Mowat–Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions
Abstract
1. Introduction
2. Detailed Case Description
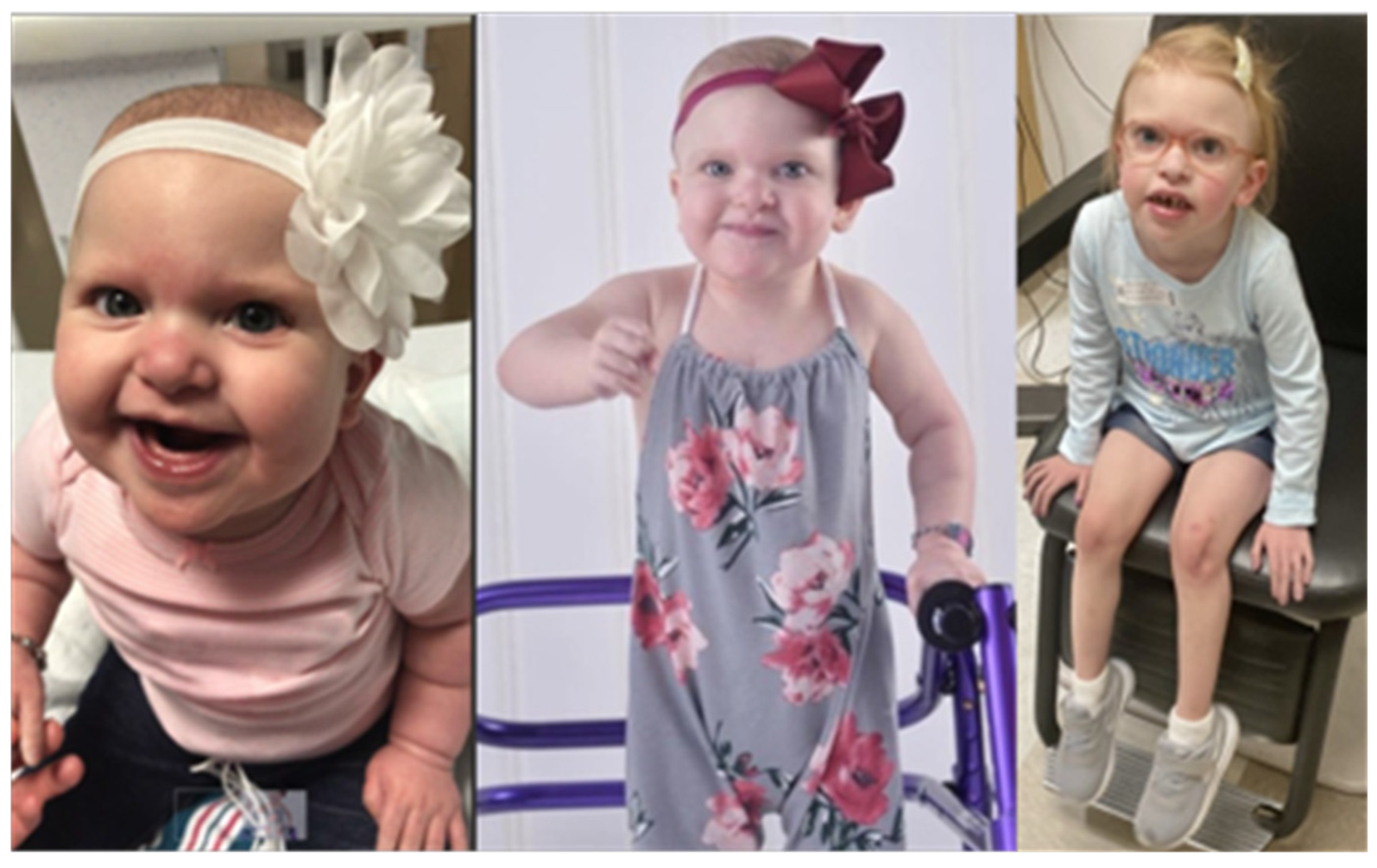
2.1. Clinical Case Report
2.2. Genetic and Protein Domain Data Collection of Patients with Mowat-Wilson Syndrome
