Structured Tandem Repeats in Protein Interactions
Abstract
1. Introduction
2. Results
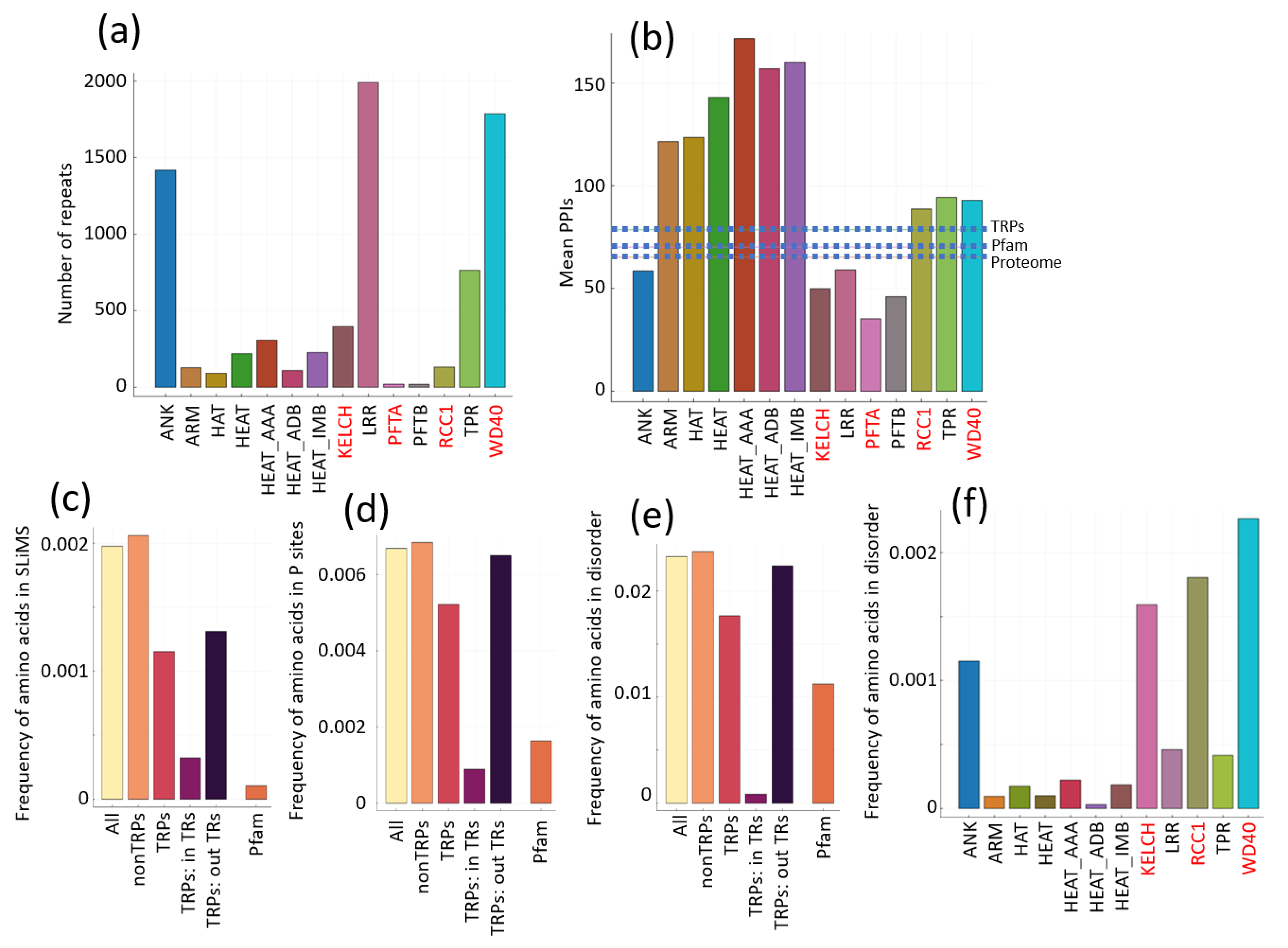
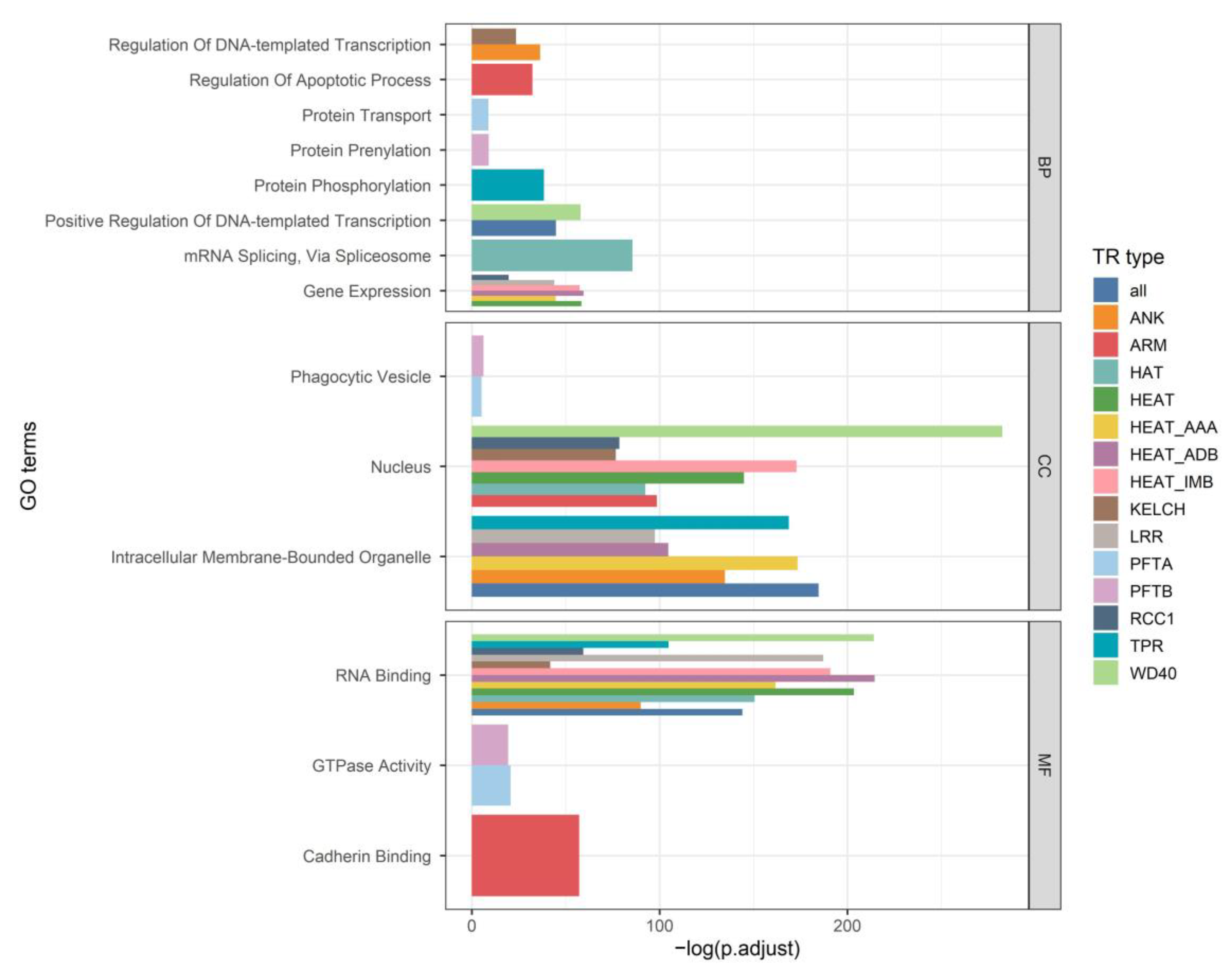
2.1. Comparison of the Properties of TRs and Other Protein Regions
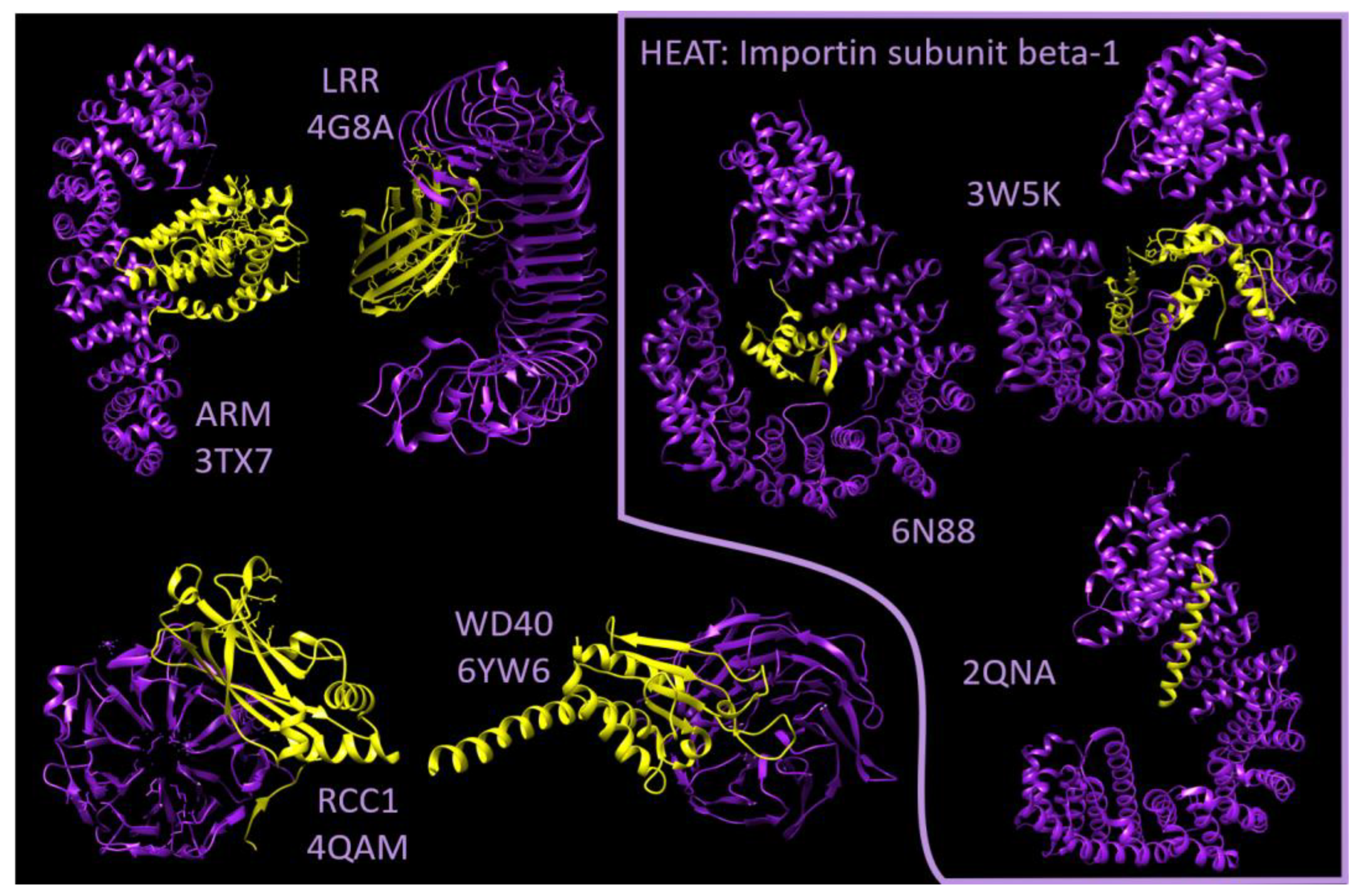
2.2. Flexibility
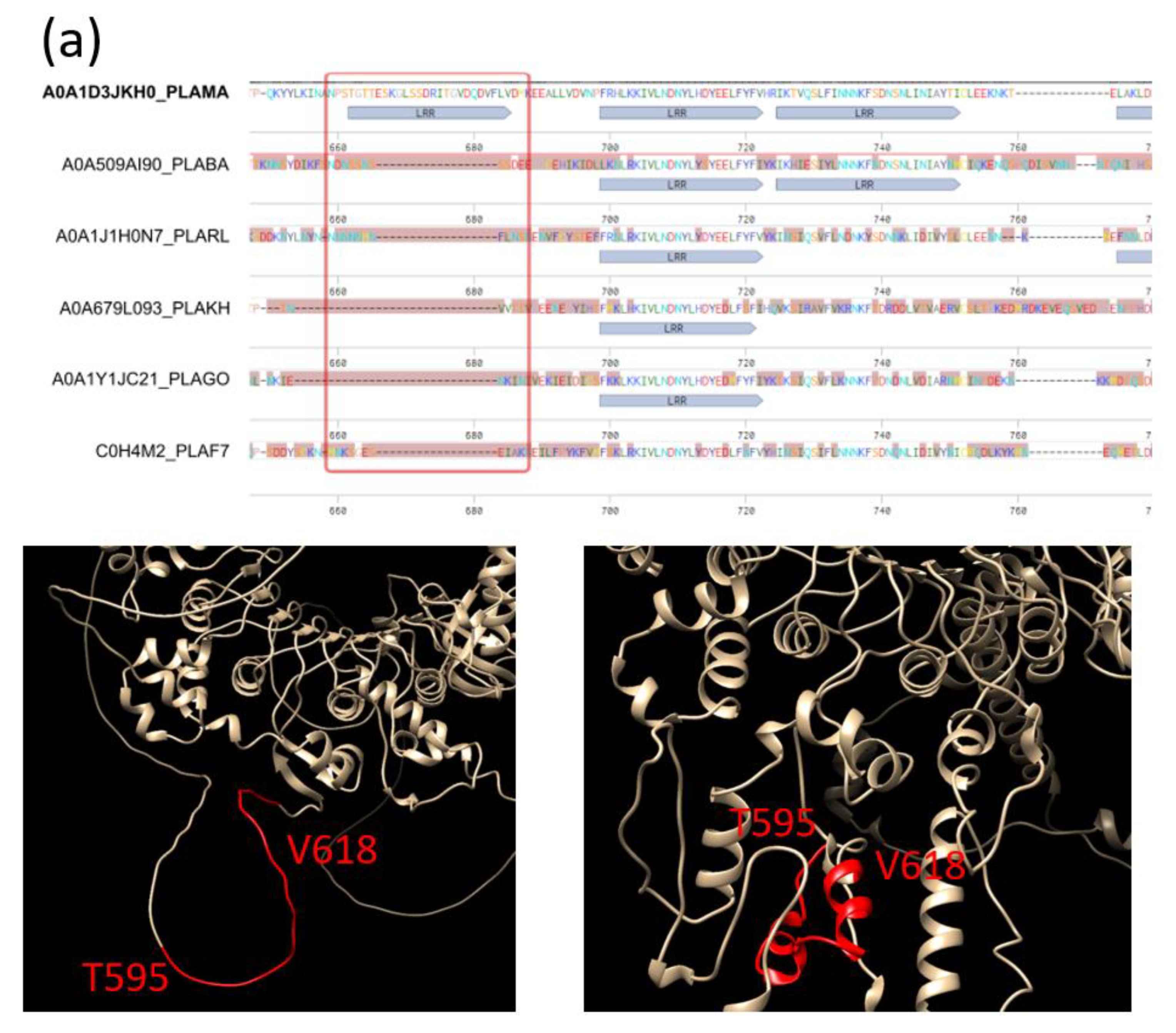
2.3. Length Variability
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobe, B.; Kajava, A.V. When protein folding is simplified to protein coiling: The continuum of solenoid protein structures. Trends Biochem. Sci. 2000, 25, 509–515. [Google Scholar] [CrossRef]
- Monzon, A.M.; Arrías, P.N.; Elofsson, A.; Mier, P.; Andrade-Navarro, M.A.; Bevilacqua, M.; Clementel, D.; Bateman, A.; Hirsh, L.; Fornasari, M.S.; et al. A STRP-ed definition of Structured Tandem Repeats in Proteins. J. Struct. Biol. 2023, 215, 108023. [Google Scholar] [CrossRef]
- Kajava, A.V. Tandem repeats in proteins: From sequence to structure. J. Struct. Biol. 2012, 179, 279–288. [Google Scholar] [CrossRef]
- Groves, M.R.; Barford, D. Topological characteristics of helical repeat proteins. Curr. Opin. Struct. Biol. 1999, 9, 383–389. [Google Scholar] [CrossRef]
- Kajava, A.V.; Steven, A.C. Beta-rolls, beta-helices, and other beta-solenoid proteins. Adv. Protein Chem. 2006, 73, 55–96. [Google Scholar] [CrossRef]
- Kobe, B.; Deisenhofer, J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature 1993, 366, 751–756. [Google Scholar] [CrossRef]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef]
