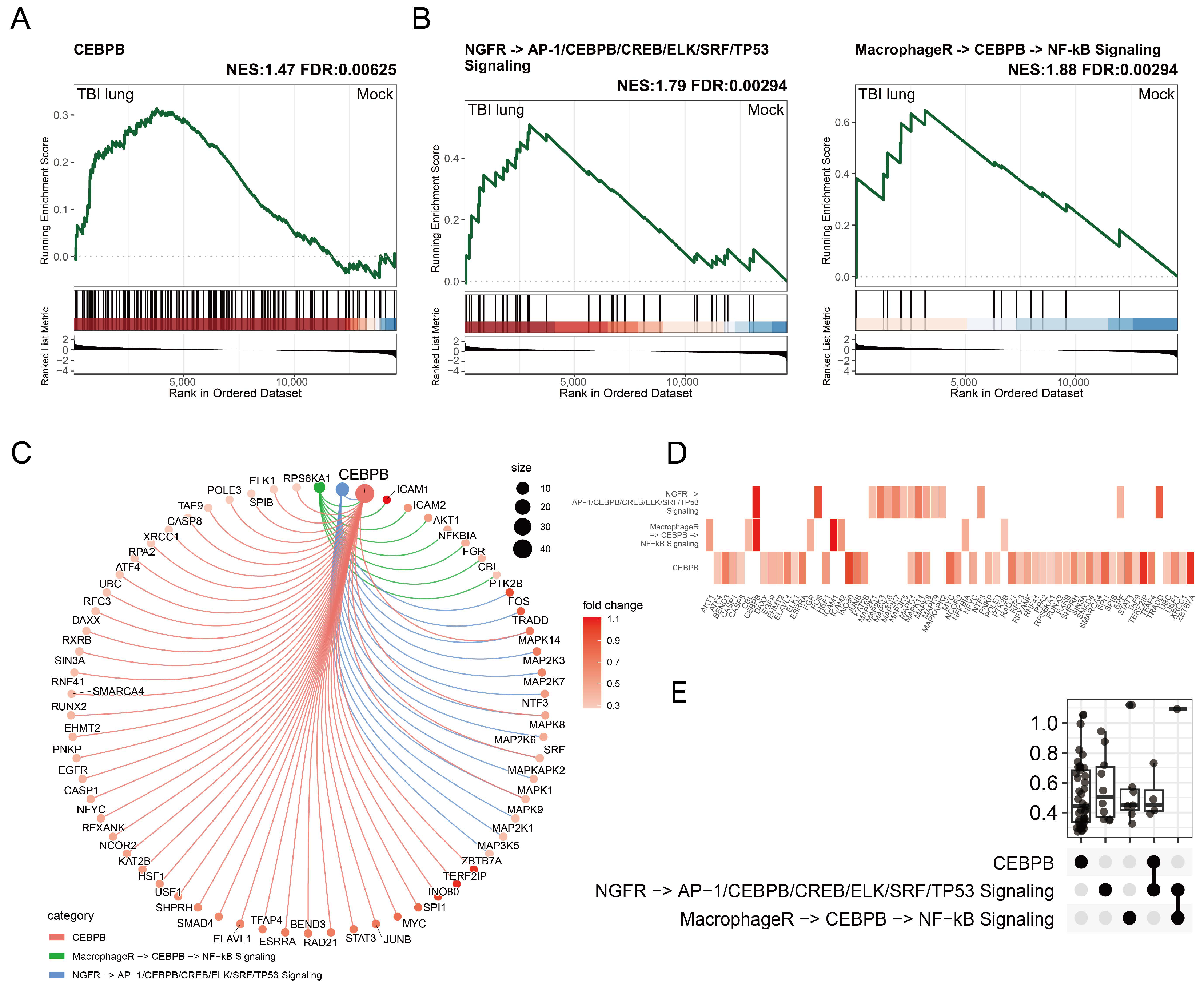
2.2. C/EBPβ-Associated Gene Enrichment in Post-TBI Lung Tissues and Implications for Macrophage Activation
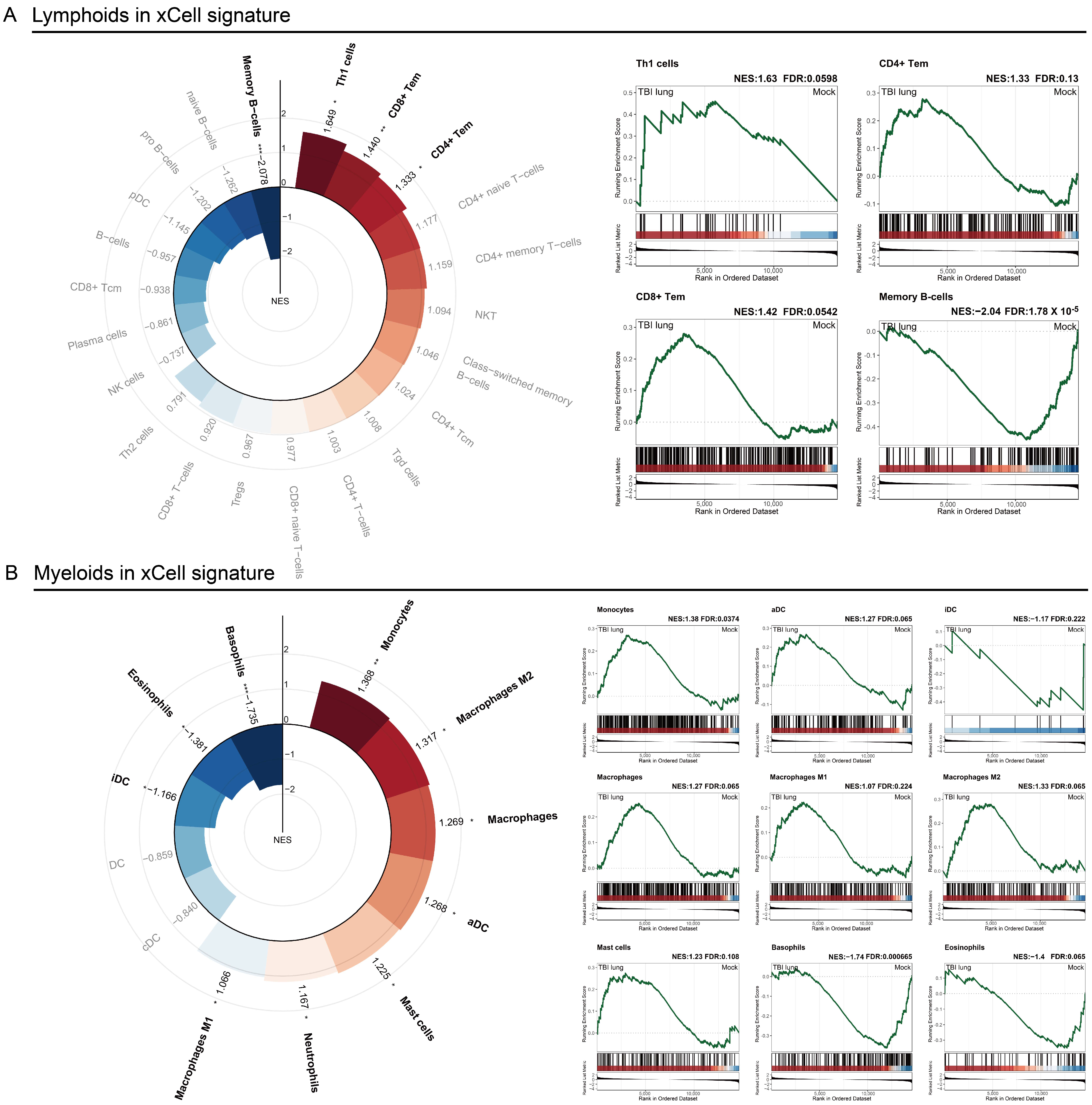
2.3. Immune Response Modulation in Post-TBI Lung Tissue: Insights from x-Cell and GSEA Analyses
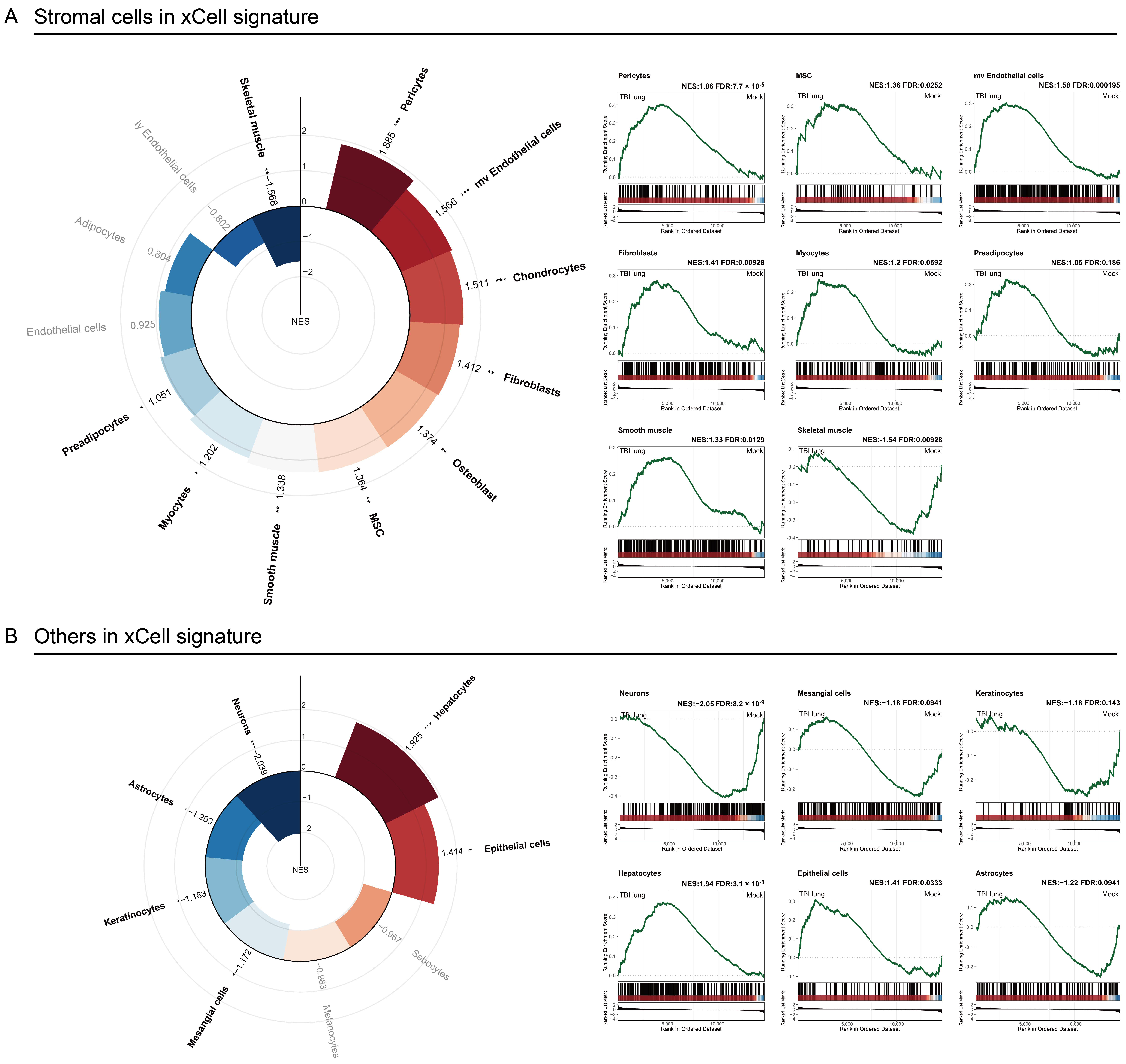
2.4. Stromal Cells Highlight TBI-Induced Pulmonary Morphogenesis Perturbations with a Notable Emphasis on Fibrosis
2.5. Exploring the Systemic Impact of Brain Injury on Lung Associated Cells Using Tabula Sapiens Database
2.6. Unraveling the Potential Association between Traumatic Brain Injury and Lung Diseases through GSEA
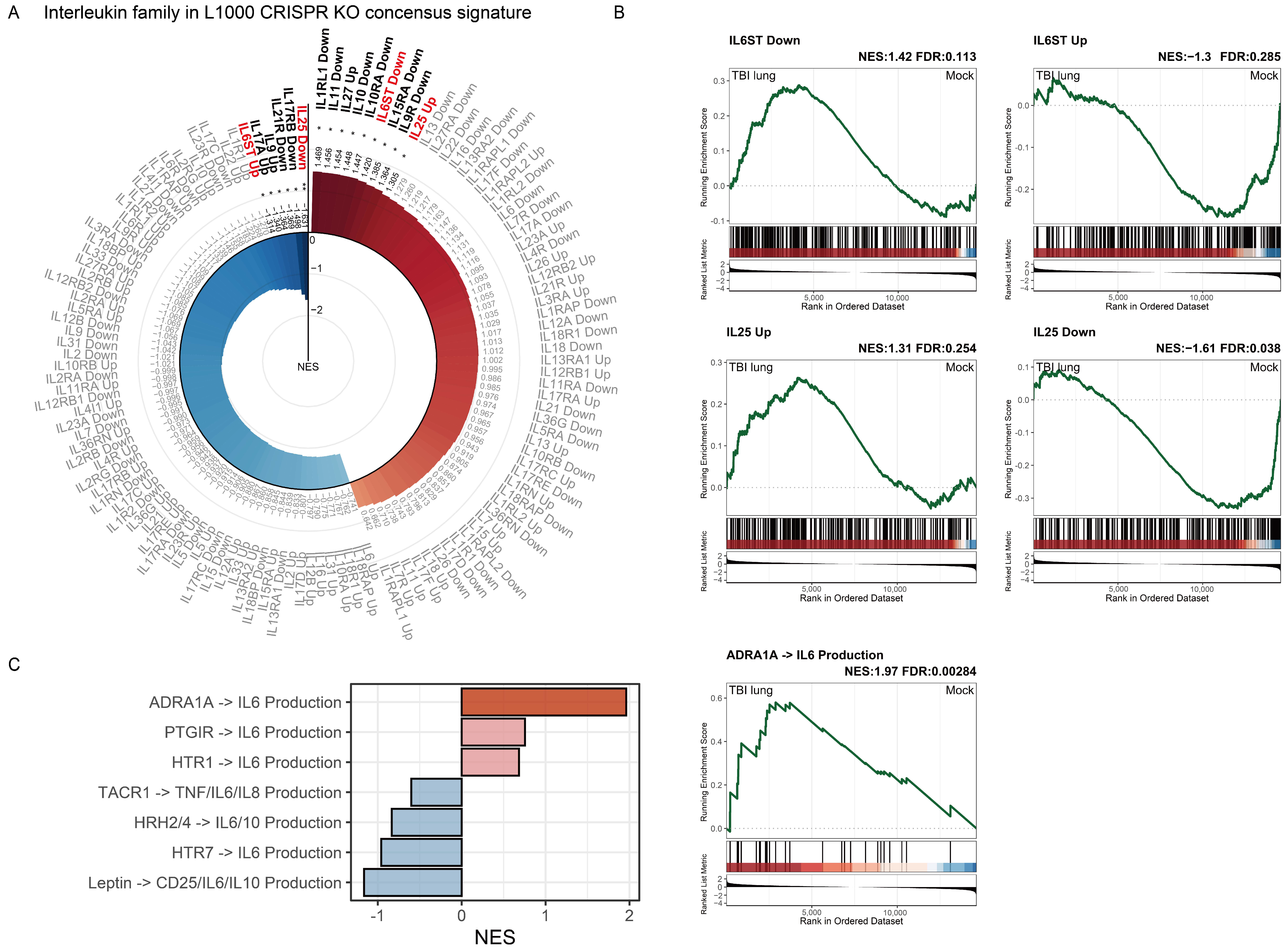
2.7. Interleukin Family Enrichment Analysis Reveals Key Regulatory Genes in Pulmonary Tissue Post-TBI Using L1000 CRISPR KO Consensus Signature
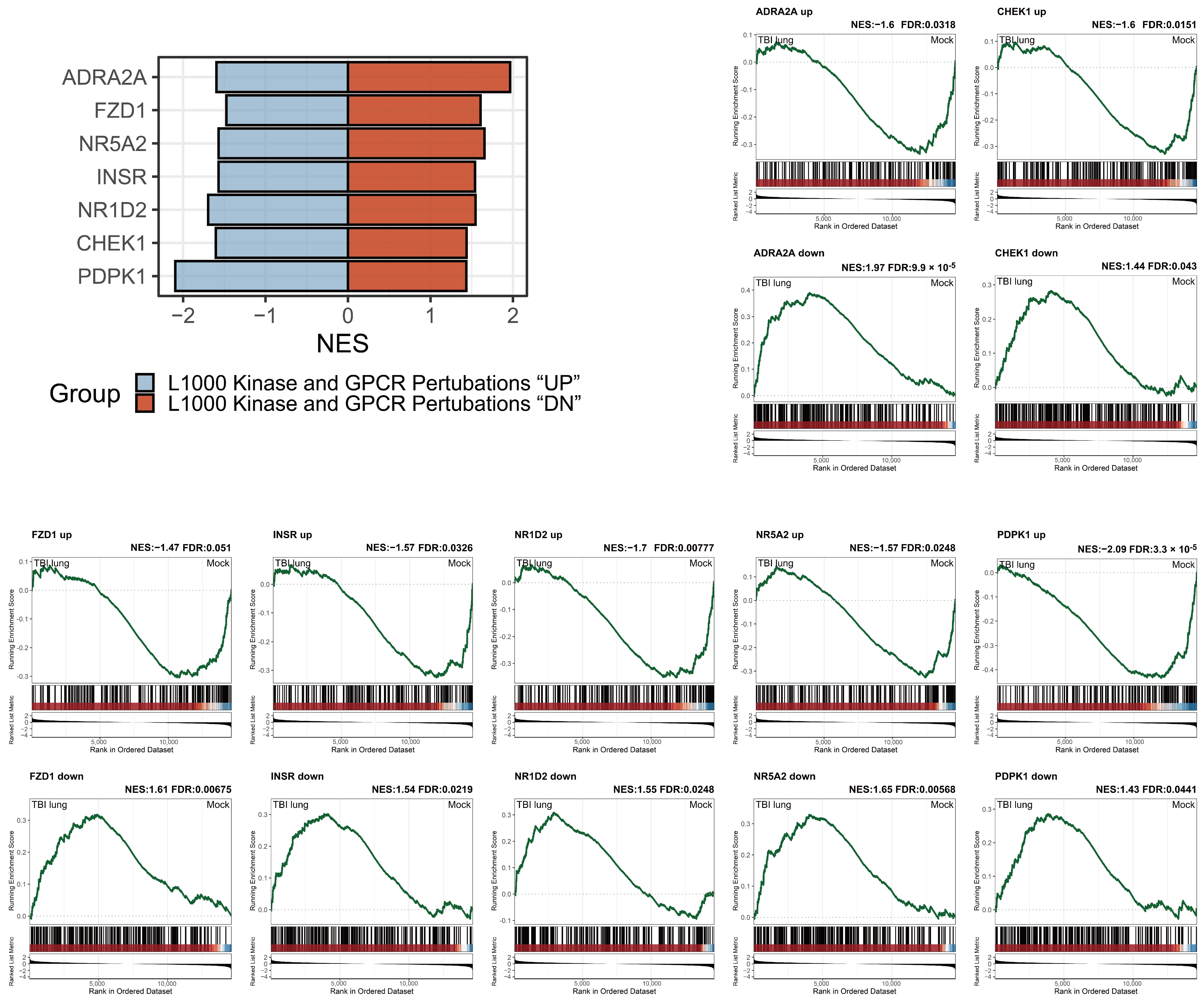
2.8. Adrenergic Receptor Enrichment and TBI-mediated Pulmonary Impacts in the L1000 Kinase and GPCR Perturbations Database
3. Discussion
4. Materials and Methods
4.1. Traumatic Brain Injury Animal Model
4.2. mRNA Expression Profiling
4.3. Differential Expression Analysis
4.4. Enrichment Analysis
4.5. Visualization Techniques
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Delage, C.; Taib, T.; Mamma, C.; Lerouet, D.; Besson, V.C. Traumatic Brain Injury: An Age-Dependent View of Post-Traumatic Neuroinflammation and Its Treatment. Pharmaceutics 2021, 13, 1624. [Google Scholar] [CrossRef]
- Summers, C.R.; Ivins, B.; Schwab, K.A. Traumatic brain injury in the United States: An epidemiologic overview. Mt. Sinai J. Med. 2009, 76, 105–110. [Google Scholar] [CrossRef]
- Young, J.T.; Hughes, N. Traumatic brain injury and homelessness: From prevalence to prevention. Lancet Public Health 2020, 5, e4–e5. [Google Scholar] [CrossRef]
- Allen, B.C.; Cummer, E.; Sarma, A.K. Traumatic Brain Injury in Select Low- and Middle-Income Countries: A Narrative Review of the Literature. J. Neurotrauma 2023, 40, 602–619. [Google Scholar] [CrossRef]
