Single Cell High Dimensional Analysis of Human Peripheral Blood Mononuclear Cells Reveals Unique Intermediate Monocyte Subsets Associated with Sex Differences in Coronary Artery Disease
Abstract
1. Introduction
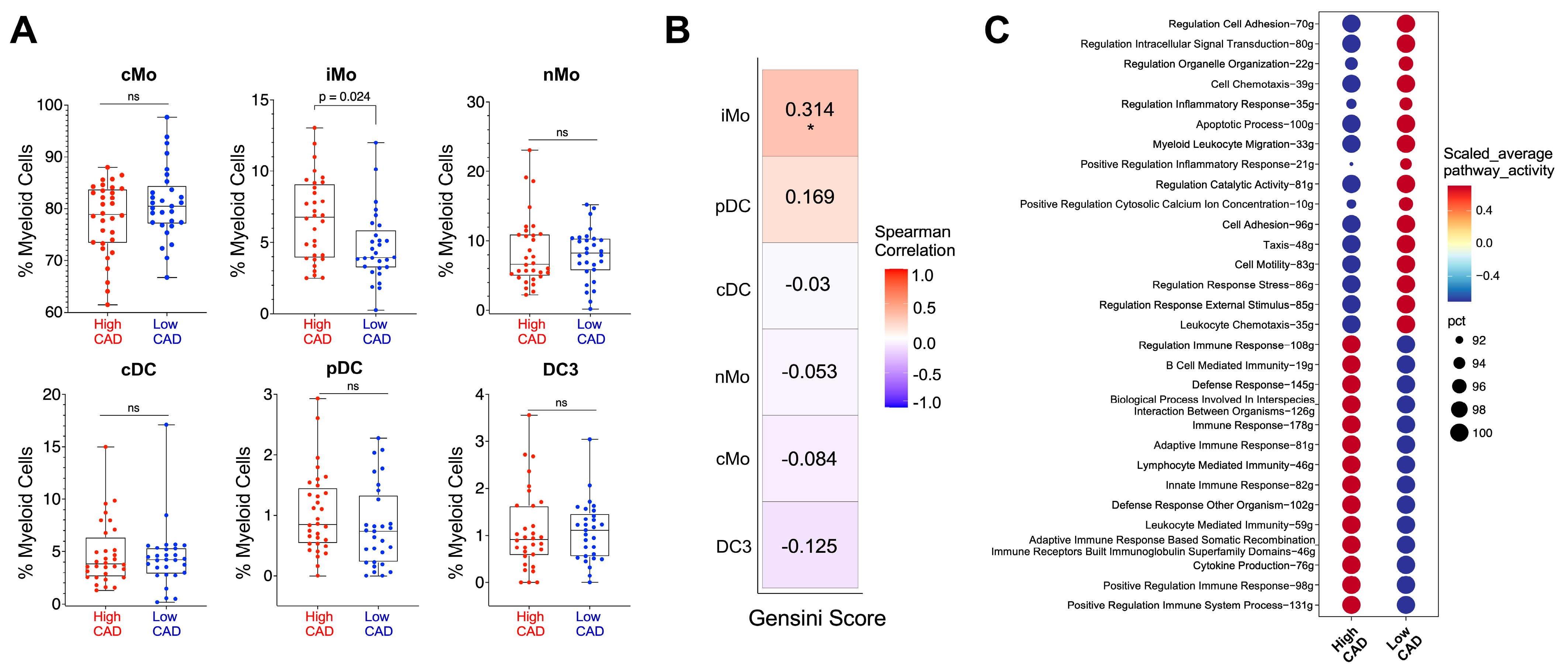
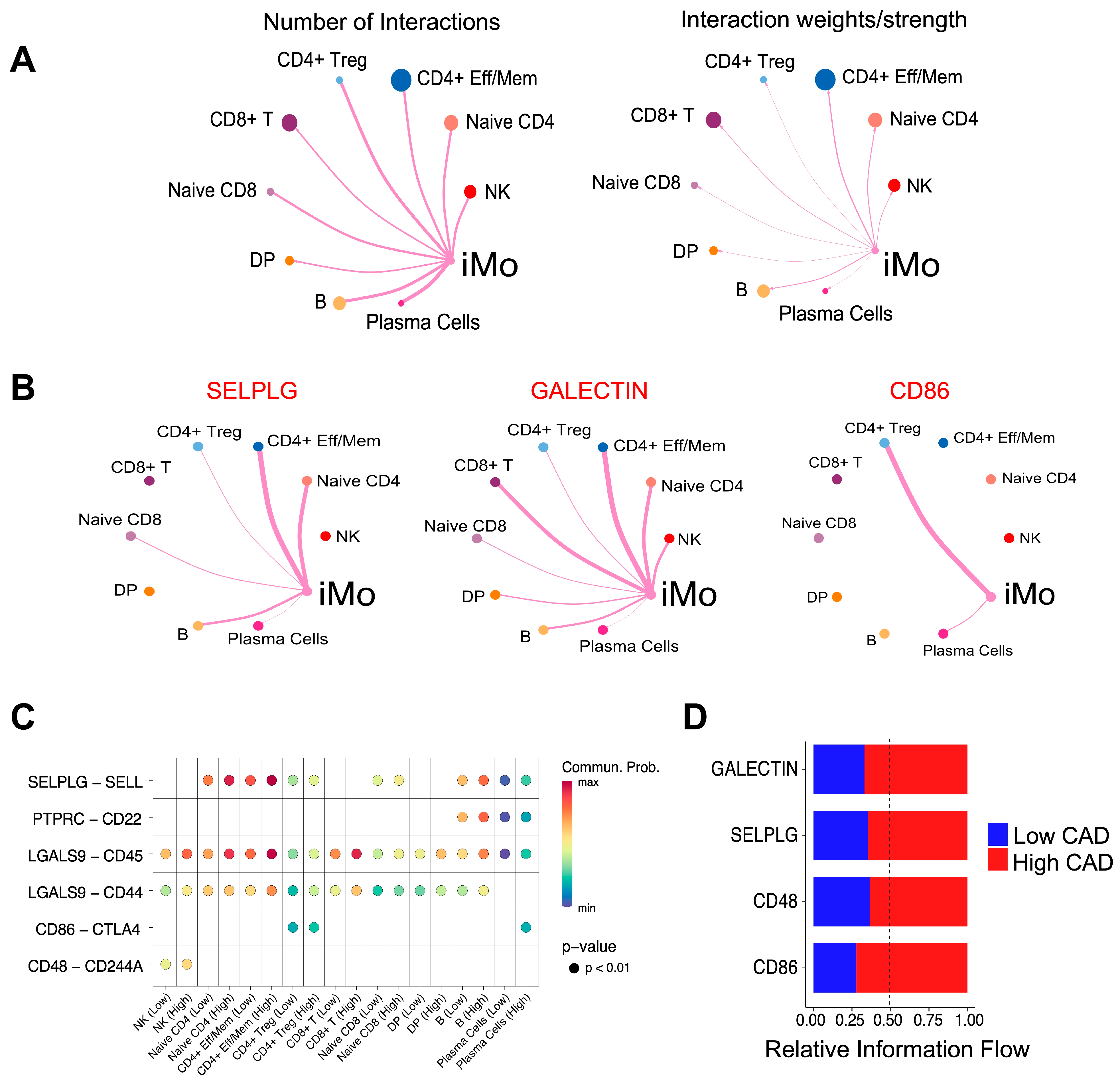
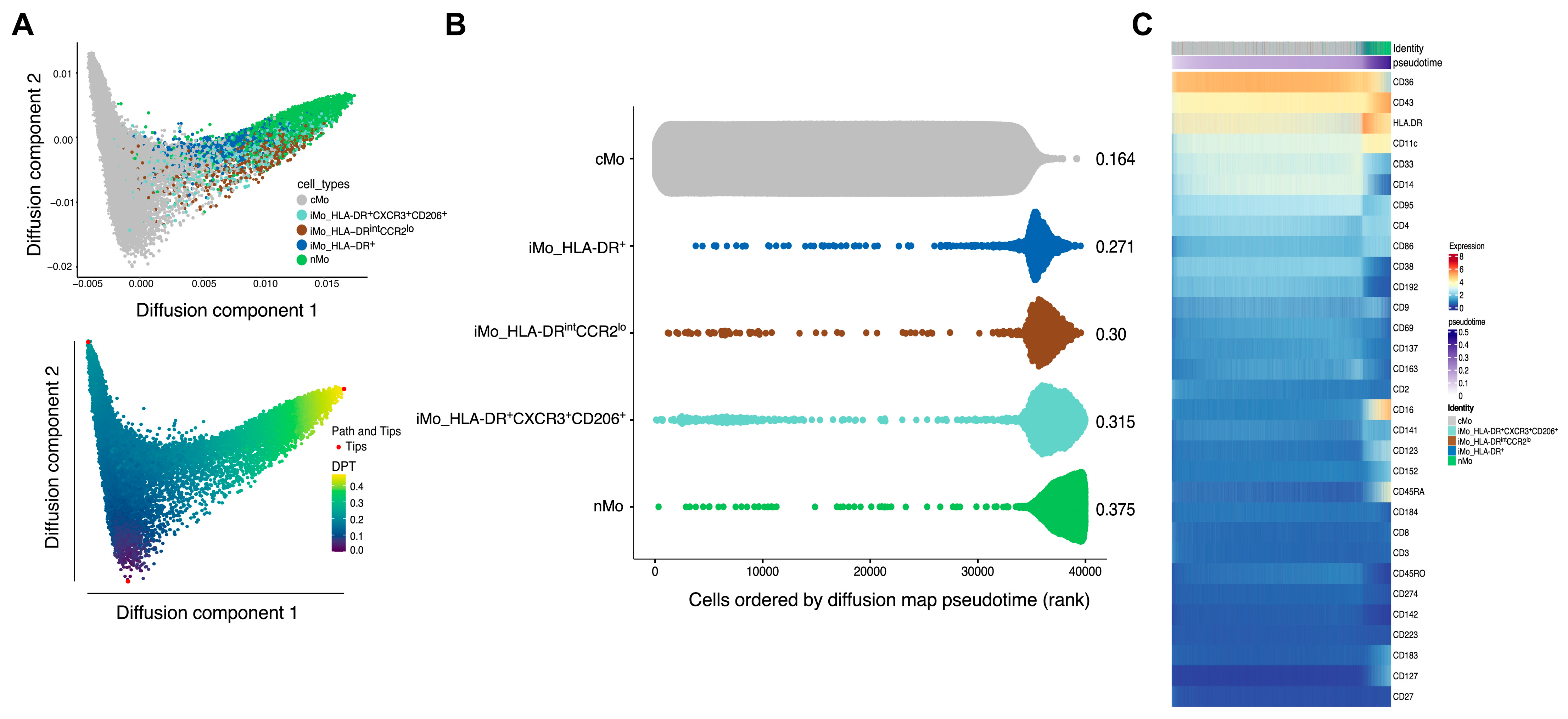
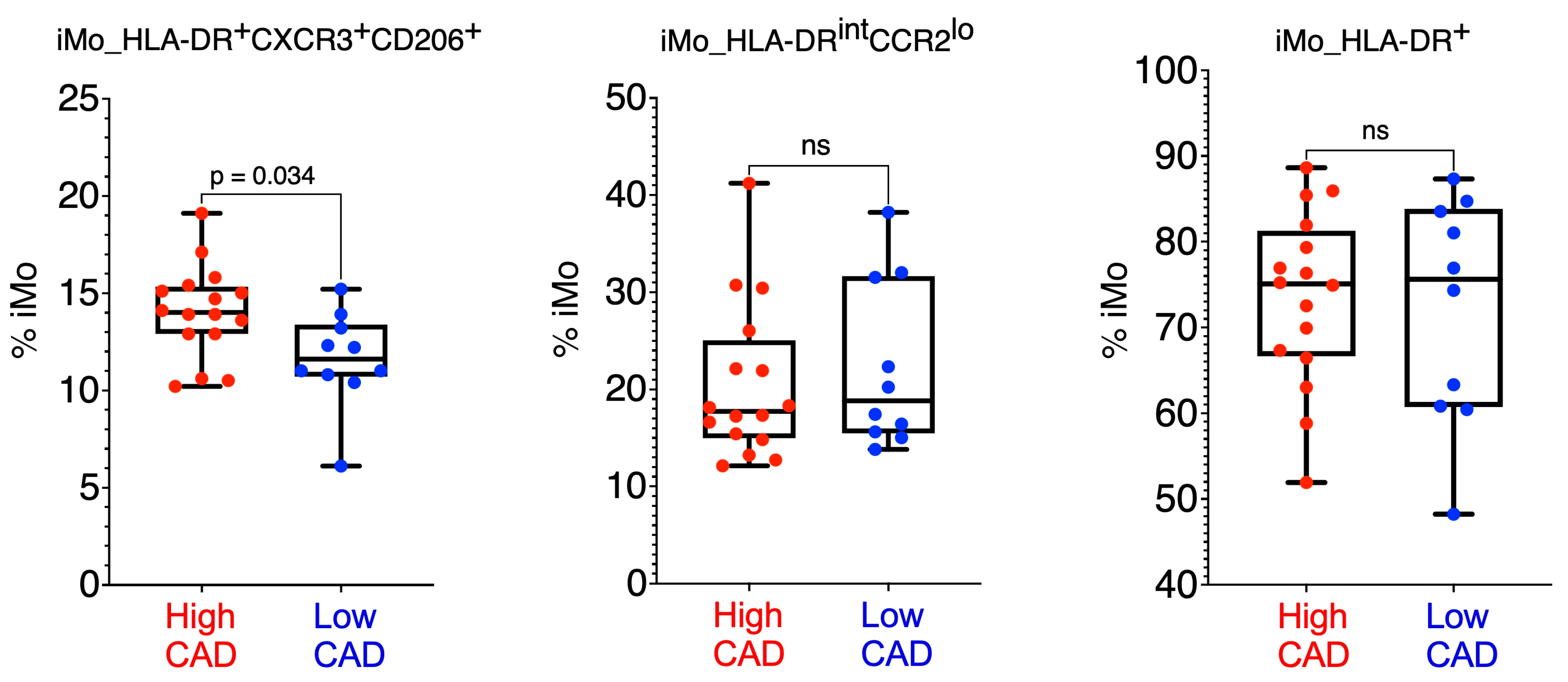
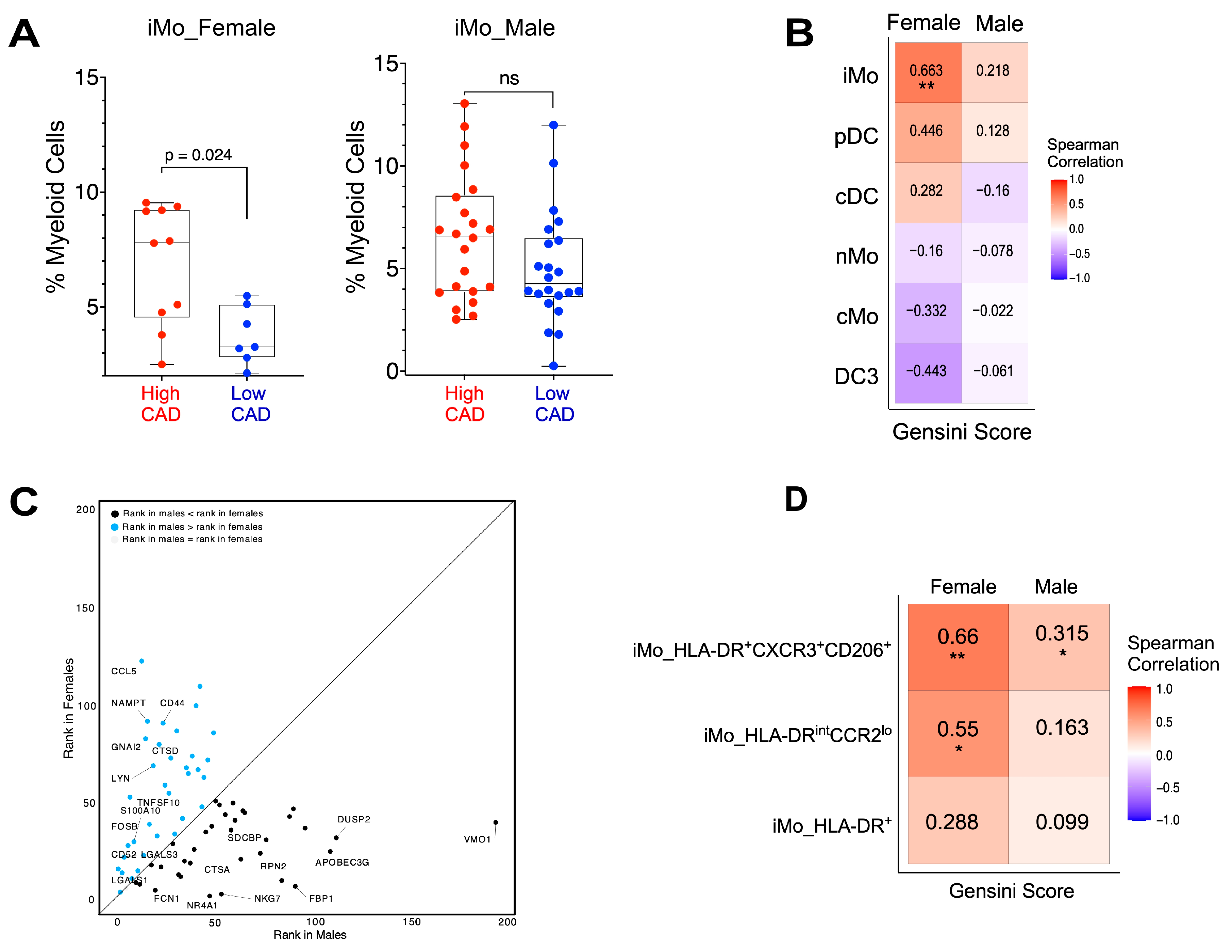
2. Results
3. Discussion
4. Methods
4.1. Human Subjects
4.2. Quantitative Coronary Angiography (QCA)
4.3. PBMC Sample Preparation for Antibody-Seq
4.4. Library Preparation
4.5. Sampling
4.6. Seurat Workflow for Targeted Data
4.7. Differentially Expressed Gene (DEG) Analysis
4.8. Ingenuity and AUCell Pathway Analyses
4.9. CellChat Analysis
4.10. Diffusion Pseudotime
4.11. Random Forest Machine Learning Algorithm
4.12. Flow Cytometry
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ray, K.K.; Laufs, U.; Cosentino, F.; Lobo, M.D.; Landmesser, U. The Year in Cardiology: Cardiovascular Prevention. Eur. Heart J. 2020, 41, 1157–1163. [Google Scholar] [CrossRef]
- Regitz-Zagrosek, V.; Gebhard, C. Gender Medicine: Effects of Sex and Gender on Cardiovascular Disease Manifestation and Outcomes. Nat. Rev. Cardiol. 2023, 20, 236–247. [Google Scholar] [CrossRef]
- Moroni, F.; Ammirati, E.; Norata, G.D.; Magnoni, M.; Camici, P.G. The Role of Monocytes and Macrophages in Human Atherosclerosis, Plaque Neoangiogenesis, and Atherothrombosis. Mediat. Inflamm. 2019, e7434376. [Google Scholar] [CrossRef]
- SahBandar, I.N.; Ndhlovu, L.C.; Saiki, K.; Kohorn, L.B.; Peterson, M.M.; D’Antoni, M.L.; Shiramizu, B.; Shikuma, C.M.; Chow, D.C. Relationship between Circulating Inflammatory Monocytes and Cardiovascular Disease Measures of Carotid Intimal Thickness. J. Atheroscler. Thromb. 2020, 27, 441–448. [Google Scholar] [CrossRef]
- Ozaki, Y.; Imanishi, T.; Taruya, A.; Aoki, H.; Masuno, T.; Shiono, Y.; Komukai, K.; Tanimoto, T.; Kitabata, H.; Akasaka, T. Circulating CD14+CD16+ Monocyte Subsets as Biomarkers of the Severity of Coronary Artery Disease in Patients with Stable Angina Pectoris. Circ. J. 2012, 76, 2412–2418. [Google Scholar] [CrossRef] [PubMed]