- Grove, T.Z.; Cortajarena, A.L.; Regan, L. Ligand binding by repeat proteins: Natural and designed. Curr. Opin. Struct. Biol. 2008, 18, 507–515. [Google Scholar] [CrossRef]
- Linke, W.A. Sense and stretchability: The role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 2008, 77, 637–648. [Google Scholar] [CrossRef]
- Dosztanyi, Z.; Chen, J.; Dunker, A.K.; Simon, I.; Tompa, P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J. Proteome Res. 2006, 5, 2985–2995. [Google Scholar] [CrossRef]
- Delucchi, M.; Schaper, E.; Sachenkova, O.; Elofsson, A.; Anisimova, M. A New Census of Protein Tandem Repeats and Their Relationship with Intrinsic Disorder. Genes 2020, 11, 407. [Google Scholar] [CrossRef]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef]
- Oka, M.; Yoneda, Y. Importin alpha: Functions as a nuclear transport factor and beyond. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2018, 94, 259–274. [Google Scholar] [CrossRef]
- Dickson, K.A.; Haigis, M.C.; Raines, R.T. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid. Res. Mol. Biol. 2005, 80, 349–374. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- Marcotte, E.M.; Pellegrini, M.; Yeates, T.O.; Eisenberg, D. A census of protein repeats. J. Mol. Biol. 1999, 293, 151–160. [Google Scholar] [CrossRef]
- Bjorklund, A.K.; Ekman, D.; Elofsson, A. Expansion of protein domain repeats. PLoS Comput. Biol. 2006, 2, e114. [Google Scholar] [CrossRef]
