Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress
Abstract
1. Introduction
2. Results
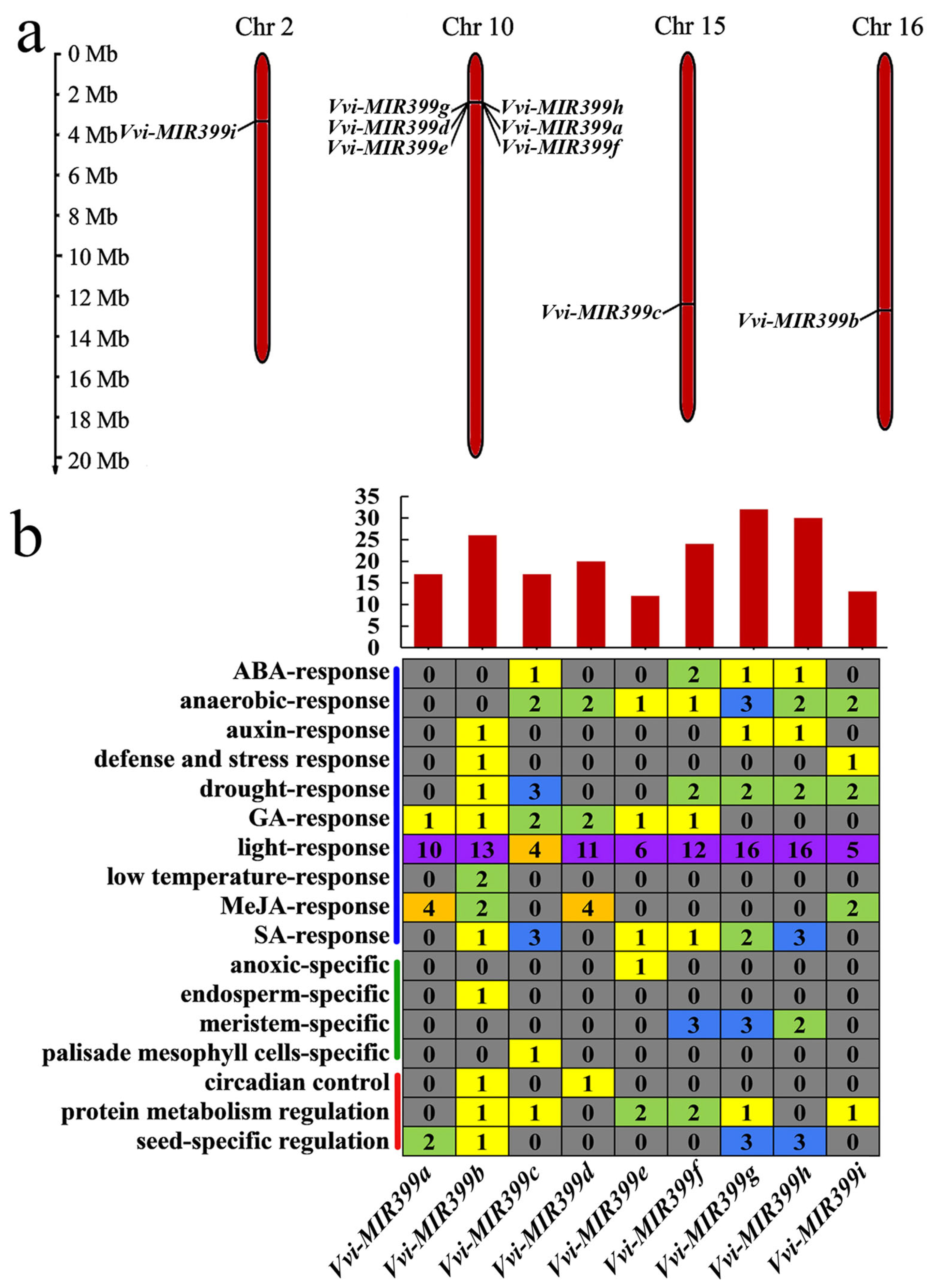
2.1. Distribution of miR399 Family Members in Grapevine
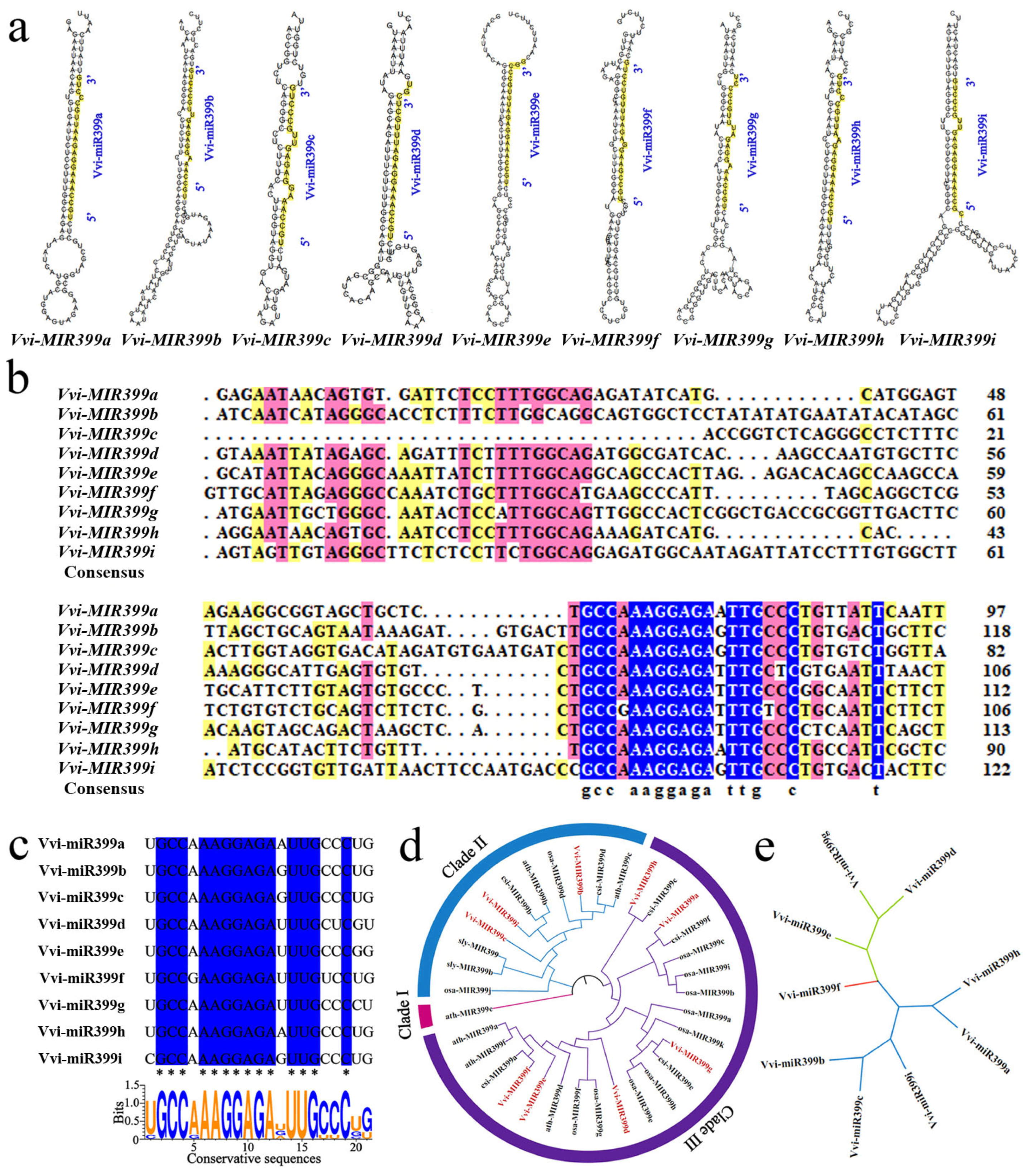
2.2. Analysis of the Vvi-miR399 Family Precursor and Mature Sequences
2.3. Prediction of Cis-Acting Elements on the Promoters of the Vvi-MIR399 Family
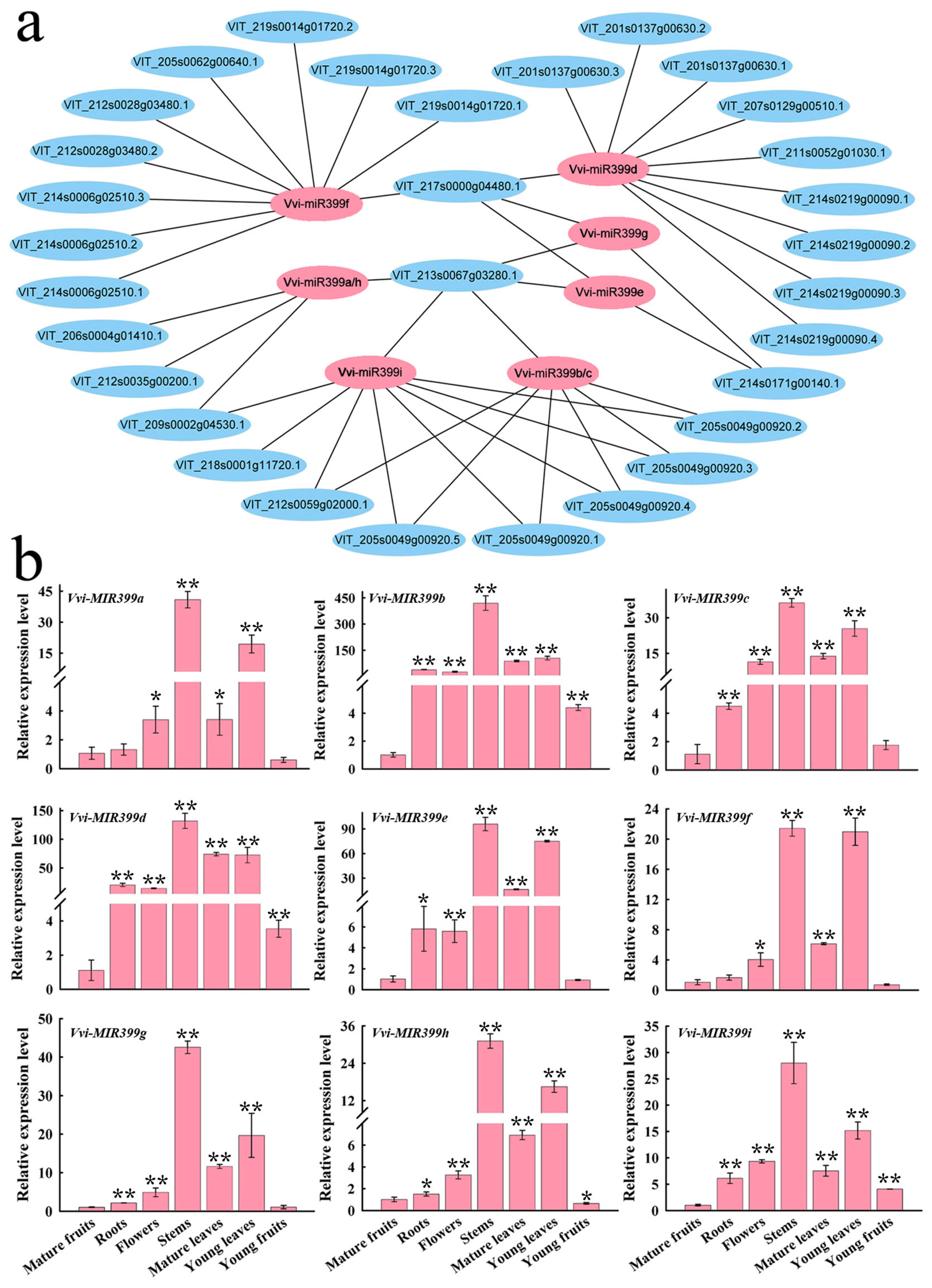
2.4. Prediction of Target Genes and Tissue-Specific Expression Analysis
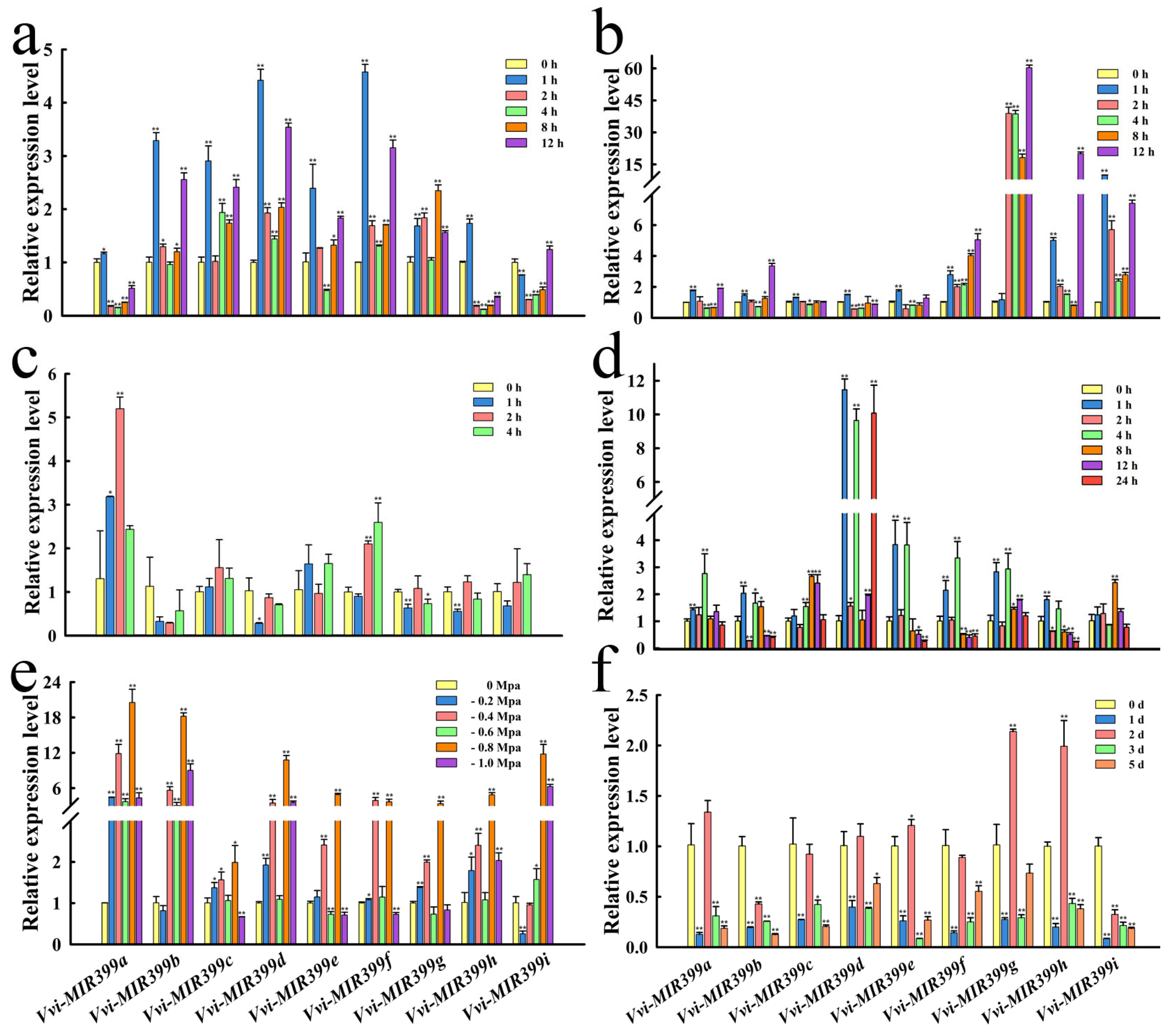
2.5. Expression Analysis of Vvi-MIR399 Family Members under Abiotic Stress
2.6. Expression Patterns of Target Genes in Response to Abiotic Stresses
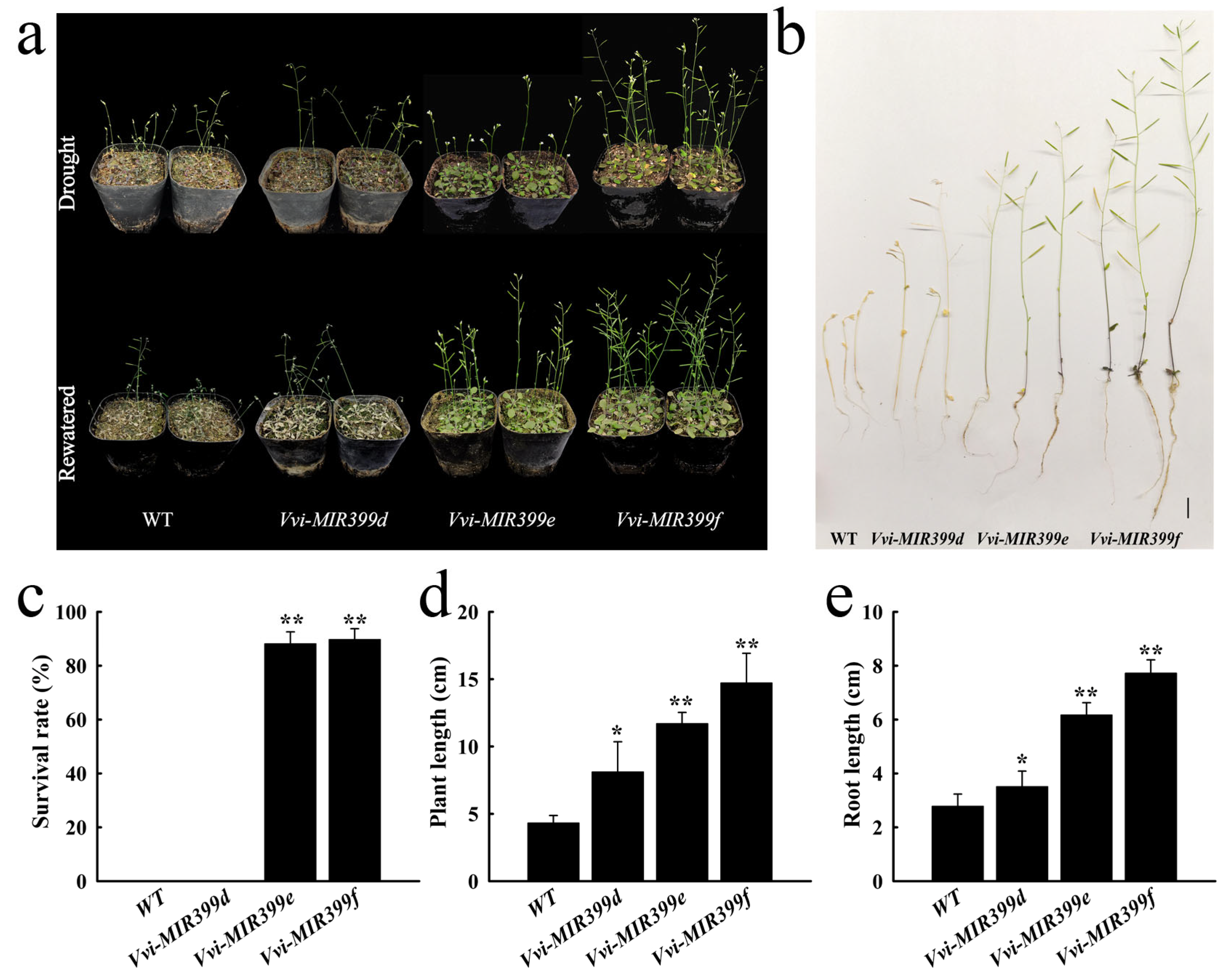
2.7. Overexpression of Vvi-MIR399 Enhanced the Drought Tolerance in Arabidopsis
3. Discussion
3.1. Evolutionary Characteristics of the MIR399 Family
3.2. The MIR399 Family May Participate in Plant Growth and Development
3.3. The MIR399 Family May Respond to Abiotic Stress in Plants
4. Materials and Methods
4.1. Plant Materials and Stress Treatments
4.2. Identification of the miR399 Family in Grapevine
4.3. Experimental Verifications of the Vvi-MIR399 Family Members
4.4. Structure Prediction of the Vvi-miR399 Precursors
4.5. Phylogenetic Analyses of the Vvi-MIR399 Family Members
4.6. Conservation Analysis of the Vvi-miR399 Family Members
4.7. Chromosome Localization of the Vvi-MIR399 Family
4.8. Regulatory Element Prediction on the Promoter Regions of the Vvi-MIR399 Family
4.9. Prediction of Vvi-miR399-Targeted Genes
4.10. Expression Analysis of the Vvi-MIR399 Family and Target Genes in Grapevine
4.11. Vector Construction
4.12. Arabidopsis Transformation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Thakur, V.; Wanchana, S.; Xu, M.; Bruskiewich, R.; Quick, W.P.; Mosig, A.; Zhu, X.G. Characterization of statistical features for plant microRNA prediction. BMC Genom. 2011, 12, 108. [Google Scholar] [CrossRef]
- Fang, L.; Wang, Y. MicroRNAs in woody plants. Front. Plant Sci. 2021, 12, 686831. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.W.; Zhang, S.; Etheridge, A.; Ma, L.; Martin, D.; Galas, D.; Wang, K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA 2010, 16, 2170–2180. [Google Scholar] [CrossRef] [PubMed]