- Wildgruber, M.; Aschenbrenner, T.; Wendorff, H.; Czubba, M.; Glinzer, A.; Haller, B.; Schiemann, M.; Zimmermann, A.; Berger, H.; Eckstein, H.-H.; et al. The “Intermediate” CD14++CD16+ Monocyte Subset Increases in Severe Peripheral Artery Disease in Humans. Sci. Rep. 2016, 6, 39483. [Google Scholar] [CrossRef] [PubMed]
- Wildgruber, M.; Czubba, M.; Aschenbrenner, T.; Wendorff, H.; Hapfelmeier, A.; Glinzer, A.; Schiemann, M.; Zimmermann, A.; Eckstein, H.-H.; Berger, H.; et al. Increased Intermediate CD14++CD16++ Monocyte Subset Levels Associate with Restenosis after Peripheral Percutaneous Transluminal Angioplasty. Atherosclerosis 2016, 253, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Rogacev, K.S.; Cremers, B.; Zawada, A.M.; Seiler, S.; Binder, N.; Ege, P.; Große-Dunker, G.; Heisel, I.; Hornof, F.; Jeken, J.; et al. CD14++CD16+ Monocytes Independently Predict Cardiovascular Events. J. Am. Coll. Cardiol. 2012, 60, 1512–1520. [Google Scholar] [CrossRef] [PubMed]
- Hamers, A.A.J.; Vos, M.; Rassam, F.; Marinković, G.; Kurakula, K.; van Gorp, P.J.; de Winther, M.P.J.; Gijbels, M.J.J.; de Waard, V.; de Vries, C.J.M. Bone Marrow-Specific Deficiency of Nuclear Receptor Nur77 Enhances Atherosclerosis. Circ. Res. 2012, 110, 428–438. [Google Scholar] [CrossRef]
- Cole, J.E.; Park, I.; Ahern, D.J.; Kassiteridi, C.; Danso Abeam, D.; Goddard, M.E.; Green, P.; Maffia, P.; Monaco, C. Immune Cell Census in Murine Atherosclerosis: Cytometry by Time of Flight Illuminates Vascular Myeloid Cell Diversity. Cardiovasc. Res. 2018, 114, 1360–1371. [Google Scholar] [CrossRef] [PubMed]
- Zernecke, A.; Erhard, F.; Weinberger, T.; Schulz, C.; Ley, K.; Saliba, A.-E.; Cochain, C. Integrated Single-Cell Analysis-Based Classification of Vascular Mononuclear Phagocytes in Mouse and Human Atherosclerosis. Cardiovasc. Res. 2023, 119, 1676–1689. [Google Scholar] [CrossRef]