- Leibson, C.L.; Brown, A.W.; Hall Long, K.; Ransom, J.E.; Mandrekar, J.; Osler, T.M.; Malec, J.F. Medical care costs associated with traumatic brain injury over the full spectrum of disease: A controlled population-based study. J. Neurotrauma 2012, 29, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Maas, A.I.R.; Menon, D.K.; Adelson, P.D.; Andelic, N.; Bell, M.J.; Belli, A.; Bragge, P.; Brazinova, A.; Büki, A.; Chesnut, R.M.; et al. Traumatic brain injury: Integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 2017, 16, 987–1048. [Google Scholar] [CrossRef]
- Bramlett, H.M.; Dietrich, W.D. Long-Term Consequences of Traumatic Brain Injury: Current Status of Potential Mechanisms of Injury and Neurological Outcomes. J. Neurotrauma 2015, 32, 1834–1848. [Google Scholar] [CrossRef]
- Corral, L.; Javierre, C.F.; Ventura, J.L.; Marcos, P.; Herrero, J.I.; Mañez, R. Impact of non-neurological complications in severe traumatic brain injury outcome. Crit. Care 2012, 16, R44. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, V.; Komisarow, J.M.; Laskowitz, D.T.; Vavilala, M.S. Multiorgan Dysfunction After Severe Traumatic Brain Injury: Epidemiology, Mechanisms, and Clinical Management. Chest 2021, 160, 956–964. [Google Scholar] [CrossRef]
- Li, N.; Zhao, W.G.; Zhang, W.F. Acute kidney injury in patients with severe traumatic brain injury: Implementation of the acute kidney injury network stage system. Neurocrit. Care 2011, 14, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Kerr, N.A.; de Rivero Vaccari, J.P.; Abbassi, S.; Kaur, H.; Zambrano, R.; Wu, S.; Dietrich, W.D.; Keane, R.W. Traumatic Brain Injury-Induced Acute Lung Injury: Evidence for Activation and Inhibition of a Neural-Respiratory-Inflammasome Axis. J. Neurotrauma 2018, 35, 2067–2076. [Google Scholar] [CrossRef]
- Blennow, K.; Brody, D.L.; Kochanek, P.M.; Levin, H.; McKee, A.; Ribbers, G.M.; Yaffe, K.; Zetterberg, H. Traumatic brain injuries. Nat. Rev. Dis. Primers. 2016, 2, 16084. [Google Scholar] [CrossRef]