- Neilsen, C.T.; Goodall, G.J.; Bracken, C.P. IsomiRs-the overlooked repertoire in the dynamic microRNAome. Trends Genet. 2012, 28, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Ruan, C.; Shah, A.H.; Li, D.; Li, H.; Ding, J.; Li, J.; Du, W. Identification of miRNA-mRNA regulatory modules involved in lipid metabolism and seed development in a woody oil tree (Camellia oleifera). Cells 2021, 11, 71. [Google Scholar] [CrossRef]
- Zhou, C.; Han, L.; Zhao, Y.; Wang, H.; Nakashima, J.; Tong, J.; Xiao, L.; Wang, Z.Y. Transforming compound leaf patterning by manipulating REVOLUTA in Medicago truncatula. Plant J. 2019, 100, 562–571. [Google Scholar] [CrossRef]
- Zhu, J.; Li, Y.; Lin, J.; Wu, Y.; Guo, H.; Shao, Y.; Wang, F.; Wang, X.; Mo, X.; Zheng, S.; et al. CRD1, an Xpo1 domain protein, regulates miRNA accumulation and crown root development in rice. Plant J. 2019, 100, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, Y.; Li, P.; Huang, X.; Chen, M.; Wu, J.; Wang, L.; Liu, X.; Li, Y. MircroRNA profiles of early rice inflorescence revealed a specific miRNA5506 regulating development of floral organs and female megagametophyte in rice. Int. J. Mol. Sci. 2021, 22, 6610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yin, L.; Wang, H.; Wang, G.; Ma, X.; Li, M.; Wu, H.; Fu, Q.; Zhang, Y.; Yi, H. Genome-wide identification of Hami melon miRNAs with putative roles during fruit development. PLoS ONE 2017, 12, e0180600. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zeng, Z.; Liu, Z.; Xia, R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 2018, 5, 63. [Google Scholar] [CrossRef]
- Wang, X.; Hong, Z.; Yang, A.; He, Y.; Zhu, Z.; Xu, Y. Systematic analysis of the CsmiR396-CsGRFs/CsGIFs module and the opposite role of CsGRF3 and CsGRF5 in regulating cell proliferation in cucumber. Plant Sci. 2022, 323, 111407. [Google Scholar] [CrossRef] [PubMed]
- Ferreira e Silva, G.F.; Silva, E.M.; Azevedo Mda, S.; Guivin, M.A.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.; Nogueira, F.T. MicroRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Vishwakarma, A.; Kenea, H.D.; Galsurker, O.; Cohen, H.; Aharoni, A.; Arazi, T. CRISPR/Cas9 mutants of tomato MICRORNA164 genes uncover their functional specialization in development. Plant Physiol. 