- Saigusa, R.; Vallejo, J.; Gulati, R.; Suthahar, S.S.A.; Suryawanshi, V.; Alimadadi, A.; Makings, J.; Durant, C.P.; Freuchet, A.; Roy, P.; et al. Sex Differences in Coronary Artery Disease and Diabetes Revealed by scRNA-Seq and CITE-Seq of Human CD4+ T Cells. IJMS 2022, 23, 9875. [Google Scholar] [CrossRef]
- Pattarabanjird, T.; Wilson, J.M.; Erickson, L.D.; Workman, L.J.; Qiao, H.; Ghosheh, Y.; Gulati, R.; Durant, C.; Vallejo, J.; Saigusa, R.; et al. Chemokine Receptor Activation Enhances Memory B Cell Class Switching Linked to IgE Sensitization to Alpha Gal and Cardiovascular Disease. Front. Cardiovasc. Med. 2022, 8, 791028. [Google Scholar] [CrossRef]
- Dib, L.; Koneva, L.A.; Edsfeldt, A.; Zurke, Y.-X.; Sun, J.; Nitulescu, M.; Attar, M.; Lutgens, E.; Schmidt, S.; Lindholm, M.W.; et al. Lipid-Associated Macrophages Transition to an Inflammatory State in Human Atherosclerosis, Increasing the Risk of Cerebrovascular Complications. Nat. Cardiovasc. Res. 2023, 2, 656–672. [Google Scholar] [CrossRef]
- Smit, V.; de Mol, J.; Schaftenaar, F.H.; Depuydt, M.A.C.; Postel, R.J.; Smeets, D.; Verheijen, F.W.M.; Bogers, L.; van Duijn, J.; Verwilligen, R.A.F.; et al. Single-Cell Profiling Reveals Age-Associated Immunity in Atherosclerosis. Cardiovasc. Res. 2023, 119, 2508–2521. [Google Scholar] [CrossRef]
- Depuydt, M.A.C.; Prange, K.H.M.; Slenders, L.; Örd, T.; Elbersen, D.; Boltjes, A.; De Jager, S.C.A.; Asselbergs, F.W.; De Borst, G.J.; Aavik, E.; et al. Microanatomy of the Human Atherosclerotic Plaque by Single-Cell Transcriptomics. Circ. Res. 2020, 127, 1437–1455. [Google Scholar] [CrossRef]
- Hamers, A.A.J.; Dinh, H.Q.; Thomas, G.D.; Marcovecchio, P.; Blatchley, A.; Nakao, C.S.; Kim, C.; McSkimming, C.; Taylor, A.M.; Nguyen, A.T.; et al. Human Monocyte Heterogeneity as Revealed by High-Dimensional Mass Cytometry. ATVB 2019, 39, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, C.-A.; Becht, E.; Irac, S.E.; Khalilnezhad, A.; Narang, V.; Khalilnezhad, S.; Ng, P.Y.; van den Hoogen, L.L.; Leong, J.Y.; Lee, B.; et al. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity 2019, 51, 573–589.e8. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.N.; Carlin, L.M.; Hubbeling, H.G.; Nackiewicz, D.; Green, A.M.; Punt, J.A.; Geissmann, F.; Hedrick, C.C. The Transcription Factor NR4A1 (Nur77) Controls Bone Marrow Differentiation and the Survival of Ly6C-Monocytes. Nat. Immunol. 2011, 12, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.E.; Ljungcrantz, I.; Andersson, L.; Bryngelsson, C.; Hedblad, B.; Fredrikson, G.N.; Nilsson, J.; Björkbacka, H. Elevated CD14++ CD16− Monocytes Predict Cardiovascular Events. Circ. Cardiovasc. Genet. 2012, 5, 122–131. [Google Scholar] [CrossRef]
- Vallejo, J.; Saigusa, R.; Gulati, R.; Armstrong Suthahar, S.S.; Suryawanshi, V.; Alimadadi, A.; Durant, C.P.; Ghosheh, Y.; Roy, P.; Ehinger, E.; et al. Combined Protein and Transcript Single-Cell RNA Sequencing in Human Peripheral Blood Mononuclear Cells. BMC Biol. 2022, 20, 193. [Google Scholar] [CrossRef]
- Guilliams, M.; Mildner, A.; Yona, S. Developmental and Functional Heterogeneity of Monocytes. Immunity 2018, 49, 595–613. [Google Scholar] [CrossRef] [PubMed]
- Yona, S.; Kim, K.-W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate Mapping Reveals Origins and Dynamics of Monocytes and Tissue Macrophages under Homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef]
- Olingy, C.E.; Dinh, H.Q.; Hedrick, C.C. Monocyte Heterogeneity and Functions in Cancer. J. Leukoc. Biol. 2019, 106, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Ruder, A.V.; Wetzels, S.M.W.; Temmerman, L.; Biessen, E.A.L.; Goossens, P. Monocyte Heterogeneity in Cardiovascular Disease. Cardiovasc. Res. 2023, 119, 2033–2045. [Google Scholar] [CrossRef] [PubMed]
- Cappellari, R.; D’Anna, M.; Bonora, B.M.; Rigato, M.; Cignarella, A.; Avogaro, A.; Fadini, G.P. Shift of Monocyte Subsets along Their Continuum Predicts Cardiovascular Outcomes. Atherosclerosis 2017, 266, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Nahrendorf, M.; Swirski, F.K. Monocyte Heterogeneity in Cardiovascular Disease. Semin. Immunopathol. 2013, 35, 553–562. [Google Scholar] [CrossRef]
- Swirski, F.K.; Pittet, M.J.; Kircher, M.F.; Aikawa, E.; Jaffer, F.A.; Libby, P.; Weissleder, R. Monocyte Accumulation in Mouse Atherogenesis Is Progressive and Proportional to Extent of Disease. Proc. Natl. Acad. Sci. USA 2006, 103, 10340–10345. [Google Scholar] [CrossRef]