- Schaper, E.; Gascuel, O.; Anisimova, M. Deep conservation of human protein tandem repeats within the eukaryotes. Mol. Biol. Evol. 2014, 31, 1132–1148. [Google Scholar] [CrossRef]
- Kamel, M.; Kastano, K.; Mier, P.; Andrade-Navarro, M.A. REP2: A Web Server to Detect Common Tandem Repeats in Protein Sequences. J. Mol. Biol. 2021, 433, 166895. [Google Scholar] [CrossRef]
- Andrade, M.A.; Ponting, C.P.; Gibson, T.J.; Bork, P. Homology-based method for identification of protein repeats using statistical significance estimates. J. Mol. Biol. 2000, 298, 521–537. [Google Scholar] [CrossRef]
- Do Viet, P.; Roche, D.B.; Kajava, A.V. TAPO: A combined method for the identification of tandem repeats in protein structures. FEBS Lett. 2015, 589, 2611–2619. [Google Scholar] [CrossRef]
- Hirsh, L.; Paladin, L.; Piovesan, D.; Tosatto, S.C.E. RepeatsDB-lite: A web server for unit annotation of tandem repeat proteins. Nucleic Acids Res. 2018, 46, W402–W407. [Google Scholar] [CrossRef]
- Bliven, S.E.; Lafita, A.; Rose, P.W.; Capitani, G.; Prlic, A.; Bourne, P.E. Analyzing the symmetrical arrangement of structural repeats in proteins with CE-Symm. PLoS Comput. Biol. 2019, 15, e1006842. [Google Scholar] [CrossRef]
- Richard, F.D.; Kajava, A.V. TRDistiller: A rapid filter for enrichment of sequence datasets with proteins containing tandem repeats. J. Struct. Biol. 2014, 186, 386–391. [Google Scholar] [CrossRef]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A million peptide motifs for the molecular biologist. Mol. Cell. 2014, 55, 161–169. [Google Scholar] [CrossRef]