2021, 187, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, C.; Wang, Y.; Dong, Q.; Gai, Y.; Ji, X. MicroRNA profiling during mulberry (Morus atropurpurea Roxb) fruit development and regulatory pathway of miR477 for anthocyanin accumulation. Front. Plant Sci. 2021, 12, 687364. [Google Scholar] [CrossRef]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Chai, W.; Song, N.; Su, A.; Wang, J.; Si, W.; Cheng, B.; Jiang, H. ZmmiR190 and its target regulate plant responses to drought stress through an ABA-dependent pathway. Plant Sci. 2021, 312, 111034. [Google Scholar] [CrossRef]
- Li, R.; Fan, T.; Wang, T.; Dominic, K.; Hu, F.; Liu, L.; Zhang, L.; Fang, R.; Pan, G.; Li, L.; et al. Characterization and functional analysis of miR166f in drought stress tolerance in mulberry (Morus multicaulis). Mol. Breed. 2018, 38, 132. [Google Scholar] [CrossRef]
- Hong, Y.; Meng, J.; He, X.; Zhang, Y.; Liu, Y.; Zhang, C.; Qi, H.; Luan, Y. Editing miR482b and miR482c simultaneously by CRISPR/Cas9 enhanced tomato resistance to Phytophthora infestans. Phytopathology 2021, 111, 1008–1016. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Medina, C.; da Rocha, M.; Magliano, M.; Ratpopoulo, A.; Revel, B.; Marteu, N.; Magnone, V.; Lebrigand, K.; Cabrera, J.; Barcala, M.; et al. Characterization of microRNAs from Arabidopsis galls highlights a role for miR159 in the plant response to the root-knot nematode Meloidogyne incognita. New Phytol. 2017, 216, 882–896. [Google Scholar] [CrossRef]
- Leng, X.; Fang, J.; Pervaiz, T.; Li, Y.; Wang, X.; Liu, D.; Zhu, X.; Fang, J. Characterization of expression patterns of grapevine microRNA family members using microRNA rapid amplification of complementary DNA ends. Plant Genome 2015, 8, plantgenome2014.10.0069. [Google Scholar] [CrossRef]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef]
- Bari, R.; Datt Pant, B.; Stitt, M.; Scheible, W.R. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Khanna, K.; Ruan, S. Expression of microRNAs and its regulation in plants. Semin. Cell Dev. Biol. 2010, 21, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chiou, T.J.; Lin, S.I.; Aung, K.; Zhu, J.K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 2005, 15, 2038–2043. [Google Scholar] [CrossRef]
- Aung, K.; Lin, S.I.; Wu, C.C.; Huang, Y.T.; Su, C.L.; Chiou, T.J. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef]