- Lo, S.-C.; Lee, W.-J.; Chen, C.-Y.; Lee, B.-C. Intermediate CD14++CD16+ Monocyte Predicts Severe Coronary Stenosis and Extensive Plaque Involvement in Asymptomatic Individuals. Int. J. Cardiovasc. Imaging 2017, 33, 1223–1236. [Google Scholar] [CrossRef]
- Thomas, G.D.; Hamers, A.A.J.; Nakao, C.; Marcovecchio, P.; Taylor, A.M.; McSkimming, C.; Nguyen, A.T.; McNamara, C.A.; Hedrick, C.C. Human Blood Monocyte Subsets: A New Gating Strategy Defined Using Cell Surface Markers Identified by Mass Cytometry. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1548–1558. [Google Scholar] [CrossRef] [PubMed]
- Abdul-Salam, V.B.; Ramrakha, P.; Krishnan, U.; Owen, D.R.; Shalhoub, J.; Davies, A.H.; Tang, T.Y.; Gillard, J.H.; Boyle, J.J.; Wilkins, M.R.; et al. Identification and Assessment of Plasma Lysozyme as a Putative Biomarker of Atherosclerosis. ATVB 2010, 30, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Woollard, K.J.; Geissmann, F. Monocytes in Atherosclerosis: Subsets and Functions. Nat. Rev. Cardiol. 2010, 7, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Blanda, V.; Bracale, U.M.; Di Taranto, M.D.; Fortunato, G. Galectin-3 in Cardiovascular Diseases. IJMS 2020, 21, 9232. [Google Scholar] [CrossRef] [PubMed]
- Fulton, D.J.R.; Li, X.; Bordan, Z.; Wang, Y.; Mahboubi, K.; Rudic, R.D.; Haigh, S.; Chen, F.; Barman, S.A. Galectin-3: A Harbinger of Reactive Oxygen Species, Fibrosis, and Inflammation in Pulmonary Arterial Hypertension. Antioxid. Redox Signal. 2019, 31, 1053–1069. [Google Scholar] [CrossRef] [PubMed]
- Gucuk Ipek, E.; Akin Suljevic, S.; Kafes, H.; Basyigit, F.; Karalok, N.; Guray, Y.; Dinc Asarcikli, L.; Acar, B.; Demirel, H. Evaluation of Galectin-3 Levels in Acute Coronary Syndrome. Ann. Cardiol. D’angéiologie 2016, 65, 26–30. [Google Scholar] [CrossRef]
- Chung, A.W.; Sieling, P.A.; Schenk, M.; Teles, R.M.B.; Krutzik, S.R.; Hsu, D.K.; Liu, F.-T.; Sarno, E.N.; Rea, T.H.; Stenger, S.; et al. Galectin-3 Regulates the Innate Immune Response of Human Monocytes. J. Infect. Dis. 2013, 207, 947–956. [Google Scholar] [CrossRef]