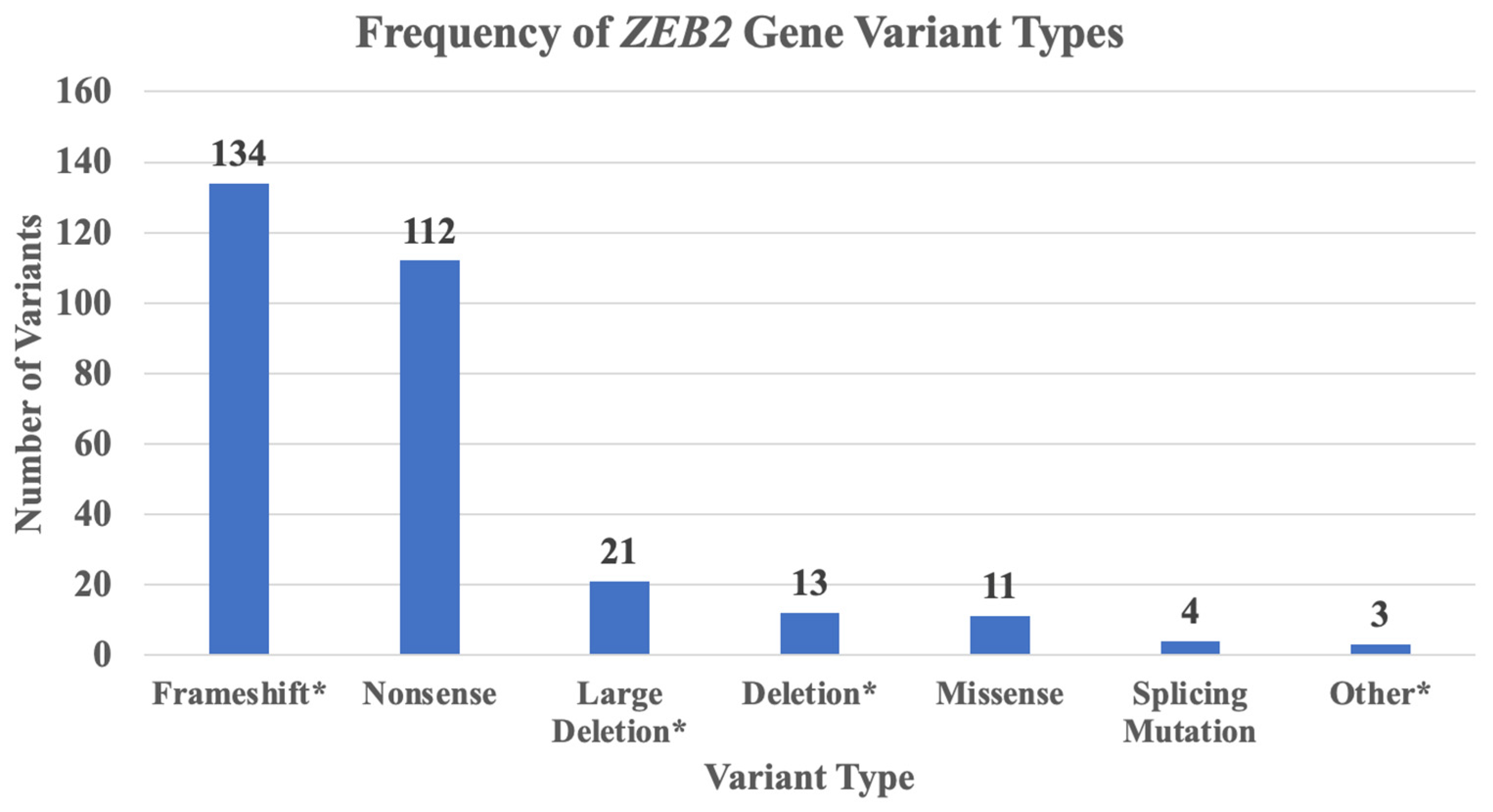
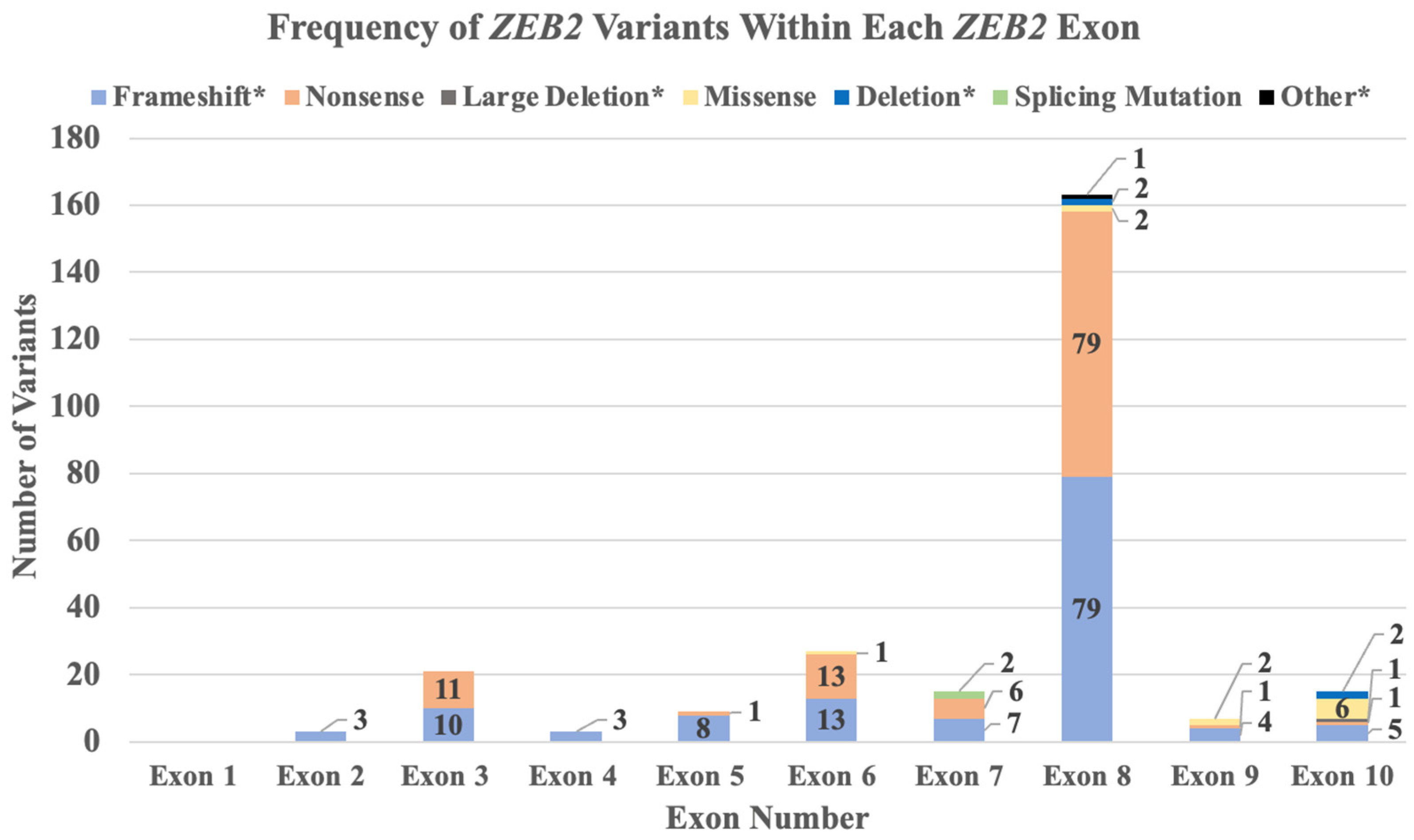
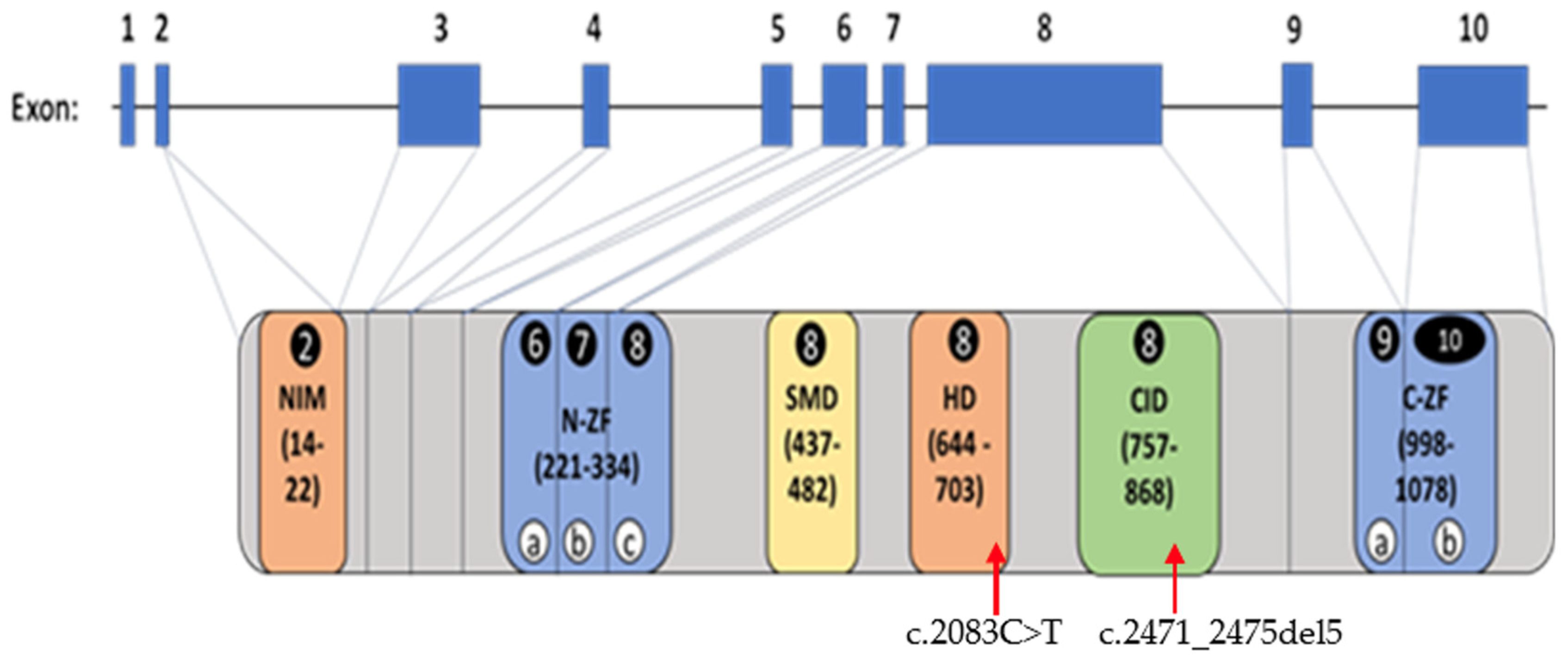
2.3. ZEB2 Gene Variant Types and Frequencies
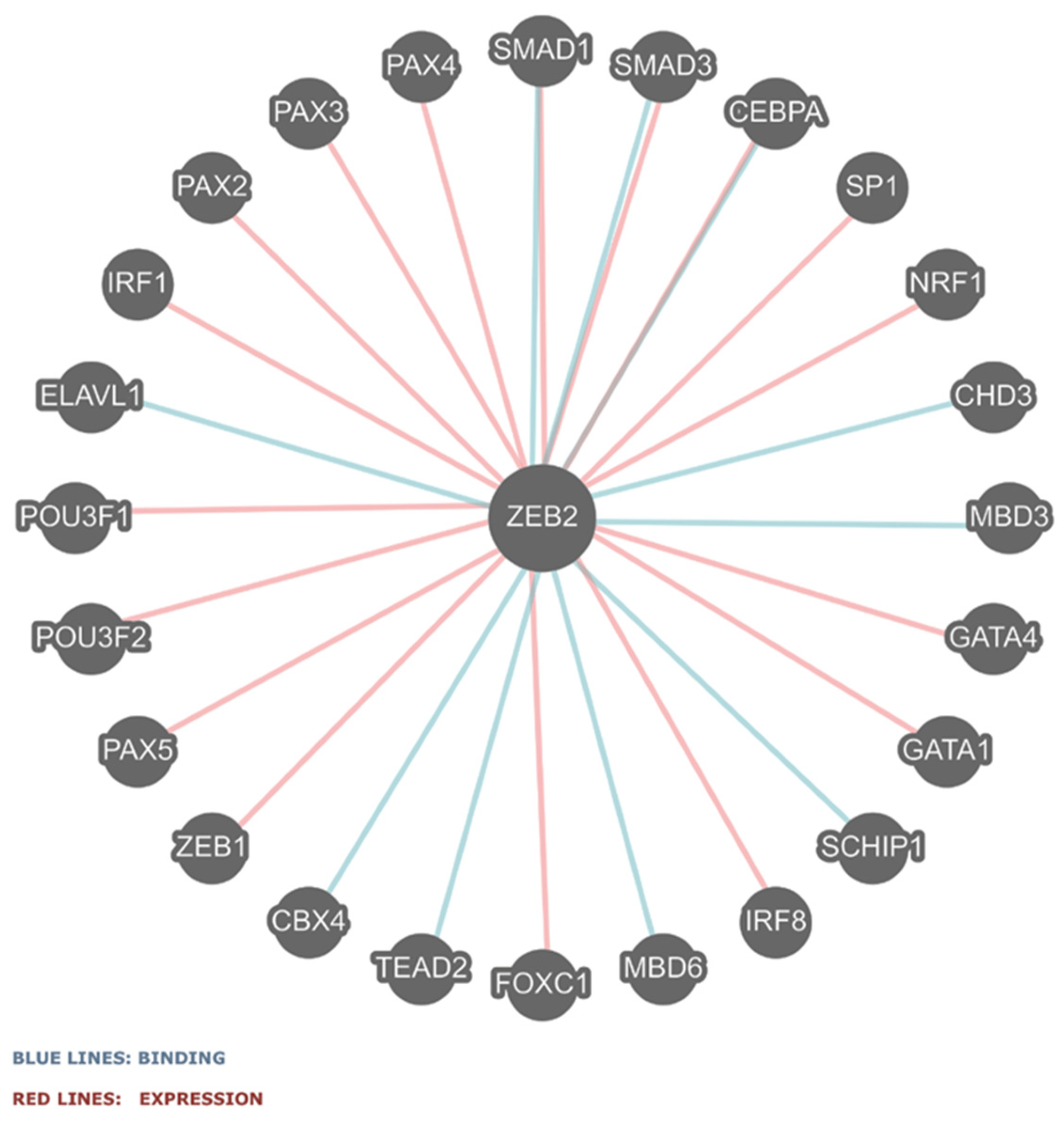
2.4. ZEB2 Gene–Gene or Protein Interactions
