Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential
Abstract
1. Introduction
1.1. General Introduction to Depression
1.2. Current Therapeutic Approaches
1.3. General Introduction to miRNAs
2. Role of miRNAs in Neurobiology
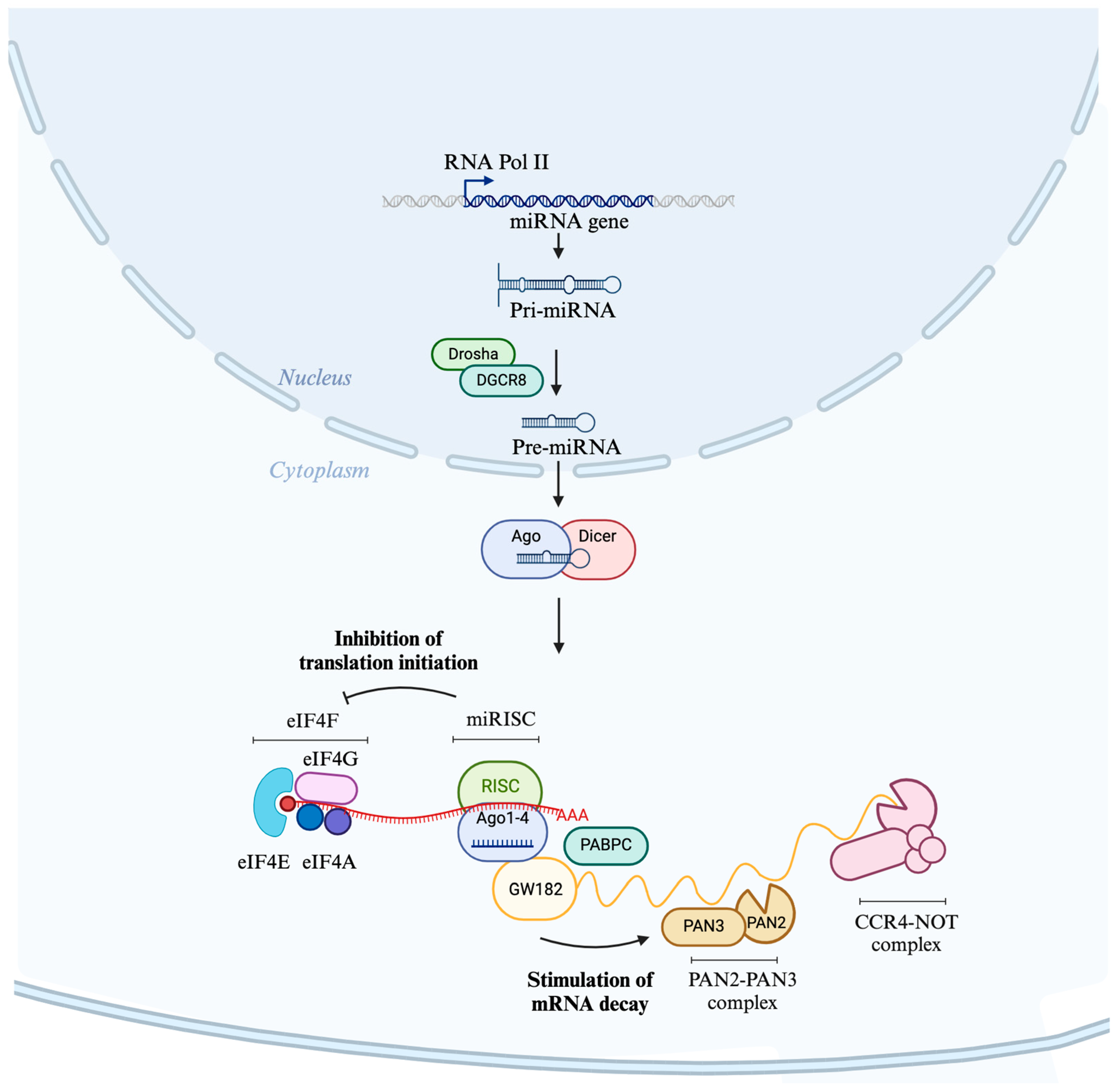
2.1. Biogenesis of miRNAs
2.2. Mode of Action of miRNAs
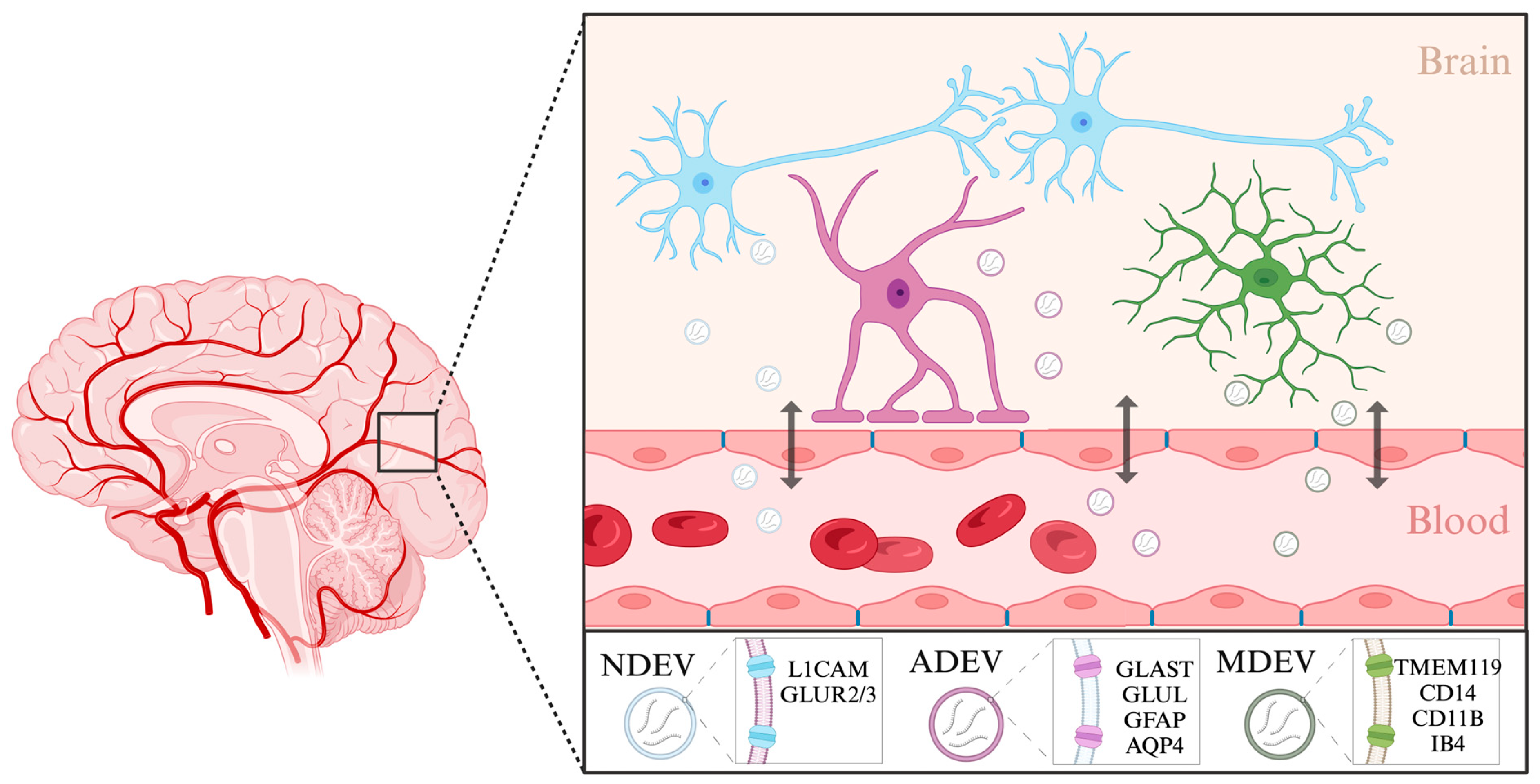
2.3. Circulation of miRNAs
2.4. Role of miRNAs in Neural Function: From Synaptic Architecture to Plasticity
2.4.1. Synaptic Localization and Functional Roles of miRNAs in Neuronal Cells
2.4.2. Regulatory Influence of miRNAs on Synaptic Transmission and Neural Plasticity
3. MicroRNAs and Depression
3.1. miRNAs in the Pathophysiology of Depression
3.2. miRNAs as Diagnostic Biomarkers
3.2.1. Blood-Derived miRNAs: Refining the Complex Landscape of Depression Biomarkers
3.2.2. Fluid-Based miRNA Landscapes: Serum, Plasma, and Cerebrospinal Fluid
3.2.3. Extracellular Vesicles: Emerging Protagonists in the Biomarker Landscape of Depression
4. Therapeutic Potentials of miRNAs in Depression
4.1. Current Therapeutic Strategies
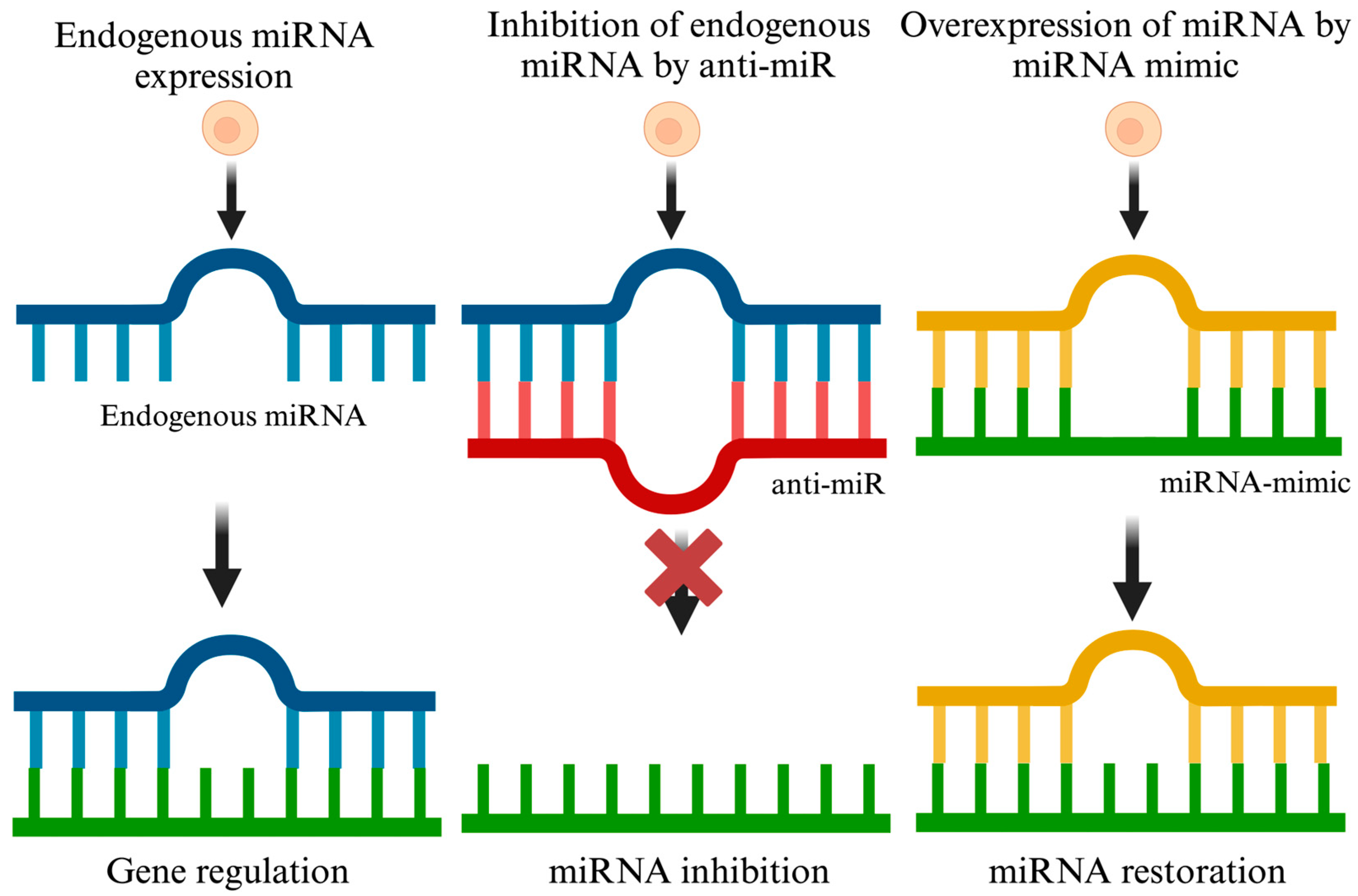
4.1.1. miRNA Inhibition Therapy
4.1.2. miRNA Restoration Therapy
4.2. Future Directions
4.2.1. Emerging Targets in the miRNA Landscape
4.2.2. Drug Delivery—Crossing the Biobarriers
4.2.3. Augmenting Existing Strategies
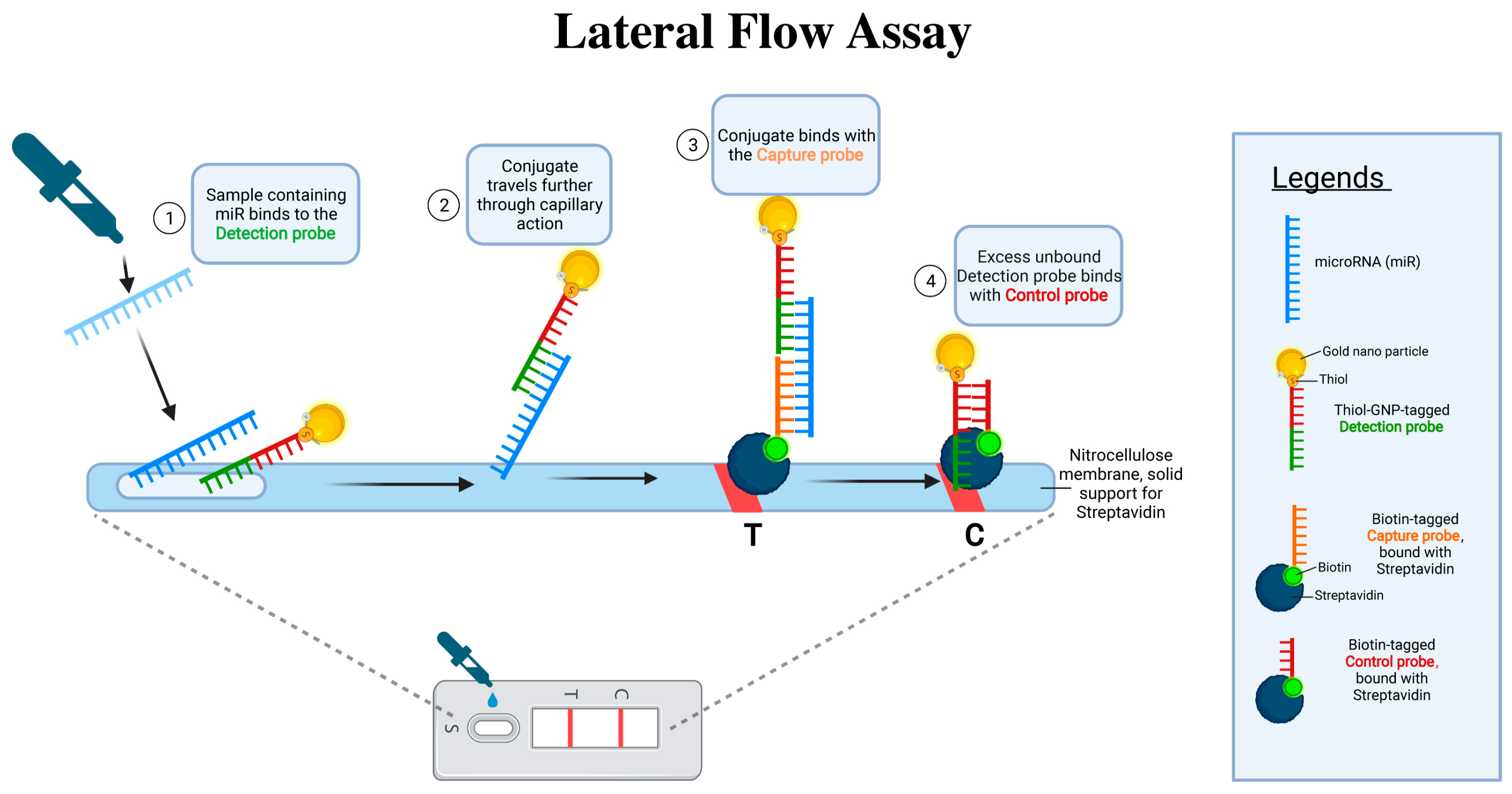
4.2.4. Lateral Flow Assay for miRNA Detection
5. Challenges and Limitations
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Institute of Health Metrics and Evaluation. Global Health Data Exchange (GHDx); Institute of Health Metrics and Evaluation: Seattle, WA, USA, 2021. [Google Scholar]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Kessler, R.C.; Bromet, E.J. The Epidemiology of Depression across Cultures. Annu. Rev. Public Health 2013, 34, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Bromet, E.; Andrade, L.H.; Hwang, I.; Sampson, N.A.; Alonso, J.; de Girolamo, G.; de Graaf, R.; Demyttenaere, K.; Hu, C.; Iwata, N.; et al. Cross-National Epidemiology of DSM-IV Major Depressive Episode. BMC Med. 2011, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Nihalani, N.; Simionescu, M.; Dunlop, B.W. Depression: Phenomenology, Epidemiology, and Pathophysiology. In Depression; CRC Press: Boca Raton, FL, USA, 2010; ISBN 978-0-429-24993-8. [Google Scholar]
- Kuehner, C. Why Is Depression More Common among Women than among Men? Lancet Psychiatry 2017, 4, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Van de Velde, S.; Bracke, P.; Levecque, K. Gender Differences in Depression in 23 European Countries. Cross-National Variation in the Gender Gap in Depression. Soc. Sci. Med. 2010, 71, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Heim, C.; Binder, E.B. Current Research Trends in Early Life Stress and Depression: Review of Human Studies on Sensitive Periods, Gene–Environment Interactions, and Epigenetics. Exp. Neurol. 2012, 233, 102–111. [Google Scholar] [CrossRef]
- Belmaker, R.H.; Agam, G. Major Depressive Disorder. N. Engl. J. Med. 2008, 358, 55–68. [Google Scholar] [CrossRef]
- Hasin, D.S.; Sarvet, A.L.; Meyers, J.L.; Saha, T.D.; Ruan, W.J.; Stohl, M.; Grant, B.F. Epidemiology of Adult DSM-5 Major Depressive Disorder and Its Specifiers in the United States. JAMA Psychiatry 2018, 75, 336–346. [Google Scholar] [CrossRef]
- Cuijpers, P.; Noma, H.; Karyotaki, E.; Vinkers, C.H.; Cipriani, A.; Furukawa, T.A. A Network Meta-Analysis of the Effects of Psychotherapies, Pharmacotherapies and Their Combination in the Treatment of Adult Depression. World Psychiatry 2020, 19, 92–107. [Google Scholar] [CrossRef]
- Cuijpers, P.; Stringaris, A.; Wolpert, M. Treatment Outcomes for Depression: Challenges and Opportunities. Lancet Psychiatry 2020, 7, 925–927. [Google Scholar] [CrossRef] [PubMed]
- Gurtan, A.M.; Sharp, P.A. The Role of miRNAs in Regulating Gene Expression Networks. J. Mol. Biol. 2013, 425, 3582. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian mRNAs Are Conserved Targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread Changes in Protein Synthesis Induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An Integrated Expression Atlas of miRNAs and Their Promoters in Human and Mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Smalheiser, N.R.; Lugli, G.; Zhang, H.; Rizavi, H.; Cook, E.H.; Dwivedi, Y. Expression of microRNAs and Other Small RNAs in Prefrontal Cortex in Schizophrenia, Bipolar Disorder and Depressed Subjects. PLoS ONE 2014, 9, e86469. [Google Scholar] [CrossRef]
- Wang, W.; Kwon, E.J.; Tsai, L.-H. MicroRNAs in Learning, Memory, and Neurological Diseases. Learn. Mem. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Issler, O.; Chen, A. Determining the Role of microRNAs in Psychiatric Disorders. Nat. Rev. Neurosci. 2015, 16, 201–212. [Google Scholar] [CrossRef]
- Dwivedi, Y. MicroRNAs in Depression and Suicide: Recent Insights and Future Perspectives. J. Affect. Disord. 2018, 240, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, Y.; Tian, C.; Wang, J.; Li, W.; Zhong, C. Differential Exosomal microRNA Profile in the Serum of a Patient with Depression. Eur. J. Psychiatry 2018, 32, 105–112. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The Widespread Regulation of microRNA Biogenesis, Function and Decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental Evaluation of Novel and Previously Annotated Genes. Genes. Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The Nuclear RNase III Drosha Initiates microRNA Processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting Effective microRNA Target Sites in Mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a Molecular Understanding of microRNA-Mediated Gene Silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs Predominantly Act to Decrease Target mRNA Levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Iwakawa, H.-O.; Tomari, Y. MicroRNAs Block Assembly of eIF4F Translation Initiation Complex in Drosophila. Mol. Cell 2014, 56, 67–78. [Google Scholar] [CrossRef]