- Menon, D.K.; Schwab, K.; Wright, D.W.; Maas, A.I. Position statement: Definition of traumatic brain injury. Arch. Phys. Med. Rehabil. 2010, 91, 1637–1640. [Google Scholar] [CrossRef]
- Yang, W.H.; Chen, P.C.; Wang, T.C.; Kuo, T.Y.; Cheng, C.Y.; Yang, Y.H. Endocrine dysfunction following traumatic brain injury: A 5-year follow-up nationwide-based study. Sci. Rep. 2016, 6, 32987. [Google Scholar] [CrossRef]
- Kourbeti, I.S.; Vakis, A.F.; Papadakis, J.A.; Karabetsos, D.A.; Bertsias, G.; Filippou, M.; Ioannou, A.; Neophytou, C.; Anastasaki, M.; Samonis, G. Infections in traumatic brain injury patients. Clin. Microbiol. Infect. 2012, 18, 359–364. [Google Scholar] [CrossRef]
- Kourbeti, I.S.; Papadakis, J.A.; Neophytou, C.; Filippou, M.; Ioannou, A.; Karabetsos, D.A.; Bertsias, G.; Anastasaki, M.; Vakis, A.F. Infections in patients with traumatic brain injury who undergo neurosurgery. Br. J. Neurosurg. 2011, 25, 9–15. [Google Scholar] [CrossRef]
- Rincon, F.; Ghosh, S.; Dey, S.; Maltenfort, M.; Vibbert, M.; Urtecho, J.; McBride, W.; Moussouttas, M.; Bell, R.; Ratliff, J.K.; et al. Impact of acute lung injury and acute respiratory distress syndrome after traumatic brain injury in the United States. Neurosurgery 2012, 71, 795–803. [Google Scholar] [CrossRef]
- Lopez-Aguilar, J.; Blanch, L. Brain injury requires lung protection. Ann. Transl. Med. 2015, 3 (Suppl. 1), S5. [Google Scholar]
- Holland, M.C.; Mackersie, R.C.; Morabito, D.; Campbell, A.R.; Kivett, V.A.; Patel, R.; Erickson, V.R.; Pittet, J.F. The development of acute lung injury is associated with worse neurologic outcome in patients with severe traumatic brain injury. J. Trauma 2003, 55, 106–111. [Google Scholar] [CrossRef]
- Della Torre, V.; Badenes, R.; Corradi, F.; Racca, F.; Lavinio, A.; Matta, B.; Bilotta, F.; Robba, C. Acute respiratory distress syndrome in traumatic brain injury: How do we manage it? J. Thorac. Dis. 2017, 9, 5368–5381. [Google Scholar] [CrossRef]