- Jin, Q.; Li, R.; Hu, N.; Xin, T.; Zhu, P.; Hu, S.; Ma, S.; Zhu, H.; Ren, J.; Zhou, H. DUSP1 Alleviates Cardiac Ischemia/Reperfusion Injury by Suppressing the Mff-Required Mitochondrial Fission and Bnip3-Related Mitophagy via the JNK Pathways. Redox Biol. 2018, 14, 576–587. [Google Scholar] [CrossRef]
- Xiao, X.; Yang, C.; Qu, S.-L.; Shao, Y.-D.; Zhou, C.-Y.; Chao, R.; Huang, L.; Zhang, C. S100 Proteins in Atherosclerosis. Clin. Chim. Acta 2020, 502, 293–304. [Google Scholar] [CrossRef]
- Cao, Z.; Sun, X.; Icli, B.; Wara, A.K.; Feinberg, M.W. Role of Krüppel-like Factors in Leukocyte Development, Function, and Disease. Blood 2010, 116, 4404–4414. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.D.; Hanna, R.N.; Vasudevan, N.T.; Hamers, A.A.; Romanoski, C.E.; McArdle, S.; Ross, K.D.; Blatchley, A.; Yoakum, D.; Hamilton, B.A.; et al. Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6Clow Monocytes While Preserving Macrophage Gene Function. Immunity 2016, 45, 975–987. [Google Scholar] [CrossRef]
- Altara, R.; Manca, M.; Brandão, R.D.; Zeidan, A.; Booz, G.W.; Zouein, F.A. Emerging Importance of Chemokine Receptor CXCR3 and Its Ligands in Cardiovascular Diseases. Clin. Sci. 2016, 130, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Segers, D.; Lipton, J.A.; Leenen, P.J.M.; Cheng, C.; Tempel, D.; Pasterkamp, G.; Moll, F.L.; de Crom, R.; Krams, R. Atherosclerotic Plaque Stability Is Affected by the Chemokine CXCL10 in Both Mice and Humans. Int. J. Inflam. 2011, 2011, 936109. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Den Borne, P.; Quax, P.H.A.; Hoefer, I.E.; Pasterkamp, G. The Multifaceted Functions of CXCL10 in Cardiovascular Disease. BioMed Res. Int. 2014, 2014, 173–181. [Google Scholar] [CrossRef]