- Fukao, A.; Mishima, Y.; Takizawa, N.; Oka, S.; Imataka, H.; Pelletier, J.; Sonenberg, N.; Thoma, C.; Fujiwara, T. MicroRNAs Trigger Dissociation of eIF4AI and eIF4AII from Target mRNAs in Humans. Mol. Cell 2014, 56, 79–89. [Google Scholar] [CrossRef]
- Liu, J.; Rivas, F.V.; Wohlschlegel, J.; Yates, J.R.; Parker, R.; Hannon, G.J. A Role for the P-Body Component GW182 in microRNA Function. Nat. Cell Biol. 2005, 7, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 Proteins Directly Recruit Cytoplasmic Deadenylase Complexes to miRNA Targets. Mol. Cell 2011, 44, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; EL Andaloussi, S.; Wood, M.J.A. Exosomes and Microvesicles: Extracellular Vesicles for Genetic Information Transfer and Gene Therapy. Hum. Mol. Genet. 2012, 21, R125–R134. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-Mediated Transfer of mRNAs and microRNAs Is a Novel Mechanism of Genetic Exchange between Cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- van Niel, G.; Carter, D.R.F.; Clayton, A.; Lambert, D.W.; Raposo, G.; Vader, P. Challenges and Directions in Studying Cell–Cell Communication by Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2022, 23, 369–382. [Google Scholar] [CrossRef]
- Anand, S.; Foot, N.; Ang, C.-S.; Gembus, K.M.; Keerthikumar, S.; Adda, C.G.; Mathivanan, S.; Kumar, S. Arrestin-Domain Containing Protein 1 (Arrdc1) Regulates the Protein Cargo and Release of Extracellular Vesicles. Proteomics 2018, 18, e1800266. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-Syntenin-ALIX Regulates the Biogenesis of Exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT Functions in Exosome Biogenesis, Composition and Secretion Highlights the Heterogeneity of Extracellular Vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Saint-Pol, J.; Gosselet, F.; Duban-Deweer, S.; Pottiez, G.; Karamanos, Y. Targeting and Crossing the Blood-Brain Barrier with Extracellular Vesicles. Cells 2020, 9, 851. [Google Scholar] [CrossRef]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef]
- Cao, L.; Jiao, X.; Zuzga, D.S.; Liu, Y.; Fong, D.M.; Young, D.; During, M.J. VEGF Links Hippocampal Activity with Neurogenesis, Learning and Memory. Nat. Genet. 2004, 36, 827–835. [Google Scholar] [CrossRef]
- Esteller, M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Lugli, G.; Torvik, V.I.; Larson, J.; Smalheiser, N.R. Expression of microRNAs and Their Precursors in Synaptic Fractions of Adult Mouse Forebrain. J. Neurochem. 2008, 106, 650–661. [Google Scholar] [CrossRef]
- Hu, Z.; Li, Z. miRNAs in Synapse Development and Synaptic Plasticity. Curr. Opin. Neurobiol. 2017, 45, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Bicker, S.; Khudayberdiev, S.; Weiß, K.; Zocher, K.; Baumeister, S.; Schratt, G. The DEAH-Box Helicase DHX36 Mediates Dendritic Localization of the Neuronal Precursor-microRNA-134. Genes. Dev. 2013, 27, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kim, P.; Phay, M.; Yoo, S. Identification of Precursor microRNAs within Distal Axons of Sensory Neuron. J. Neurochem. 2015, 134, 193–199. [Google Scholar] [CrossRef]
- Lugli, G.; Larson, J.; Martone, M.E.; Jones, Y.; Smalheiser, N.R. Dicer and eIF2c Are Enriched at Postsynaptic Densities in Adult Mouse Brain and Are Modified by Neuronal Activity in a Calpain-Dependent Manner. J. Neurochem. 2005, 94, 896–905. [Google Scholar] [CrossRef]
- Kumar, S.; Orlov, E.; Gowda, P.; Bose, C.; Swerdlow, R.H.; Lahiri, D.K.; Reddy, P.H. Synaptosome microRNAs Regulate Synapse Functions in Alzheimer’s Disease. NPJ Genom. Med. 2022, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.H.; Seyedmoalemi, S.; Moghanlou, M.; Akhlagh, S.A.; Talaei Zavareh, S.A.; Hamblin, M.R.; Jafari, A.; Mirzaei, H. MicroRNAs and Synaptic Plasticity: From Their Molecular Roles to Response to Therapy. Mol. Neurobiol. 2022, 59, 5084–5102. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Yu, D.; Gu, Q.; Yang, Y.; Tu, K.; Zhu, J.; Li, Z. miR-191 and miR-135 Are Required for Long-Lasting Spine Remodelling Associated with Synaptic Long-Term Depression. Nat. Commun. 2014, 5, 3263. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhao, J.; Hu, T.; Luo, Y.; Zhu, J.; Li, Z. miR-501-3p Mediates the Activity-Dependent Regulation of the Expression of AMPA Receptor Subunit GluA1. J. Cell Biol. 2015, 208, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Aksoy-Aksel, A.; Zampa, F.; Schratt, G. MicroRNAs and Synaptic Plasticity—A Mutual Relationship. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130515. [Google Scholar] [CrossRef]
- Natera-Naranjo, O.; Aschrafi, A.; Gioio, A.E.; Kaplan, B.B. Identification and Quantitative Analyses of microRNAs Located in the Distal Axons of Sympathetic Neurons. RNA 2010, 16, 1516–1529. [Google Scholar] [CrossRef] [PubMed]
- Epple, R.; Krüger, D.; Berulava, T.; Brehm, G.; Ninov, M.; Islam, R.; Köster, S.; Fischer, A. The Coding and Small Non-Coding Hippocampal Synaptic RNAome. Mol. Neurobiol. 2021, 58, 2940–2953. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Xu, H.; Su, X.; He, X. Role of MicroRNA in Governing Synaptic Plasticity. Neural Plast. 2016, 2016, 4959523. [Google Scholar] [CrossRef]
- Zhou, L.; Zhu, Y.; Chen, W.; Tang, Y. Emerging Role of microRNAs in Major Depressive Disorder and Its Implication on Diagnosis and Therapeutic Response. J. Affect. Disord. 2021, 286, 80–86. [Google Scholar] [CrossRef]