- Chiou, T.J.; Aung, K.; Lin, S.I.; Wu, C.C.; Chiang, S.F.; Su, C.L. Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 2006, 18, 412–421. [Google Scholar] [CrossRef]
- Hu, B.; Zhu, C.; Li, F.; Tang, J.; Wang, Y.; Lin, A.; Liu, L.; Che, R.; Chu, C. LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 2011, 156, 1101–1115. [Google Scholar] [CrossRef]
- Valdes-Lopez, O.; Arenas-Huertero, C.; Ramirez, M.; Girard, L.; Sanchez, F.; Vance, C.P.; Luis Reyes, J.; Hernandez, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Q.; Allan, D.L.; Vance, C.P. Systemic signaling and local sensing of phosphate in common bean: Cross-talk between photosynthate and microRNA399. Mol. Plant 2010, 3, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Kuo, H.F.; Chiou, T.J. The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 2011, 156, 1016–1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fang, Y.N.; Wu, X.M.; Qing, M.; Li, C.C.; Xie, K.D.; Deng, X.X.; Guo, W.W. The miR399-CsUBC24 module regulates reproductive development and male fertility in citrus. Plant Physiol. 2020, 183, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Tian, Y.; Sun, X.; Song, C.; Ren, Z.; Bao, Y.; Xing, J.; Li, Y.; Xu, Q.; Yu, J.; et al. tae-miR399-UBC24 module enhances freezing tolerance in winter wheat via a CBF signaling pathway. J. Agric. Food Chem. 2021, 69, 13398–13415. [Google Scholar] [CrossRef] [PubMed]
- Starega-Roslan, J.; Koscianska, E.; Kozlowski, P.; Krzyzosiak, W.J. The role of the precursor structure in the biogenesis of microRNA. Cell. Mol. Life Sci. 2011, 68, 2859–2871. [Google Scholar] [CrossRef]
- Bologna, N.G.; Schapire, A.L.; Zhai, J.; Chorostecki, U.; Boisbouvier, J.; Meyers, B.C.; Palatnik, J.F. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013, 23, 1675–1689. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xiong, X.; Wang, Q.; Zhu, L.; Wang, L.; He, Y.; Zeng, H. Integrated analysis of small RNA, transcriptome, and degradome sequencing reveals the miR156, miR5488 and miR399 are involved in the regulation of male sterility in PTGMS rice. Int. J. Mol. Sci. 2021, 22, 2260. [Google Scholar] [CrossRef]
- Jones-Rhoades, M.W. Conservation and divergence in plant microRNAs. Plant Mol. Biol. 2012, 80, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural basis for target-directed microRNA degradation. Mol. Cell 2019, 75, 1243–1255.e1247. [Google Scholar] [CrossRef]