- Van Roey, K.; Dinkel, H.; Weatheritt, R.J.; Gibson, T.J.; Davey, N.E. The switches.ELM resource: A compendium of conditional regulatory interaction interfaces. Sci. Signal. 2013, 6, rs7. [Google Scholar] [CrossRef]
- Neduva, V.; Russell, R.B. Peptides mediating interaction networks: New leads at last. Curr. Opin. Biotechnol. 2006, 17, 465–471. [Google Scholar] [CrossRef]
- Yaffe, M.B. Phosphotyrosine-binding domains in signal transduction. Nat. Rev. Mol. Cell Biol. 2002, 3, 177–186. [Google Scholar] [CrossRef]
- Jin, J.; Pawson, T. Modular evolution of phosphorylation-based signalling systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012, 367, 2540–2555. [Google Scholar] [CrossRef]
- Davey, N.E.; Cyert, M.S.; Moses, A.M. Short linear motifs—Ex nihilo evolution of protein regulation. Cell Commun. Signal. 2015, 13, 43. [Google Scholar] [CrossRef]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef]
- Van der Lee, R.; Lang, B.; Kruse, K.; Gsponer, J.; de Groot, N.S.; Huynen, M.A.; Matouschek, A.; Fuxreiter, M.; Babu, M.M. Intrinsically disordered segments affect protein half-life in the cell and during evolution. Cell Rep. 2014, 8, 1832–1844. [Google Scholar] [CrossRef]
- Burgi, J.; Ekal, L.; Wilmanns, M. Versatile allosteric properties in Pex5-like tetratricopeptide repeat proteins to induce diverse downstream function. Traffic 2021, 22, 140–152. [Google Scholar] [CrossRef]