- Banerjee, S.; Neveu, P.; Kosik, K.S. A Coordinated Local Translational Control Point at the Synapse Involving Relief from Silencing and MOV10 Degradation. Neuron 2009, 64, 871–884. [Google Scholar] [CrossRef]
- Sosanya, N.M.; Huang, P.P.C.; Cacheaux, L.P.; Chen, C.J.; Nguyen, K.; Perrone-Bizzozero, N.I.; Raab-Graham, K.F. Degradation of High Affinity HuD Targets Releases Kv1.1 mRNA from miR-129 Repression by mTORC1. J. Cell Biol. 2013, 202, 53–69. [Google Scholar] [CrossRef]
- Gao, Y.-N.; Zhang, Y.-Q.; Wang, H.; Deng, Y.-L.; Li, N.-M. A New Player in Depression: MiRNAs as Modulators of Altered Synaptic Plasticity. Int. J. Mol. Sci. 2022, 23, 4555. [Google Scholar] [CrossRef] [PubMed]
- McNeill, E.; Van Vactor, D. MicroRNAs Shape the Neuronal Landscape. Neuron 2012, 75, 363–379. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.-F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of Synaptic Structure and Function by FMRP-Associated MicroRNAs miR-125b and miR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Harraz, M.M.; Eacker, S.M.; Wang, X.; Dawson, T.M.; Dawson, V.L. MicroRNA-223 Is Neuroprotective by Targeting Glutamate Receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 18962–18967. [Google Scholar] [CrossRef] [PubMed]
- Olde Loohuis, N.F.M.; Ba, W.; Stoerchel, P.H.; Kos, A.; Jager, A.; Schratt, G.; Martens, G.J.M.; van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA-137 Controls AMPA-Receptor-Mediated Transmission and mGluR-Dependent LTD. Cell Rep. 2015, 11, 1876–1884. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-L.; Shen, C.-K.J. Modulation of mGluR-Dependent MAP1B Translation and AMPA Receptor Endocytosis by microRNA miR-146a-5p. J. Neurosci. 2013, 33, 9013–9020. [Google Scholar] [CrossRef]
- Lee, K.; Kim, J.-H.; Kwon, O.-B.; An, K.; Ryu, J.; Cho, K.; Suh, Y.-H.; Kim, H.-S. An Activity-Regulated microRNA, miR-188, Controls Dendritic Plasticity and Synaptic Transmission by Downregulating Neuropilin-2. J. Neurosci. 2012, 32, 5678–5687. [Google Scholar] [CrossRef]
- Verma, P.; Augustine, G.J.; Ammar, M.-R.; Tashiro, A.; Cohen, S.M. A Neuroprotective Role for microRNA miR-1000 Mediated by Limiting Glutamate Excitotoxicity. Nat. Neurosci. 2015, 18, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.-H.; Yu, D.; Hu, Z.; Liu, X.; Yang, Y.; Luo, Y.; Zhu, J.; Li, Z. miR-26a and miR-384-5p Are Required for LTP Maintenance and Spine Enlargement. Nat. Commun. 2015, 6, 6789. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shu, X.; Liu, D.; Shang, Y.; Wu, Y.; Pei, L.; Xu, X.; Tian, Q.; Zhang, J.; Qian, K.; et al. EPAC Null Mutation Impairs Learning and Social Interactions via Aberrant Regulation of miR-124 and Zif268 Translation. Neuron 2012, 73, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Kohen, R.; Dobra, A.; Tracy, J.H.; Haugen, E. Transcriptome Profiling of Human Hippocampus Dentate Gyrus Granule Cells in Mental Illness. Transl. Psychiatry 2014, 4, e366. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-J.; Xu, M.; Gao, Z.-H.; Wang, Y.-Q.; Yue, Z.; Zhang, Y.-X.; Li, X.-X.; Zhang, C.; Xie, S.-Y.; Wang, P.-Y. Alterations of Serum Levels of BDNF-Related miRNAs in Patients with Depression. PLoS ONE 2013, 8, e63648. [Google Scholar] [CrossRef] [PubMed]
- Yuta, Y.; Roy, B.; Dwivedi, Y. Altered miRNA Landscape of the Anterior Cingulate Cortex Is Associated with Potential Loss of Key Neuronal Functions in Depressed Brain. Eur. Neuropsychopharmacol. J. Eur. Coll. Neuropsychopharmacol. 2020, 40, 70. [Google Scholar] [CrossRef]
- Lopez, J.P.; Lim, R.; Cruceanu, C.; Crapper, L.; Fasano, C.; Labonte, B.; Maussion, G.; Yang, J.P.; Yerko, V.; Vigneault, E.; et al. miR-1202 Is a Primate-Specific and Brain-Enriched microRNA Involved in Major Depression and Antidepressant Treatment. Nat. Med. 2014, 20, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Issler, O.; Haramati, S.; Paul, E.D.; Maeno, H.; Navon, I.; Zwang, R.; Gil, S.; Mayberg, H.S.; Dunlop, B.W.; Menke, A.; et al. MicroRNA 135 Is Essential for Chronic Stress Resiliency, Antidepressant Efficacy, and Intact Serotonergic Activity. Neuron 2014, 83, 344–360. [Google Scholar] [CrossRef]
- Belzeaux, R.; Bergon, A.; Jeanjean, V.; Loriod, B.; Formisano-Tréziny, C.; Verrier, L.; Loundou, A.; Baumstarck-Barrau, K.; Boyer, L.; Gall, V.; et al. Responder and Nonresponder Patients Exhibit Different Peripheral Transcriptional Signatures during Major Depressive Episode. Transl. Psychiatry 2012, 2, e185. [Google Scholar] [CrossRef]
- Kaurani, L.; Besse, M.; Methfessel, I.; Methi, A.; Zhou, J.; Pradhan, R.; Burkhardt, S.; Kranaster, L.; Sartorius, A.; Habel, U.; et al. Baseline Levels of miR-223-3p Correlate with the Effectiveness of Electroconvulsive Therapy in Patients with Major Depression. Transl. Psychiatry 2023, 13, 294. [Google Scholar] [CrossRef]
- Burrows, K.; Figueroa-Hall, L.; Stewart, J.; Alarbi, A.; Kuplicki, R.; Hannafon, B.; Tan, C.; Risbrough, V.; McKinney, B.; Ramesh, R.; et al. Exploring the Role of Neuronal-Enriched Extracellular Vesicle miR-93 and Interoception in Major Depressive Disorder. Res. Sq. 2023; preprint. [Google Scholar] [CrossRef]
- Saeedi, S.; Nagy, C.; Ibrahim, P.; Théroux, J.-F.; Wakid, M.; Fiori, L.M.; Yang, J.; Rotzinger, S.; Foster, J.A.; Mechawar, N.; et al. Neuron-Derived Extracellular Vesicles Enriched from Plasma Show Altered Size and miRNA Cargo as a Function of Antidepressant Drug Response. Mol. Psychiatry 2021, 26, 7417–7424. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, S.; Yan, J.; Wang, D.; Yin, R.; Zhao, L.; Zhu, Y.; Zhu, X. miR-182 (microRNA-182) Suppression in the Hippocampus Evokes Antidepressant-like Effects in Rats. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 65, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.P.; Fiori, L.M.; Cruceanu, C.; Lin, R.; Labonte, B.; Cates, H.M.; Heller, E.A.; Vialou, V.; Ku, S.M.; Gerald, C.; et al. MicroRNAs 146a/b-5 and 425-3p and 24-3p Are Markers of Antidepressant Response and Regulate MAPK/Wnt-System Genes. Nat. Commun. 2017, 8, 15497. [Google Scholar] [CrossRef] [PubMed]