- Baek, D.; Chun, H.J.; Kang, S.; Shin, G.; Park, S.J.; Hong, H.; Kim, C.; Kim, D.H.; Lee, S.Y.; Kim, M.C.; et al. A role for Arabidopsis miR399f in salt, drought, and ABA signaling. Mol. Cells 2016, 39, 111–118. [Google Scholar] [CrossRef]
- Gu, M.; Xu, K.; Chen, A.; Zhu, Y.; Tang, G.; Xu, G. Expression analysis suggests potential roles of microRNAs for phosphate and arbuscular mycorrhizal signaling in Solanum lycopersicum. Physiol. Plant 2010, 138, 226–237. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, R.; Albrecht, U.; Padmanabhan, C.; Wang, A.; Coffey, M.D.; Girke, T.; Wang, Z.; Close, T.J.; Roose, M.; et al. Small RNA profiling reveals phosphorus deficiency as a contributing factor in symptom expression for citrus huanglongbing disease. Mol. Plant 2013, 6, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Hsu, J.Y.; Theisen, J.W.; Kadonaga, J.T. The RNA polymerase II core promoter—The gateway to transcription. Curr. Opin. Cell Biol. 2008, 20, 253–259. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Ren, B. The gateway to transcription: Identifying, characterizing and understanding promoters in the eukaryotic genome. Cell Mol. Life Sci. 2007, 64, 386–400. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Cui, W.; Guan, C.; Mao, W.; Zhang, Z. Improvement in fruit quality by overexpressing miR399a in woodland strawberry. J. Agric. Food Chem. 2017, 65, 7361–7370. [Google Scholar] [CrossRef]
- Peng, Z.; Tian, J.; Luo, R.; Kang, Y.; Lu, Y.; Hu, Y.; Liu, N.; Zhang, J.; Cheng, H.; Niu, S.; et al. MiR399d and epigenetic modification comodulate anthocyanin accumulation in Malus leaves suffering from phosphorus deficiency. Plant Cell Environ. 2020, 43, 1148–1159. [Google Scholar] [CrossRef]
- Ma, L.; Mu, J.; Grierson, D.; Wang, Y.; Gao, L.; Zhao, X.; Zhu, B.; Luo, Y.; Shi, K.; Wang, Q.; et al. Noncoding RNAs: Functional regulatory factors in tomato fruit ripening. Theor. Appl. Genet. 2020, 133, 1753–1762. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yuan, D.; Liu, Y.; Liang, Y.; He, J.; Yang, X.; Hang, R.; Jia, H.; Mo, B.; Tian, F.; et al. INDETERMINATE1 autonomously regulates phosphate homeostasis upstream of the miR399-ZmPHO2 signaling module in maize. Plant Cell 2023, 35, 2208–2231. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-I.; Chiang, S.-F.; Lin, W.-Y.; Chen, J.-W.; Tseng, C.-Y.; Wu, P.-C.; Chiou, T.-J. Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 2008, 147, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Lu, X.Y.; Huang, X.L. Plant miRNAs and abiotic stress responses. BioChem. Biophys. Res. Commun. 2008, 368, 458–462. [Google Scholar] [CrossRef]
- Joanne, C. Light modulation of vegetative development. Plant Cell 1997, 9, 1225–1234. [Google Scholar] [CrossRef][Green Version]
- Zhang, H.; Wang, J.; Tian, S.; Hao, W.; Du, L. Two B-box proteins, MaBBX20 and MaBBX51, coordinate light-induced anthocyanin biosynthesis in grape hyacinth. Int. J. Mol. Sci. 2022, 23, 5678. [Google Scholar] [CrossRef] [PubMed]
- Folta, K.M.; Carvalho, S.D. Photoreceptors and control of horticultural plant traits. Hort. Sci. 2015, 50, 1274–1280. [Google Scholar] [CrossRef]
- Paradiso, R.; Proietti, S. Light-quality manipulation to control plant growth and photomorphogenesis in greenhouse horticulture: The state of the art and the opportunities of modern LED systems. J. Plant Growth Regul. 2021, 41, 742–780. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, W.; Wang, Y.; Yong, T.; Yang, F.; Liu, W.; Wu, X.; Du, J.; Shu, K.; Liu, J.; et al. Leaf area and photosynthesis of newly emerged trifoliolate leaves are regulated by mature leaves in soybean. J. Plant Res. 2018, 131, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Prerostova, S.; Dobrev, P.I.; Knirsch, V.; Jarosova, J.; Gaudinova, A.; Zupkova, B.; Prasil, I.T.; Janda, T.; Brzobohaty, B.; Skalak, J.; et al. Light quality and intensity modulate cold acclimation in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 2736. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Saha, S.; Vangaru, S.; Purkayastha, S.; Das, D.; Saha, A.K.; Roy, A.; Das, S.; Bhattacharyya, P.K.; Mukherjee, S.; et al. Identification and analysis of low light tolerant rice genotypes in field conditions and their SSR-based diversity in various abiotic stress tolerant lines. J. Genet. 2020, 99, 24. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, M.; Ranga, A.; Das, J.; Panigrahi, K.C.S. Shade tolerance in Swarnaprabha rice is associated with higher rate of panicle emergence and positively regulated by genes of ethylene and cytokinin pathway. Sci. Rep. 2019, 9, 6817. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, W.; Yang, W. Shade inhibits leaf size by controlling cell proliferation and enlargement in soybean. Sci. Rep. 2017, 7, 9259. [Google Scholar] [CrossRef]