- Ramya, L.; Gautham, N.; Chaloin, L.; Kajava, A.V. Restricted mobility of side chains on concave surfaces of solenoid proteins may impart heightened potential for intermolecular interactions. Proteins 2015, 83, 1654–1664. [Google Scholar] [CrossRef]
- Ma, P.; Li, D.W.; Bruschweiler, R. Predicting protein flexibility with AlphaFold. Proteins 2023, 91, 847–855. [Google Scholar] [CrossRef]
- Paladin, L.; Bevilacqua, M.; Errigo, S.; Piovesan, D.; Mičetić, I.; Necci, M.; Monzon, A.M.; Fabre, M.L.; Lopez, J.L.; Nilsson, J.F.; et al. RepeatsDB in 2021: Improved data and extended classification for protein tandem repeat structures. Nucleic Acids Res. 2021, 49, D452–D457. [Google Scholar] [CrossRef]
- Li, J.; Mahajan, A.; Tsai, M.D. Ankyrin repeat: A unique motif mediating protein-protein interactions. Biochemistry 2006, 45, 15168–15178. [Google Scholar] [CrossRef]
- Vlassi, M.; Brauns, K.; Andrade-Navarro, M.A. Short tandem repeats in the inhibitory domain of the mineralocorticoid receptor: Prediction of a beta-solenoid structure. BMC Struct. Biol. 2013, 13, 17. [Google Scholar] [CrossRef][Green Version]
- Reinar, W.B.; Greulich, A.; Stø, I.M.; Knutsen, J.B.; Reitan, T.; Tørresen, O.K.; Jentoft, S.; Butenko, M.A.; Jakobsen, K.S. Adaptive protein evolution through length variation of short tandem repeats in Arabidopsis. Sci. Adv. 2023, 9, eadd6960. [Google Scholar] [CrossRef]