- Robba, C.; Camporota, L.; Citerio, G. Acute respiratory distress syndrome complicating traumatic brain injury. Can opposite strategies converge? Intensive Care Med. 2023, 49, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Elmaleh, Y.; Yavchitz, A.; Léguillier, T.; Squara, P.-A.; Palpacuer, C.; Grégoire, C. Feasibility of Prone Positioning for Brain-injured Patients with Severe Acute Respiratory Distress Syndrome: A Systematic Review and Pilot Study (ProBrain). Anesthesiology 2024, 140, 495–512. [Google Scholar] [CrossRef] [PubMed]
- Borsellino, B.; Schultz, M.J.; Gama de Abreu, M.; Robba, C.; Bilotta, F. Mechanical ventilation in neurocritical care patients: A systematic literature review. Expert Rev. Respir. Med. 2016, 10, 1123–1132. [Google Scholar] [CrossRef]
- Bouras, M.; Asehnoune, K.; Roquilly, A. Immune modulation after traumatic brain injury. Front. Med. 2022, 9, 995044. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.F.; Li, B.; Shen, J.; Wang, Q.F.; Chen, S.S.; Jiang, X.C.; Qiang, D. Ghrelin alleviates traumatic brain injury-induced acute lung injury through pyroptosis/NF-κB pathway. Int. Immunopharmacol. 2020, 79, 106175. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, J.; Wang, X.; Chen, L.; Liu, S.; Zhang, Z.; Yao, W. Erythropoietin-Derived Peptide Protects against Acute Lung Injury after Rat Traumatic Brain Injury. Cell. Physiol. Biochem. 2017, 41, 2037–2044. [Google Scholar] [CrossRef] [PubMed]
- Balu, R. Inflammation and immune system activation after traumatic brain injury. Curr. Neurol. Neurosci. Rep. 2014, 14, 484. [Google Scholar] [CrossRef]
- Hu, P.J.; Pittet, J.F.; Kerby, J.D.; Bosarge, P.L.; Wagener, B.M. Acute brain trauma, lung injury, and pneumonia: More than just altered mental status and decreased airway protection. Am. J. Physiol. Lung Cell Mol. Physiol. 2017, 313, L1–L15. [Google Scholar] [CrossRef]
- Chan, W.H.; Hsu, Y.J.; Cheng, C.P.; Chou, K.N.; Chen, C.L.; Huang, S.M.; Kan, W.C.; Chiu, Y.L. Assessing the Global Impact on the Mouse Kidney After Traumatic Brain Injury: A Transcriptomic Study. J. Inflamm. Res. 2022, 15, 4833–4851. [Google Scholar] [CrossRef]