- Tavakolian Ferdousie, V.; Mohammadi, M.; Hassanshahi, G.; Khorramdelazad, H.; Khanamani Falahati-Pour, S.; Mirzaei, M.; Allah Tavakoli, M.; Kamiab, Z.; Ahmadi, Z.; Vazirinejad, R.; et al. Serum CXCL10 and CXCL12 Chemokine Levels Are Associated with the Severity of Coronary Artery Disease and Coronary Artery Occlusion. Int. J. Cardiol. 2017, 233, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, L.; Liu, H.; Xie, Y. Innate Immune Memory in Monocytes and Macrophages: The Potential Therapeutic Strategies for Atherosclerosis. Cells 2022, 11, 4072. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Yan, S.; Zhang, K.; Sai, W.; Zhang, Q.; Ti, Y.; Wang, Z.; Zhang, W.; Zheng, C.; Zhong, M. Serum Complement C1q Level Is Associated with Acute Coronary Syndrome. Mol. Immunol. 2020, 120, 130–135. [Google Scholar] [CrossRef]
- Kishore, U.; Ghebrehiwet, B. Editorial: C1q: A Molecular Bridge to Innate and Adaptive Immunity. Front. Immunol. 2020, 11, 417. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, C.S.; Murrow, L.M.; Gartner, Z.J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019, 8, 329–337.e4. [Google Scholar] [CrossRef] [PubMed]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.; Raychaudhuri, S. Fast, Sensitive and Accurate Integration of Single-Cell Data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing Well-Connected Communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal Analysis Approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.-H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and Analysis of Cell-Cell Communication Using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Angerer, P.; Haghverdi, L.; Büttner, M.; Theis, F.J.; Marr, C.; Buettner, F. Destiny: Diffusion Maps for Large-Scale Single-Cell Data in R. Bioinformatics 2016, 32, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T. Gam: Generalized Additive Models 2023. Available online: https://cran.r-project.org/web/packages/gam/gam.pdf (accessed on 15 January 2024).
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Caret: Classification and Regression Training, ascl:1505.003; Astrophysics Source Code Library: Houghton, MI, USA, 2015. Available online: https://ui.adsabs.harvard.edu/abs/2015ascl.soft05003K/abstra (accessed on 24 February 2024).