- Davies, H.M.; Nofal, S.D.; McLaughlin, E.J.; Osborne, A.R. Repetitive sequences in malaria parasite proteins. FEMS Microbiol. Rev. 2017, 41, 923–940. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Ormazábal, A.; Carletti, M.S.; Saldaño, T.E.; Gonzalez Buitron, M.; Marchetti, J.; Palopoli, N.; Bateman, A. Expanding the repertoire of human tandem repeat RNA-binding proteins. PLoS ONE 2023, 18, e0290890. [Google Scholar] [CrossRef]
- Schaper, E.; Anisimova, M. The evolution and function of protein tandem repeats in plants. New Phytol. 2015, 206, 397–410. [Google Scholar] [CrossRef]
- Erdozain, S.; Barrionuevo, E.; Ripoll, L.; Mier, P.; Andrade-Navarro, M.A. Protein repeats evolve and emerge in giant viruses. J. Struct. Biol. 2023, 215, 107962. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Alanis-Lobato, G.; Andrade-Navarro, M.A.; Schaefer, M.H. HIPPIE v2.0: Enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017, 45, D408–D414. [Google Scholar] [CrossRef]
- Kumar, M.; Michael, S.; Alvarado-Valverde, J.; Zeke, A.; Lazar, T.; Glavina, J.; Nagy-Kanta, E.; Donagh, J.M.; Kalman, Z.E.; Pascarelli, S.; et al. ELM-the Eukaryotic Linear Motif resource-2024 update. Nucleic Acids Res. 2023, 55, D442–D455. [Google Scholar] [CrossRef]
- Dinkel, H.; Chica, C.; Via, A.; Gould, C.M.; Jensen, L.J.; Gibson, T.J.; Diella, F. Phospho.ELM: A database of phosphorylation sites--update 2011. Nucleic Acids Res. 2011, 39, D261–D267. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Piovesan, D.; Necci, M.; Escobedo, N.; Monzon, A.M.; Hatos, A.; Mičetić, I.; Quaglia, F.; Paladin, L.; Ramasamy, P.; Dosztányi, Z.; et al. MobiDB: Intrinsically disordered proteins in 2021. Nucleic Acids Res. 2021, 49, D361–D367. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mac Donagh, J.; Marchesini, A.; Spiga, A.; Fallico, M.J.; Arrías, P.N.; Monzon, A.M.; Vagiona, A.-C.; Gonçalves-Kulik, M.; Mier, P.; Andrade-Navarro, M.A. Structured Tandem Repeats in Protein Interactions. Int. J. Mol. Sci. 2024, 25, 2994. https://doi.org/10.3390/ijms25052994
Mac Donagh J, Marchesini A, Spiga A, Fallico MJ, Arrías PN, Monzon AM, Vagiona A-C, Gonçalves-Kulik M, Mier P, Andrade-Navarro MA. Structured Tandem Repeats in Protein Interactions. International Journal of Molecular Sciences. 2024; 25(5):2994. https://doi.org/10.3390/ijms25052994
Chicago/Turabian StyleMac Donagh, Juan, Abril Marchesini, Agostina Spiga, Maximiliano José Fallico, Paula Nazarena Arrías, Alexander Miguel Monzon, Aimilia-Christina Vagiona, Mariane Gonçalves-Kulik, Pablo Mier, and Miguel A. Andrade-Navarro. 2024. "Structured Tandem Repeats in Protein Interactions" International Journal of Molecular Sciences 25, no. 5: 2994. https://doi.org/10.3390/ijms25052994
APA StyleMac Donagh, J., Marchesini, A., Spiga, A., Fallico, M. J., Arrías, P. N., Monzon, A. M., Vagiona, A.-C., Gonçalves-Kulik, M., Mier, P., & Andrade-Navarro, M. A. (2024). Structured Tandem Repeats in Protein Interactions. International Journal of Molecular Sciences, 25(5), 2994. https://doi.org/10.3390/ijms25052994