- Żurawek, D.; Turecki, G. The miRNome of Depression. Int. J. Mol. Sci. 2021, 22, 11312. [Google Scholar] [CrossRef]
- He, C.; Bai, Y.; Wang, Z.; Fan, D.; Wang, Q.; Liu, X.; Zhang, H.; Zhang, H.; Zhang, Z.; Yao, H.; et al. Identification of microRNA-9 Linking the Effects of Childhood Maltreatment on Depression Using Amygdala Connectivity. Neuroimage 2021, 224, 117428. [Google Scholar] [CrossRef]
- Bocchio-Chiavetto, L.; Maffioletti, E.; Bettinsoli, P.; Giovannini, C.; Bignotti, S.; Tardito, D.; Corrada, D.; Milanesi, L.; Gennarelli, M. Blood microRNA Changes in Depressed Patients during Antidepressant Treatment. Eur. Neuropsychopharmacol. 2013, 23, 602–611. [Google Scholar] [CrossRef]
- Fang, Y.; Qiu, Q.; Zhang, S.; Sun, L.; Li, G.; Xiao, S.; Li, X. Changes in miRNA-132 and miR-124 Levels in Non-Treated and Citalopram-Treated Patients with Depression. J. Affect. Disord. 2018, 227, 745–751. [Google Scholar] [CrossRef]
- Li, J.; Meng, H.; Cao, W.; Qiu, T. MiR-335 Is Involved in Major Depression Disorder and Antidepressant Treatment through Targeting GRM4. Neurosci. Lett. 2015, 606, 167–172. [Google Scholar] [CrossRef]
- Fiori, L.M.; Lopez, J.P.; Richard-Devantoy, S.; Berlim, M.; Chachamovich, E.; Jollant, F.; Foster, J.; Rotzinger, S.; Kennedy, S.H.; Turecki, G. Investigation of miR-1202, miR-135a, and miR-16 in Major Depressive Disorder and Antidepressant Response. Int. J. Neuropsychopharmacol. 2017, 20, 619–623. [Google Scholar] [CrossRef]
- Funatsuki, T.; Ogata, H.; Tahara, H.; Shimamoto, A.; Takekita, Y.; Koshikawa, Y.; Nonen, S.; Higasa, K.; Kinoshita, T.; Kato, M. Changes in Multiple microRNA Levels with Antidepressant Treatment Are Associated with Remission and Interact with Key Pathways: A Comprehensive microRNA Analysis. Int. J. Mol. Sci. 2023, 24, 12199. [Google Scholar] [CrossRef] [PubMed]
- Belzeaux, R.; Fiori, L.M.; Lopez, J.P.; Boucekine, M.; Boyer, L.; Blier, P.; Farzan, F.; Frey, B.N.; Giacobbe, P.; Lam, R.W.; et al. Predicting Worsening Suicidal Ideation With Clinical Features and Peripheral Expression of Messenger RNA and MicroRNA During Antidepressant Treatment. J. Clin. Psychiatry 2019, 80, 18m12556. [Google Scholar] [CrossRef] [PubMed]
- Yrondi, A.; Fiori, L.M.; Frey, B.N.; Lam, R.W.; MacQueen, G.M.; Milev, R.; Müller, D.J.; Foster, J.A.; Kennedy, S.H.; Turecki, G. Association Between Side Effects and Blood microRNA Expression Levels and Their Targeted Pathways in Patients With Major Depressive Disorder Treated by a Selective Serotonin Reuptake Inhibitor, Escitalopram: A CAN-BIND-1 Report. Int. J. Neuropsychopharmacol. 2020, 23, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sundquist, K.; Hedelius, A.; Palmér, K.; Memon, A.A.; Sundquist, J. Circulating microRNA-144-5p Is Associated with Depressive Disorders. Clin. Epigenet. 2015, 7, 69. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.-H.; Dong, Z.-Q.; Tian, L.-T.; Li, J. MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as Predictors of Response to Antidepressant Treatment. Braz. J. Med. Biol. Res. 2018, 51, e7212. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.-B.; Sheng, X.-M.; Jin, X.; Guan, W. MiR-182-5p: A Novel Biomarker in the Treatment of Depression in CSDS-Induced Mice. Int. J. Neuropsychopharmacol. 2023, 27, pyad064. [Google Scholar] [CrossRef]
- Li, Y.; Wang, N.; Pan, J.; Wang, X.; Zhao, Y.; Guo, Z. Hippocampal miRNA-144 Modulates Depressive-Like Behaviors in Rats by Targeting PTP1B. Neuropsychiatr. Dis. Treat. 2021, 17, 389–399. [Google Scholar] [CrossRef]
- Gheysarzadeh, A.; Sadeghifard, N.; Afraidooni, L.; Pooyan, F.; Mofid, M.R.; Valadbeigi, H.; Bakhtiari, H.; Keikhavani, S. Serum-Based microRNA Biomarkers for Major Depression: MiR-16, miR-135a, and miR-1202. J. Res. Med. Sci. 2018, 23, 69. [Google Scholar] [CrossRef]
- Roy, B.; Ochi, S.; Dwivedi, Y. Potential of Circulating miRNAs as Molecular Markers in Mood Disorders and Associated Suicidal Behavior. Int. J. Mol. Sci. 2023, 24, 4664. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-Coding RNAs in Depression: Promising Diagnostic and Therapeutic Biomarkers. eBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, Y.; Wang, X.; Wu, J.; Liu, K.; Zhou, J.; Liu, L.; Zhang, C. Identification of Differential microRNAs in Cerebrospinal Fluid and Serum of Patients with Major Depressive Disorder. PLoS ONE 2015, 10, e0121975. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The Emerging Role of Exosomes in Mental Disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Buzas, E.I.; György, B.; Nagy, G.; Falus, A.; Gay, S. Emerging Role of Extracellular Vesicles in Inflammatory Diseases. Nat. Rev. Rheumatol. 2014, 10, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.T.; Iqbal, A.J.; Norling, L.V. The Role and Impact of Extracellular Vesicles in the Modulation and Delivery of Cytokines during Autoimmunity. Int. J. Mol. Sci. 2020, 21, 7096. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; McAndrews, K.M. The Role of Extracellular Vesicles in Cancer. Cell 2023, 186, 1610–1626. [Google Scholar] [CrossRef]
- Raghav, A.; Singh, M.; Jeong, G.-B.; Giri, R.; Agarwal, S.; Kala, S.; Gautam, K.A. Extracellular Vesicles in Neurodegenerative Diseases: A Systematic Review. Front. Mol. Neurosci. 2022, 15, 1061076. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, D.; Huang, S.; Lai, J.; Lu, L.; Zhang, J.; Hu, S. Extracellular Vesicles in Mental Disorders: A State-of-Art Review. Int. J. Biol. Sci. 2023, 19, 1094–1109. [Google Scholar] [CrossRef]