- Kerr, N.; de Rivero Vaccari, J.P.; Dietrich, W.D.; Keane, R.W. Neural-respiratory inflammasome axis in traumatic brain injury. Exp. Neurol. 2020, 323, 113080. [Google Scholar] [CrossRef]
- Chacón-Aponte, A.A.; Durán-Vargas, É.A.; Arévalo-Carrillo, J.A.; Lozada-Martínez, I.D.; Bolaño-Romero, M.P.; Moscote-Salazar, L.R.; Grille, P.; Janjua, T. Brain-lung interaction: A vicious cycle in traumatic brain injury. Acute Crit. Care 2022, 37, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Wu, M.; Cao, J.; Tang, H.; Zhu, M.; Johnson, P.F.; Gao, H. Critical role for CCAAT/enhancer-binding protein beta in immune complex-induced acute lung injury. J. Immunol. 2012, 189, 1480–1490. [Google Scholar] [CrossRef]
- Hung, C.F.; Wilson, C.L.; Schnapp, L.M. Pericytes in the Lung. Adv. Exp. Med. Biol. 2019, 1122, 41–58. [Google Scholar] [PubMed]
- Kato, K.; Dieguez-Hurtado, R.; Park, D.Y.; Hong, S.P.; Kato-Azuma, S.; Adams, S.; Stehling, M.; Trappmann, B.; Wrana, J.L.; Koh, G.Y.; et al. Pulmonary pericytes regulate lung morphogenesis. Nat. Commun. 2018, 9, 2448. [Google Scholar] [CrossRef] [PubMed]
- Tabula Sapiens, C.; Jones, R.C.; Karkanias, J.; Krasnow, M.A.; Pisco, A.O.; Quake, S.R.; Salzman, J.; Yosef, N.; Bulthaup, B.; Brown, P.; et al. The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 2022, 376, eabl4896. [Google Scholar] [CrossRef] [PubMed]
- Ucero, A.C.; Bakiri, L.; Roediger, B.; Suzuki, M.; Jimenez, M.; Mandal, P.; Braghetta, P.; Bonaldo, P.; Paz-Ares, L.; Fustero-Torre, C.; et al. Fra-2–expressing macrophages promote lung fibrosis. J. Clin. Investig. 2019, 129, 3293–3309. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Huaux, F.; Liu, T.; McGarry, B.; Ullenbruch, M.; Phan, S.H. Dual roles of IL-4 in lung injury and fibrosis. J. Immunol. 2003, 170, 2083–2092. [Google Scholar] [CrossRef]
- Roos, A.B.; Berg, T.; Barton, J.L.; Didon, L.; Nord, M. Airway epithelial cell differentiation during lung organogenesis requires C/EBPalpha and C/EBPbeta. Dev. Dyn. 2012, 241, 911–923. [Google Scholar] [CrossRef]