- Feng, L.; Raza, M.A.; Li, Z.; Chen, Y.; Khalid, M.H.B.; Du, J.; Liu, W.; Wu, X.; Song, C.; Yu, L.; et al. The influence of light intensity and leaf movement on photosynthesis characteristics and carbon balance of soybean. Front. Plant Sci. 2019, 9, 1952. [Google Scholar] [CrossRef]
- Mallappa, C.; Yadav, V.; Negi, P.; Chattopadhyay, S. A basic leucine zipper transcription factor, G-box-binding factor 1, regulates blue light-mediated photomorphogenic growth in Arabidopsis. J. Biol. Chem. 2006, 281, 22190–22199. [Google Scholar] [CrossRef]
- Roy, S.; Choudhury, S.R.; Singh, S.K.; Das, K.P. Functional analysis of light-regulated promoter region of AtPolλ gene. Planta 2011, 235, 411–432. [Google Scholar] [CrossRef]
- Chen, J.; Jiang, T.; Jiang, J.; Deng, L.; Liu, Y.; Zhong, Z.; Fu, H.; Yang, B.; Zhang, L. The chloroplast GATA-motif of Mahonia bealei participates in alkaloid-mediated photosystem inhibition during dark to light transition. J. Plant Physiol. 2023, 280, 153894. [Google Scholar] [CrossRef]
- Kim, W.; Ahn, H.J.; Chiou, T.J.; Ahn, J.H. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Vicent, I.; Navarro, A.; Mulet, J.M.; Sharma, S.; Serrano, R. Uptake of inorganic phosphate is a limiting factor for Saccharomyces cerevisiae during growth at low temperatures. FEMS Yeast Res. 2015, 15, fov008. [Google Scholar] [CrossRef] [PubMed]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Profiling the abiotic stress responsive microRNA landscape of Arabidopsis thaliana. Plants 2019, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Pegler, J.L.; Oultram, J.M.J.; Grof, C.P.L.; Eamens, A.L. Molecular manipulation of the miR399/PHO2 expression module alters the salt stress response of Arabidopsis thaliana. Plants 2020, 10, 73. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, Y.; Mou, F.; Tian, Y.; Chen, L.; Zhang, S.; Jiang, Q.; Li, X. Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front. Plant Sci. 2016, 6, 1273. [Google Scholar] [CrossRef] [PubMed]