- Wei, Z.-X.; Xie, G.-J.; Mao, X.; Zou, X.-P.; Liao, Y.-J.; Liu, Q.-S.; Wang, H.; Cheng, Y. Exosomes from Patients with Major Depression Cause Depressive-like Behaviors in Mice with Involvement of miR-139-5p-Regulated Neurogenesis. Neuropsychopharmacology 2020, 45, 1050–1058. [Google Scholar] [CrossRef]
- Levine, A.; Strawn, J.R. Blood Tests of Brain Function: Neuronal Extracellular Vesicles. Biomark. Neuropsychiatry 2022, 7, 100058. [Google Scholar] [CrossRef]
- Mizohata, Y.; Toda, H.; Koga, M.; Saito, T.; Fujita, M.; Kobayashi, T.; Hatakeyama, S.; Morimoto, Y. Neural Extracellular Vesicle-Derived miR-17 in Blood as a Potential Biomarker of Subthreshold Depression. Hum. Cell 2021, 34, 1087–1092. [Google Scholar] [CrossRef]
- Sun, B.; Dalvi, P.; Abadjian, L.; Tang, N.; Pulliam, L. Blood Neuron-Derived Exosomes as Biomarkers of Cognitive Impairment in HIV. AIDS 2017, 31, F9–F17. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of Exosomes from Differentiated Neurons and Its Regulation by Synaptic Glutamatergic Activity. Mol. Cell Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Datta Chaudhuri, A.; Dasgheyb, R.M.; DeVine, L.R.; Bi, H.; Cole, R.N.; Haughey, N.J. Stimulus-Dependent Modifications in Astrocyte-Derived Extracellular Vesicle Cargo Regulate Neuronal Excitability. Glia 2020, 68, 128–144. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Mustapic, M.; Kapogiannis, D.; Eitan, E.; Lobach, I.V.; Goetzl, L.; Schwartz, J.B.; Miller, B.L. Cargo Proteins of Plasma Astrocyte-Derived Exosomes in Alzheimer’s Disease. FASEB J. 2016, 30, 3853–3859. [Google Scholar] [CrossRef] [PubMed]
- Wallensten, J.; Nager, A.; Åsberg, M.; Borg, K.; Beser, A.; Wilczek, A.; Mobarrez, F. Leakage of Astrocyte-Derived Extracellular Vesicles in Stress-Induced Exhaustion Disorder: A Cross-Sectional Study. Sci. Rep. 2021, 11, 2009. [Google Scholar] [CrossRef]
- Han, J.; Cho, H.-J.; Park, D.; Han, S. DICAM in the Extracellular Vesicles from Astrocytes Attenuates Microglia Activation and Neuroinflammation. Cells 2022, 11, 2977. [Google Scholar] [CrossRef]
- Long, X.; Yao, X.; Jiang, Q.; Yang, Y.; He, X.; Tian, W.; Zhao, K.; Zhang, H. Astrocyte-Derived Exosomes Enriched with miR-873a-5p Inhibit Neuroinflammation via Microglia Phenotype Modulation after Traumatic Brain Injury. J. Neuroinflamm. 2020, 17, 89. [Google Scholar] [CrossRef] [PubMed]
- Luarte, A.; Nardocci, G.; Chakraborty, A.; Batiz, L.F.; Pino-Lagos, K.; Wyneken, Ú. Astrocyte-Derived Extracellular Vesicles in Stress-Associated Mood Disorders. Does the Immune System Get Astrocytic? Pharmacol. Res. 2023, 194, 106833. [Google Scholar] [CrossRef]
- Gabrielli, M.; Raffaele, S.; Fumagalli, M.; Verderio, C. The Multiple Faces of Extracellular Vesicles Released by Microglia: Where Are We 10 Years After? Front. Cell. Neurosci. 2022, 16, 984690. [Google Scholar] [CrossRef]
- Roseborough, A.D.; Myers, S.J.; Khazaee, R.; Zhu, Y.; Zhao, L.; Iorio, E.; Elahi, F.M.; Pasternak, S.H.; Whitehead, S.N. Plasma Derived Extracellular Vesicle Biomarkers of Microglia Activation in an Experimental Stroke Model. J. Neuroinflamm. 2023, 20, 20. [Google Scholar] [CrossRef]
- Cohn, W.; Melnik, M.; Huang, C.; Teter, B.; Chandra, S.; Zhu, C.; McIntire, L.B.; John, V.; Gylys, K.H.; Bilousova, T. Multi-Omics Analysis of Microglial Extracellular Vesicles From Human Alzheimer’s Disease Brain Tissue Reveals Disease-Associated Signatures. Front. Pharmacol. 2021, 12, 766082. [Google Scholar] [CrossRef] [PubMed]
- Scaroni, F.; Visconte, C.; Serpente, M.; Golia, M.T.; Gabrielli, M.; Huiskamp, M.; Hulst, H.E.; Carandini, T.; De Riz, M.; Pietroboni, A.; et al. miR-150-5p and Let-7b-5p in Blood Myeloid Extracellular Vesicles Track Cognitive Symptoms in Patients with Multiple Sclerosis. Cells 2022, 11, 1551. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Wang, L.; Lan, T.; Gao, R.; Wang, W.; Yu, S.Y. MicroRNA-26a-3p Rescues Depression-like Behaviors in Male Rats via Preventing Hippocampal Neuronal Anomalies. J. Clin. Investig. 2021, 131, e148853. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Alvarez-Mon, M.A.; García-Montero, C.; Fraile-Martinez, O.; Lahera, G.; Monserrat, J.; Muñoz-Merida, L.; Mora, F.; Rodríguez-Jiménez, R.; Fernandez-Rojo, S.; et al. MicroRNAs as Critical Biomarkers of Major Depressive Disorder: A Comprehensive Perspective. Biomedicines 2021, 9, 1659. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.A.; Moeng, S.; Sim, S.; Kuh, H.J.; Choi, S.Y.; Park, J.K. MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies. Cells 2019, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-Y.; Xia, Q.-H.; Xia, Q.-R.; Zhang, X.-L.; Liang, J. MicroRNA-Based Biomarkers in the Diagnosis and Monitoring of Therapeutic Response in Patients with Depression. Neuropsychiatr. Dis. Treat. 2019, 15, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging Concepts of miRNA Therapeutics: From Cells to Clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
- Fu, Y.; Chen, J.; Huang, Z. Recent Progress in microRNA-Based Delivery Systems for the Treatment of Human Disease. ExRNA 2019, 1, 24. [Google Scholar] [CrossRef]
- Xia, S.; Xu, C.; Liu, F.; Chen, G. Development of microRNA-Based Therapeutics for Central Nervous System Diseases. Eur. J. Pharmacol. 2023, 956, 175956. [Google Scholar] [CrossRef]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA Oligonucleotides: A Comprehensive Guide for Design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.; O’Brien, D.; Henshall, D.C. Opportunities and Challenges for microRNA-Targeting Therapeutics for Epilepsy. Trends Pharmacol. Sci. 2021, 42, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Elmén, J.; Lindow, M.; Schütz, S.; Lawrence, M.; Petri, A.; Obad, S.; Lindholm, M.; Hedtjärn, M.; Hansen, H.F.; Berger, U.; et al. LNA-Mediated microRNA Silencing in Non-Human Primates. Nature 2008, 452, 896–899. [Google Scholar] [CrossRef]