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using Lme4. J. Stat. Softw. 2015, 67, 48. [Google Scholar] [CrossRef]
- Harrell, F.E., Jr. Package ‘Hmisc’. 2022. Available online: https://hbiostat.org/R/Hmisc/ (accessed on 15 January 2024).



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, N.; Komaravolu, R.K.; Durant, C.P.; Wu, R.; McSkimming, C.; Drago, F.; Kumar, S.; Valentin-Guillama, G.; Miller, Y.I.; McNamara, C.A.; et al. Single Cell High Dimensional Analysis of Human Peripheral Blood Mononuclear Cells Reveals Unique Intermediate Monocyte Subsets Associated with Sex Differences in Coronary Artery Disease. Int. J. Mol. Sci. 2024, 25, 2894. https://doi.org/10.3390/ijms25052894
Chatterjee N, Komaravolu RK, Durant CP, Wu R, McSkimming C, Drago F, Kumar S, Valentin-Guillama G, Miller YI, McNamara CA, et al. Single Cell High Dimensional Analysis of Human Peripheral Blood Mononuclear Cells Reveals Unique Intermediate Monocyte Subsets Associated with Sex Differences in Coronary Artery Disease. International Journal of Molecular Sciences. 2024; 25(5):2894. https://doi.org/10.3390/ijms25052894
Chicago/Turabian StyleChatterjee, Nandini, Ravi K. Komaravolu, Christopher P. Durant, Runpei Wu, Chantel McSkimming, Fabrizio Drago, Sunil Kumar, Gabriel Valentin-Guillama, Yury I. Miller, Coleen A. McNamara, and et al. 2024. "Single Cell High Dimensional Analysis of Human Peripheral Blood Mononuclear Cells Reveals Unique Intermediate Monocyte Subsets Associated with Sex Differences in Coronary Artery Disease" International Journal of Molecular Sciences 25, no. 5: 2894. https://doi.org/10.3390/ijms25052894
APA StyleChatterjee, N., Komaravolu, R. K., Durant, C. P., Wu, R., McSkimming, C., Drago, F., Kumar, S., Valentin-Guillama, G., Miller, Y. I., McNamara, C. A., Ley, K., Taylor, A., Alimadadi, A., & Hedrick, C. C. (2024). Single Cell High Dimensional Analysis of Human Peripheral Blood Mononuclear Cells Reveals Unique Intermediate Monocyte Subsets Associated with Sex Differences in Coronary Artery Disease. International Journal of Molecular Sciences, 25(5), 2894. https://doi.org/10.3390/ijms25052894