- Luesse, D.R.; DeBlasio, S.L.; Hangarter, R.P. Plastid movement impaired 2, a new gene involved in normal blue-light-induced chloroplast movements in Arabidopsis. Plant Physiol. 2006, 141, 1328–1337. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhu, C.; Wang, C.; Liu, L.; Shen, Q.; Xu, D.; Wang, Q. Maize transcription factor ZmEREB20 enhanced salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 159, 257–267. [Google Scholar] [CrossRef]
- Barciszewska-Pacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A.; et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [CrossRef]
- Ferreira, T.H.; Gentile, A.; Vilela, R.D.; Costa, G.G.; Dias, L.I.; Endres, L.; Menossi, M. MicroRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS ONE 2012, 7, e46703. [Google Scholar] [CrossRef]
- Frazier, T.P.; Sun, G.; Burklew, C.E.; Zhang, B. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol. Biotechnol. 2011, 49, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Zhang, L.; Song, Y.; He, J.; Xu, W.; Ma, C.; Ren, Y.; Liu, H. Integrative investigation of root-related mRNAs, lncRNAs and circRNAs of “Muscat Hamburg” (Vitis vinifera L.) grapevine in response to root restriction through transcriptomic analyses. Genes 2022, 13, 1547. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 2008, 16, 735–743. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Ren, Y.; Sun, Y.; Yin, Y.; Han, B.; Zhang, L.; Song, Y.; Zhang, Z.; Xu, Y.; Fan, D.; et al. Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 2979. https://doi.org/10.3390/ijms25052979
Liu J, Ren Y, Sun Y, Yin Y, Han B, Zhang L, Song Y, Zhang Z, Xu Y, Fan D, et al. Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress. International Journal of Molecular Sciences. 2024; 25(5):2979. https://doi.org/10.3390/ijms25052979
Chicago/Turabian StyleLiu, Jingjing, Yi Ren, Yan Sun, Yonggang Yin, Bin Han, Lipeng Zhang, Yue Song, Zhen Zhang, Yuanyuan Xu, Dongying Fan, and et al. 2024. "Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress" International Journal of Molecular Sciences 25, no. 5: 2979. https://doi.org/10.3390/ijms25052979
APA StyleLiu, J., Ren, Y., Sun, Y., Yin, Y., Han, B., Zhang, L., Song, Y., Zhang, Z., Xu, Y., Fan, D., Li, J., Liu, H., & Ma, C. (2024). Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress. International Journal of Molecular Sciences, 25(5), 2979. https://doi.org/10.3390/ijms25052979