- Tassone, P.; Di Martino, M.T.; Arbitrio, M.; Fiorillo, L.; Staropoli, N.; Ciliberto, D.; Cordua, A.; Scionti, F.; Bertucci, B.; Salvino, A.; et al. Safety and Activity of the First-in-Class Locked Nucleic Acid (LNA) miR-221 Selective Inhibitor in Refractory Advanced Cancer Patients: A First-in-Human, Phase 1, Open-Label, Dose-Escalation Study. J. Hematol. Oncol. 2023, 16, 68. [Google Scholar] [CrossRef]
- Vester, B.; Wengel, J. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. [Google Scholar] [CrossRef]
- Islam, M.R.; Kaurani, L.; Berulava, T.; Heilbronner, U.; Budde, M.; Centeno, T.P.; Elerdashvili, V.; Zafieriou, M.-P.; Benito, E.; Sertel, S.M.; et al. A microRNA Signature That Correlates with Cognition and Is a Target against Cognitive Decline. EMBO Mol. Med. 2021, 13, e13659. [Google Scholar] [CrossRef]
- Ebert, M.S.; Sharp, P.A. MicroRNA Sponges: Progress and Possibilities. RNA 2010, 16, 2043–2050. [Google Scholar] [CrossRef]
- Nguyen, D.-D.; Chang, S. Development of Novel Therapeutic Agents by Inhibition of Oncogenic MicroRNAs. Int. J. Mol. Sci. 2017, 19, 65. [Google Scholar] [CrossRef]
- Alberi, L.; Hoey, S.E.; Brai, E.; Scotti, A.L.; Marathe, S. Notch Signaling in the Brain: In Good and Bad Times. Ageing Res. Rev. 2013, 12, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Zhang, X.; Li, Y.; Ma, Z.; Si, S.; Gao, X. miR-9 Inhibition of Neuronal Apoptosis and Expression Levels of Apoptosis Genes Bcl-2 and Bax in Depression Model Rats through Notch Pathway. Exp. Ther. Med. 2020, 19, 551–556. [Google Scholar] [CrossRef]
- You, J.; Sun, L.; Wang, J.; Sun, F.; Wang, W.; Wang, D.; Fan, X.; Liu, D.; Xu, Z.; Qiu, C.; et al. Role of Adiponectin-Notch Pathway in Cognitive Dysfunction Associated with Depression and in the Therapeutic Effect of Physical Exercise. Aging Cell 2021, 20, e13387. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhu, Z.; Wang, G.; Cui, S.; Shen, M.; Song, Z.; Wang, J.-H. microRNA-15b Contributes to Depression-like Behavior in Mice by Affecting Synaptic Protein Levels and Function in the Nucleus Accumbens. J. Biol. Chem. 2020, 295, 6831–6848. [Google Scholar] [CrossRef] [PubMed]
- Fogaça, M.V.; Duman, R.S. Cortical GABAergic Dysfunction in Stress and Depression: New Insights for Therapeutic Interventions. Front. Cell Neurosci. 2019, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Zurawek, D.; Gruca, P.; Antkiewicz-Michaluk, L.; Dziedzicka-Wasylewska, M. Resilient Phenotype in Chronic Mild Stress Paradigm Is Associated with Altered Expression Levels of miR-18a-5p and Serotonin 5-HT1a Receptor in Dorsal Part of the Hippocampus. Mol. Neurobiol. 2019, 56, 7680–7693. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Li, M.-D.; Nie, P.-Y.; Chen, Y.; Chen, Y.-L.; Ji, L.-L. miR-132 Downregulation Alleviates Behavioral Impairment of Rats Exposed to Single Prolonged Stress, Reduces the Level of Apoptosis in PFC, and Upregulates the Expression of MeCP2 and BDNF. Neurobiol. Stress 2021, 14, 100311. [Google Scholar] [CrossRef]
- Wanet, A.; Tacheny, A.; Arnould, T.; Renard, P. miR-212/132 Expression and Functions: Within and beyond the Neuronal Compartment. Nucleic Acids Res. 2012, 40, 4742–4753. [Google Scholar] [CrossRef] [PubMed]
- Xin, C.; Xia, J.; Liu, Y.; Zhang, Y. MicroRNA-202-3p Targets Brain-Derived Neurotrophic Factor and Is Involved in Depression-Like Behaviors. Neuropsychiatr. Dis. Treat. 2020, 16, 1073–1083. [Google Scholar] [CrossRef]
- Jiang, T.; Hu, S.; Dai, S.; Yi, Y.; Wang, T.; Li, X.; Luo, M.; Li, K.; Chen, L.; Wang, H.; et al. Programming Changes of Hippocampal miR-134-5p/SOX2 Signal Mediate the Susceptibility to Depression in Prenatal Dexamethasone-Exposed Female Offspring. Cell Biol. Toxicol. 2022, 38, 69–86. [Google Scholar] [CrossRef]
- van der Zee, Y.Y.; Eijssen, L.M.T.; Mews, P.; Ramakrishnan, A.; Alvarez, K.; Lardner, C.K.; Cates, H.M.; Walker, D.M.; Torres-Berrío, A.; Browne, C.J.; et al. Blood miR-144-3p: A Novel Diagnostic and Therapeutic Tool for Depression. Mol. Psychiatry 2022, 27, 4536–4549. [Google Scholar] [CrossRef]
- Sun, L.; Bai, D.; Lin, M.; Eerdenidalai; Zhang, L.; Wang, F.; Jin, S. miR-96 Inhibits SV2C to Promote Depression-Like Behavior and Memory Disorders in Mice. Front. Behav. Neurosci. 2020, 14, 575345. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.T.B.; Clark, I.M.; Le, L.T.T. MicroRNA-Based Diagnosis and Therapy. Int. J. Mol. Sci. 2022, 23, 7167. [Google Scholar] [CrossRef] [PubMed]
- Kota, J.; Chivukula, R.R.; O’Donnell, K.A.; Wentzel, E.A.; Montgomery, C.L.; Hwang, H.-W.; Chang, T.-C.; Vivekanandan, P.; Torbenson, M.; Clark, K.R.; et al. Therapeutic microRNA Delivery Suppresses Tumorigenesis in a Murine Liver Cancer Model. Cell 2009, 137, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.B.; Liu, J.J.; Villaescusa, J.C.; Åberg, E.; Brené, S.; Wegener, G.; Mathé, A.A.; Lavebratt, C. Elevation of Il6 Is Associated with Disturbed Let-7 Biogenesis in a Genetic Model of Depression. Transl. Psychiatry 2016, 6, e869. [Google Scholar] [CrossRef]
- Bahi, A.; Dreyer, J.-L. Lentiviral-Mediated Let-7d microRNA Overexpression Induced Anxiolytic- and Anti-Depressant-like Behaviors and Impaired Dopamine D3 Receptor Expression. Eur. Neuropsychopharmacol. 2018, 28, 1394–1404. [Google Scholar] [CrossRef]