- Clark, J.G.; Madtes, D.K.; Hackman, R.C.; Chen, W.; Cheever, M.A.; Martin, P.J. Lung injury induced by alloreactive Th1 cells is characterized by host-derived mononuclear cell inflammation and activation of alveolar macrophages. J. Immunol. 1998, 161, 1913–1920. [Google Scholar] [CrossRef]
- Atif, S.M.; Mack, D.G.; McKee, A.S.; Rangel-Moreno, J.; Martin, A.K.; Getahun, A.; Maier, L.A.; Cambier, J.C.; Tuder, R.; Fontenot, A.P. Protective role of B cells in sterile particulate-induced lung injury. JCI Insight 2019, 5, e125494. [Google Scholar] [CrossRef]
- Tsymbalyuk, O.; Gerzanich, V.; Simard, J.M.; Rathinam, C.V. Traumatic brain injury alters dendritic cell differentiation and distribution in lymphoid and non-lymphoid organs. J. Neuroinflamm. 2022, 19, 238. [Google Scholar] [CrossRef]
- Bantsimba-Malanda, C.; Marchal-Somme, J.; Goven, D.; Freynet, O.; Michel, L.; Crestani, B.; Soler, P. A role for dendritic cells in bleomycin-induced pulmonary fibrosis in mice? Am. J. Respir. Crit. Care Med. 2010, 182, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Izzy, S.; Chen, P.M.; Tahir, Z.; Grashow, R.; Radmanesh, F.; Cote, D.J.; Yahya, T.; Dhand, A.; Taylor, H.; Shih, S.L.; et al. Association of Traumatic Brain Injury with the Risk of Developing Chronic Cardiovascular, Endocrine, Neurological, and Psychiatric Disorders. JAMA Netw. Open 2022, 5, e229478. [Google Scholar] [CrossRef] [PubMed]
- Tsujino, K.; Reed, N.I.; Atakilit, A.; Ren, X.; Sheppard, D. Transforming growth factor-β plays divergent roles in modulating vascular remodeling, inflammation, and pulmonary fibrosis in a murine model of scleroderma. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L22–L31. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, N.; Zhang, Y.; Xia, J.; Zhan, Q.; Wang, C. Landscape of transcription and long non-coding RNAs reveals new insights into the inflammatory and fibrotic response following ventilator-induced lung injury. Respir. Res. 2018, 19, 122. [Google Scholar] [CrossRef]