- Huang, P.; Wei, S.; Luo, M.; Tang, Z.; Lin, Q.; Wang, X.; Luo, M.; He, Y.; Wang, C.; Wei, D.; et al. MiR-139-5p Has an Antidepressant-like Effect by Targeting Phosphodiesterase 4D to Activate the cAMP/PKA/CREB Signaling Pathway. Ann. Transl. Med. 2021, 9, 1594. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y.; Wang, P.; Li, X.; Song, Z.; Wei, C.; Zhang, Q.; Luo, B.; Liu, Z.; Yang, Y.; et al. Clinical and Preclinical Evaluation of miR-144-5p as a Key Target for Major Depressive Disorder. CNS Neurosci. Ther. 2023, 29, 3598–3611. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Yuan, P.; Wang, Y.; Hunsberger, J.G.; Elkahloun, A.; Wei, Y.; Damschroder-Williams, P.; Du, J.; Chen, G.; Manji, H.K. Evidence for Selective microRNAs and Their Effectors as Common Long-Term Targets for the Actions of Mood Stabilizers. Neuropsychopharmacology 2009, 34, 1395–1405. [Google Scholar] [CrossRef]
- Lou, D.; Wang, J.; Wang, X. miR-124 Ameliorates Depressive-like Behavior by Targeting STAT3 to Regulate Microglial Activation. Mol. Cell Probes 2019, 48, 101470. [Google Scholar] [CrossRef]
- Ge, X.; Guo, M.; Hu, T.; Li, W.; Huang, S.; Yin, Z.; Li, Y.; Chen, F.; Zhu, L.; Kang, C.; et al. Increased Microglial Exosomal miR-124-3p Alleviates Neurodegeneration and Improves Cognitive Outcome after rmTBI. Mol. Ther. 2020, 28, 503–522. [Google Scholar] [CrossRef]
- Xie, L.; Chen, J.; Ding, Y.-M.; Gui, X.-W.; Wu, L.-X.; Tian, S.; Wu, W. MicroRNA-26a-2 Maintains Stress Resiliency and Antidepressant Efficacy by Targeting the Serotonergic Autoreceptor HTR1A. Biochem. Biophys. Res. Commun. 2019, 511, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, S.; Chu, Z.; Dang, Y.; Zhu, J.; Su, X. MicroRNA-101 in the Ventrolateral Orbital Cortex (VLO) Modulates Depressive-like Behaviors in Rats and Targets Dual-Specificity Phosphatase 1 (DUSP1). Brain Res. 2017, 1669, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Chang, J. The Important Roles of microRNAs in Depression: New Research Progress and Future Prospects. J. Mol. Med. 2021, 99, 619–636. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Tang, W.; Yang, J.; Peng, J.; Guo, J.; Fan, C. MicroRNA-Related Strategies to Improve Cardiac Function in Heart Failure. Front. Cardiovasc. Med. 2021, 8, 773083. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, J.; You, J.; Shi, H.; Xue, X.; Huang, J.; Xu, L.; Jiang, G.; Yuan, L.; Gong, X.; et al. HSF1 Deficiency Accelerates the Transition from Pressure Overload-Induced Cardiac Hypertrophy to Heart Failure through Endothelial miR-195a-3p-Mediated Impairment of Cardiac Angiogenesis. J. Mol. Cell Cardiol. 2018, 118, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, M.; Zhang, Y.; Diao, Y. Crossing the Blood-Brain Barrier with AAV Vectors. Metab. Brain Dis. 2021, 36, 45–52. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Li, H.; Martin-Caraballo, M.; Hsia, S.V. Establishing a Herpesvirus Quiescent Infection in Differentiated Human Dorsal Root Ganglion Neuronal Cell Line Mediated by Micro-RNA Overexpression. Pathogens 2022, 11, 803. [Google Scholar] [CrossRef]
- Rachev, E.; Nalbansky, B.; Kolarov, G.; Agrosì, M. Efficacy and Safety of Phospholipid Liposomes in the Treatment of Neuropsychological Disorders Associated with the Menopause: A Double-Blind, Randomised, Placebo-Controlled Study. Curr. Med. Res. Opin. 2001, 17, 105–110. [Google Scholar] [CrossRef]
- Alberto, M.; Paiva-Santos, A.C.; Veiga, F.; Pires, P.C. Lipid and Polymeric Nanoparticles: Successful Strategies for Nose-to-Brain Drug Delivery in the Treatment of Depression and Anxiety Disorders. Pharmaceutics 2022, 14, 2742. [Google Scholar] [CrossRef] [PubMed]
- Kahana, M.; Weizman, A.; Gabay, M.; Loboda, Y.; Segal-Gavish, H.; Gavish, A.; Barhum, Y.; Offen, D.; Finberg, J.; Allon, N.; et al. Liposome-Based Targeting of Dopamine to the Brain: A Novel Approach for the Treatment of Parkinson’s Disease. Mol. Psychiatry 2021, 26, 2626–2632. [Google Scholar] [CrossRef] [PubMed]
- Moraes, F.C.; Pichon, C.; Letourneur, D.; Chaubet, F. miRNA Delivery by Nanosystems: State of the Art and Perspectives. Pharmaceutics 2021, 13, 1901. [Google Scholar] [CrossRef]
- Crew, E.; Tessel, M.A.; Rahman, S.; Razzak-Jaffar, A.; Mott, D.; Kamundi, M.; Yu, G.; Tchah, N.; Lee, J.; Bellavia, M.; et al. MicroRNA Conjugated Gold Nanoparticles and Cell Transfection. Anal. Chem. 2012, 84, 26–29. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Shoorei, H.; Noferesti, L.; Hussen, B.M.; Moghadam, M.H.B.; Taheri, M.; Rashnoo, F. Nanoparticle-Mediated Delivery of microRNAs-Based Therapies for Treatment of Disorders. Pathol. Res. Pract. 2023, 248, 154667. [Google Scholar] [CrossRef]
- O’neill, C.P.; Dwyer, R.M. Nanoparticle-Based Delivery of Tumor Suppressor microRNA for Cancer Therapy. Cells 2020, 9, 521. [Google Scholar] [CrossRef]
- Khaliq, N.U.; Lee, J.; Kim, J.; Kim, Y.; Yu, S.; Kim, J.; Kim, S.; Sung, D.; Kim, H. Mesoporous Silica Nanoparticles as a Gene Delivery Platform for Cancer Therapy. Pharmaceutics 2023, 15, 1432. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Mathiyalagan, P.; Sahoo, S. Exosomes-Based Gene Therapy for MicroRNA Delivery. Methods Mol. Biol. 2017, 1521, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Rubio, D.; Martin-Burriel, I.; Gil, A.; Cubero, P.; Forner, M.; Khalyfa, A.; Marin, J.M. Stability of Circulating Exosomal miRNAs in Healthy Subjects. Sci. Rep. 2018, 8, 10306. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, X.; Chen, X.; Wang, L.; Yang, G. Exosome Mediated Delivery of miR-124 Promotes Neurogenesis after Ischemia. Mol. Ther. Nucleic Acids 2017, 7, 278–287. [Google Scholar] [CrossRef]
- Kandeel, M.; Morsy, M.A.; Alkhodair, K.M.; Alhojaily, S. Mesenchymal Stem Cell-Derived Extracellular Vesicles: An Emerging Diagnostic and Therapeutic Biomolecules for Neurodegenerative Disabilities. Biomolecules 2023, 13, 1250. [Google Scholar] [CrossRef] [PubMed]
- Kou, M.; Huang, L.; Yang, J.; Chiang, Z.; Chen, S.; Liu, J.; Guo, L.; Zhang, X.; Zhou, X.; Xu, X.; et al. Mesenchymal Stem Cell-Derived Extracellular Vesicles for Immunomodulation and Regeneration: A next Generation Therapeutic Tool? Cell Death Dis. 2022, 13, 580. [Google Scholar] [CrossRef] [PubMed]