- Xu, F.; Wang, S.; Wang, Y.; Hu, L.; Zhu, L. Inhibition of gp130 alleviates LPS-induced lung injury by attenuating apoptosis and inflammation through JAK1/STAT3 signaling pathway. Inflamm. Res. 2023, 72, 493–507. [Google Scholar] [CrossRef]
- Li, Z.G.; Scott, M.J.; Brzoska, T.; Sundd, P.; Li, Y.H.; Billiar, T.R.; Wilson, M.A.; Wang, P.; Fan, J. Lung epithelial cell-derived IL-25 negatively regulates LPS-induced exosome release from macrophages. Mil. Med. Res. 2018, 5, 24. [Google Scholar] [CrossRef]
- Ciryam, P.; Gerzanich, V.; Simard, J.M. Interleukin-6 in Traumatic Brain Injury: A Janus-Faced Player in Damage and Repair. J. Neurotrauma 2023, 40, 2249–2269. [Google Scholar] [CrossRef]
- Aisiku, I.P.; Yamal, J.-M.; Doshi, P.; Benoit, J.S.; Gopinath, S.; Goodman, J.C.; Robertson, C.S. Plasma cytokines IL-6, IL-8, and IL-10 are associated with the development of acute respiratory distress syndrome in patients with severe traumatic brain injury. Crit. Care 2016, 20, 288. [Google Scholar] [CrossRef] [PubMed]
- Perez, D.M.; Papay, R.S.; Shi, T. alpha1-Adrenergic receptor stimulates interleukin-6 expression and secretion through both mRNA stability and transcriptional regulation: Involvement of p38 mitogen-activated protein kinase and nuclear factor-kappaB. Mol. Pharmacol. 2009, 76, 144–152. [Google Scholar] [CrossRef]
- Davison, D.L.; Terek, M.; Chawla, L.S. Neurogenic pulmonary edema. Crit. Care 2012, 16, 212. [Google Scholar] [CrossRef]
- Yang, A.; Liu, B.; Inoue, T. Role of autonomic system imbalance in neurogenic pulmonary oedema. Eur. J. Neurosci. 2022, 55, 1645–1657. [Google Scholar] [CrossRef]
- Henriquez, A.R.; Snow, S.J.; Schladweiler, M.C.; Miller, C.N.; Dye, J.A.; Ledbetter, A.D.; Richards, J.E.; Mauge-Lewis, K.; McGee, M.A.; Kodavanti, U.P. Adrenergic and glucocorticoid receptor antagonists reduce ozone-induced lung injury and inflammation. Toxicol. Appl. Pharmacol. 2018, 339, 161–171. [Google Scholar] [CrossRef]
- Ziaka, M.; Exadaktylos, A. Brain–lung interactions and mechanical ventilation in patients with isolated brain injury. Crit. Care 2021, 25, 358. [Google Scholar] [CrossRef]