- Hade, M.D.; Suire, C.N.; Suo, Z. An Effective Peptide-Based Platform for Efficient Exosomal Loading and Cellular Delivery of a microRNA. ACS Appl. Mater. Interfaces 2023, 15, 3851–3866. [Google Scholar] [CrossRef] [PubMed]
- Ruseska, I.; Zimmer, A. Cellular Uptake and Trafficking of Peptide-Based Drug Delivery Systems for miRNA. Eur. J. Pharm. Biopharm. 2023, 191, 189–204. [Google Scholar] [CrossRef]
- Dos Santos Rodrigues, B.; Arora, S.; Kanekiyo, T.; Singh, J. Efficient Neuronal Targeting and Transfection Using RVG and Transferrin-Conjugated Liposomes. Brain Res. 2020, 1734, 146738. [Google Scholar] [CrossRef]
- Hao, R.; Sun, B.; Yang, L.; Ma, C.; Li, S. RVG29-Modified microRNA-Loaded Nanoparticles Improve Ischemic Brain Injury by Nasal Delivery. Drug Deliv. 2020, 27, 772–781. [Google Scholar] [CrossRef]
- Koczula, K.M.; Gallotta, A. Lateral Flow Assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, J.; Xiao, B.; Sun, X.; Xie, R.; Chen, A. Recent Advances in the Rapid Detection of microRNA with Lateral Flow Assays. Biosens. Bioelectron. 2022, 211, 114345. [Google Scholar] [CrossRef] [PubMed]

| Refs. | Sample | Data Source | miRNA (Upregulated) | miRNA (Downregulated) | Gene Target | Effect on Target |
|---|---|---|---|---|---|---|
| [76,77,85] | Dentate gyrus granule cells | Postmortem | hsa-miR-182 | - | BDNF | BDNF downregulation |
| [78] | Anterior cingulate cortex | Postmortem | hsa-miR-6077, hsa-miR-4632-3p, hsa-miR-6789-3p, hsa-miR-648, hsa-miR-4498, hsa-miR-6084, hsa-miR-4433a-3p, hsa-miR-4638-5p, hsa-miR-4258, hsa-miR-6850-5p, hsa-miR-3651, hsa-miR-4497, hsa-miR-668-5p, hsa-miR-7108-3p, hsa-miR-4761-3p, hsa-miR-4426, hsa-miR-572, hsa-miR-1470, hsa-miR-520f-3p, hsa-miR-4746-3p, hsa-miR-1193, hsa-miR-6075, hsa-miR-6737-5p, hsa-miR-6879-3p, hsa-miR-4701-3p | hsa-miR-367-3p, hsa-miR-5590-5p, hsa-miR-20b-3p, hsa-miR-548ar-3p, hsa-miR-3689a-5p | ||
| [79] | Prefrontal cortex | Postmortem | - | hsa-miR-1202 | GRM4 | GRM4 upregulation |
| [80] | Blood | MDD vs. controls | - | hsa-miR-135a | Serotonin transporter | |
| MDD 0 weeks vs. MDD 12 weeks | hsa-miR-135a | - | Increased expression of related genes after selective serotonin reuptake inhibitors | |||
| [81,86] | Peripheral blood mononuclear cells | MDD vs. controls | hsa-miR-589, hsa-miR-579, hsa-miR-941, hsa-miR-133a, hsa-miR-494, hsa-miR-107, hsa-miR-148a, hsa-miR-652, hsa-miR-425-3p | hsa-miR-517b, hsa-miR-636, hsa-miR-1243, hsa-miR-381, hsa-miR-200c | hsa-miR-425-3p targets MAPK/Wnt signaling pathway | |
| MDD 0 weeks vs. MDD 8 weeks | hsa-miR-20b-3p, hsa-miR-433, hsa-miR-409-3p, hsa-miR-410, hsa-miR-485-3p, hsa-miR-133a, hsa-miR-145 | hsa-miR-331-5p | ||||
| [82] | Blood | MDD 0 weeks vs. MDD 4 weeks (responders vs. non-responders) | - | hsa-miR-223-3p | IL-6, IL-1b, NLRP3 and TNF-α | Upregulation of these genes in ECT responders at baseline |
| [83] | Plasma NDEV | MDD vs. controls | - | hsa-miR-93 | ||
| [84,87] | Plasma NDEV | MDD 0 weeks vs. MDD 8 weeks (responders vs. non-responders) | hsa-miR-30d-5p, hsa-miR-486-5p | hsa-miR-21-5p | NR3C1, SIRT1, SERPINE1, RPS6KB1, ATF6, PSEN1 |
| Refs. | Tissue Source | miRNAs | Status (Patient vs. Control) | Antidepressant Treatment | Regulation by Treatment | miRNAs |
|---|---|---|---|---|---|---|
| [79,92] | Whole blood | miR-1202 | Down | Citalopram | Up | miR-1202 |
| [87,89] | Whole blood | - | - | Escitalopram (12 weeks) | Up | miR-130b, miR-505, miR-29b-2, miR-26b, miR-22, miR-26a, miR-664, miR-494, let-7d, let-7g, let-7e, let-7f, miR-629, miR-106b, miR-103, miR-191, miR-128, miR-502-3p, miR-374b, miR-132, miR-30d, miR-500, miR-589, miR-183, miR-574-3p, miR-140-3p, miR-335, miR-361-5p |
| Down | miR-34c-5p, miR-770-5p | |||||
| [90] | Whole blood | miR-132-3p, miR-124-3p | Up | Citalopram (8 weeks) | Up | miR-124 |
| Down | miR-132-3p | |||||
| [94] | Whole blood | - | - | Duloxetine (8 weeks) | Up | miR-3688, miR-5695 |
| [95] | Whole blood | - | - | Escitalopram (2 weeks) | Up | miR-103a-3p, miR-103b, miR-106a-5p, miR-106b-3p, miR-140-3p, miR-145-5p, miR-148b-3p, miR-151a-5p, miR-15a-5p, miR-15b-5p, miR-17-5p, miR-182-5p, miR-185-3p, miR-185-5p, miR-186-5p, miR-20a-5p, miR-20b-5p, miR-210-3p, miR-25-3p, miR-30a-5p, miR-30b-5p, miR-3158-3p, miR-3158-5p, miR-324-5p, miR-331-5p, miR-500a-3p, miR-502-3p, miR-532-5p, miR-550a-3p, miR-584-5p, miR-589-5p, miR-660-5p, miR-93-5p |
| Down | miR-1301-3p, miR-191-3p, miR-200b-3p, miR-222-3p, miR-25-5p, miR-27a-3p, miR-30c-1-3p, miR-3168, miR-328-3p, miR-505-5p, miR-744-5p, miR-92a-1-5p | |||||
| [96] | Plasma | miR-144-5p | Down | Personalized (8 weeks) | Up | miR-144-5p |
| [97] | Serum | miRNA-34a-5p, miRNA-221-3p | Up | Paroxetine (8 weeks) | Down | miRNA-34a-5p, miRNA-221-3p |
| miRNA-451a | Down | Up | miRNA-451a | |||
| [84] | Neuron-derived EV | - | - | Escitalopram (8 weeks) | Up (responders) | miR-30d-5p and miR-486-5p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaurani, L. Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential. Int. J. Mol. Sci. 2024, 25, 2866. https://doi.org/10.3390/ijms25052866
Kaurani L. Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential. International Journal of Molecular Sciences. 2024; 25(5):2866. https://doi.org/10.3390/ijms25052866
Chicago/Turabian StyleKaurani, Lalit. 2024. "Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential" International Journal of Molecular Sciences 25, no. 5: 2866. https://doi.org/10.3390/ijms25052866
APA StyleKaurani, L. (2024). Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential. International Journal of Molecular Sciences, 25(5), 2866. https://doi.org/10.3390/ijms25052866