- Konigshoff, M.; Balsara, N.; Pfaff, E.M.; Kramer, M.; Chrobak, I.; Seeger, W.; Eickelberg, O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS ONE 2008, 3, e2142. [Google Scholar] [CrossRef]
- Jung, S.M.; Park, K.S.; Kim, K.J. Integrative analysis of lung molecular signatures reveals key drivers of systemic sclerosis-associated interstitial lung disease. Ann. Rheum. Dis. 2022, 81, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sundar, I.K.; Lucas, J.H.; Park, J.G.; Nogales, A.; Martinez-Sobrido, L.; Rahman, I. Circadian clock molecule REV-ERBalpha regulates lung fibrotic progression through collagen stabilization. Nat. Commun. 2023, 14, 1295. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, B.; Chen, J. Knockdown of phosphoinositide-dependent kinase 1 (PDK1) inhibits fibrosis and inflammation in lipopolysaccharide-induced acute lung injury rat model by attenuating NF-kappaB/p65 pathway activation. Ann. Transl. Med. 2021, 9, 1671. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Walter, W.; Sanchez-Cabo, F.; Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 2015, 31, 2912–2914. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, W.-H.; Huang, S.-M.; Chiu, Y.-L. Pulmonary Effects of Traumatic Brain Injury in Mice: A Gene Set Enrichment Analysis. Int. J. Mol. Sci. 2024, 25, 3018. https://doi.org/10.3390/ijms25053018
Chan W-H, Huang S-M, Chiu Y-L. Pulmonary Effects of Traumatic Brain Injury in Mice: A Gene Set Enrichment Analysis. International Journal of Molecular Sciences. 2024; 25(5):3018. https://doi.org/10.3390/ijms25053018
Chicago/Turabian StyleChan, Wei-Hung, Shih-Ming Huang, and Yi-Lin Chiu. 2024. "Pulmonary Effects of Traumatic Brain Injury in Mice: A Gene Set Enrichment Analysis" International Journal of Molecular Sciences 25, no. 5: 3018. https://doi.org/10.3390/ijms25053018
APA StyleChan, W.-H., Huang, S.-M., & Chiu, Y.-L. (2024). Pulmonary Effects of Traumatic Brain Injury in Mice: A Gene Set Enrichment Analysis. International Journal of Molecular Sciences, 25(5), 3018. https://doi.org/10.3390/ijms25053018