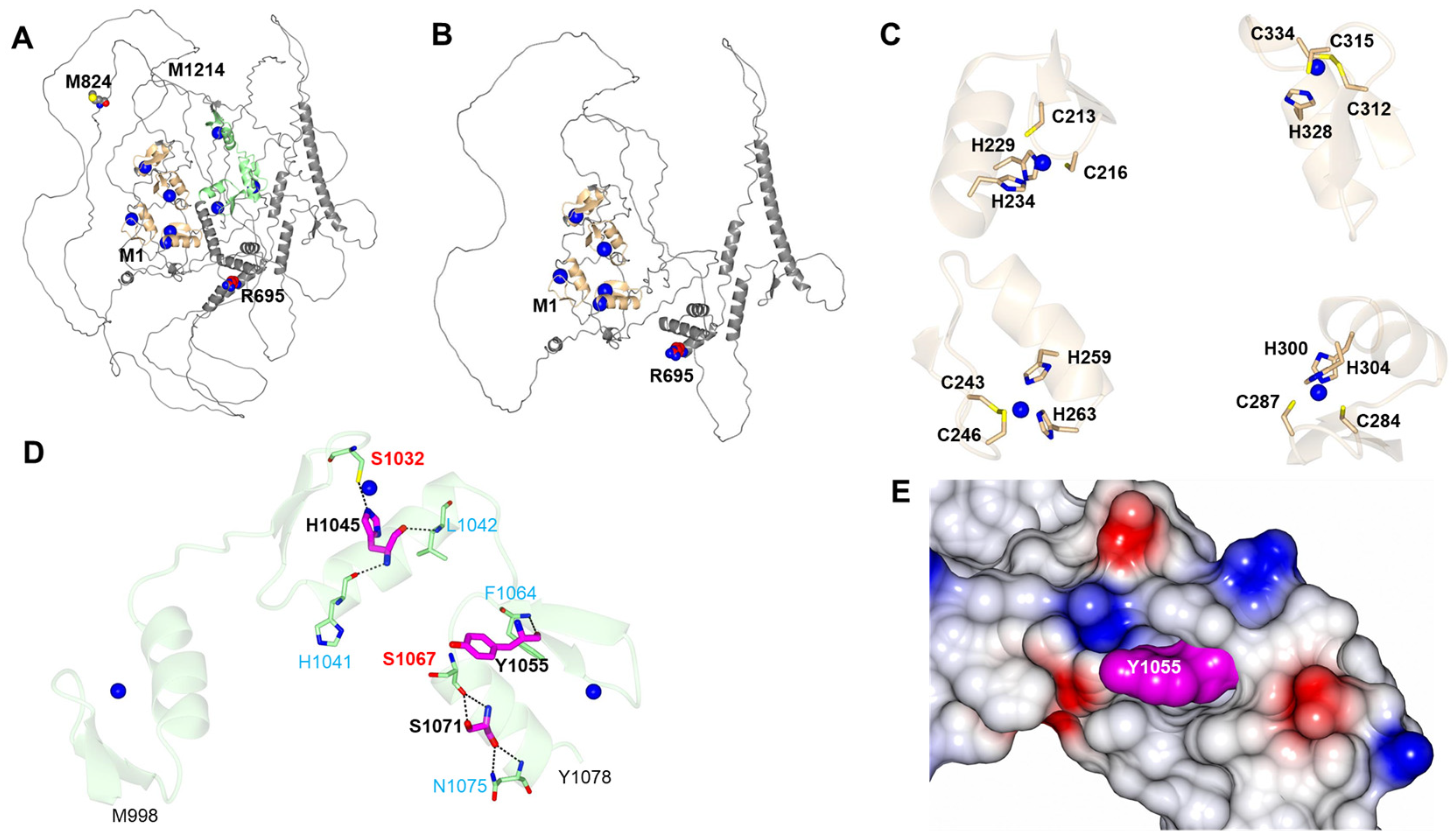
2.5. ZEB2 Structural Protein Domains, Functions and Models
3. Discussion
3.1. Mowat–Wilson Syndrome (MWS) and Clinical Findings
3.2. ZEB2 Gene Structure and Variants
3.3. ZEB2 Gene Functions and Interactions
3.4. Genes That Interact with ZEB2 and Potential Relationship with Mowat–Wilson Syndrome
3.5. ZEB2 Protein Domains and Functions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haendel, M.; Vasilevsky, N.; Unni, D.; Bologa, C.; Harris, N.; Rehm, H.; Hamosh, A.; Baynam, G.; Groza, T.; McMurry, J.; et al. How many rare diseases are there? Nat. Rev. Drug Discov. 2020, 19, 77–78. [Google Scholar] [CrossRef]
- Yang, Y.; Muzny, D.M.; Reid, J.G.; Bainbridge, M.N.; Willis, A.; Ward, P.A.; Braxton, A.; Beuten, J.; Xia, F.; Niu, Z.; et al. Clinical whole-exome sequencing for the diagnosis of Mendelian Disorders. N. Engl. J. Med. 2013, 369, 1502–1511. [Google Scholar] [CrossRef]
- Mowat, D.R.; Croaker, G.D.; Cass, D.T.; Kerr, B.A.; Chaitow, J.; Adès, L.C.; Chia, N.L.; Wilson, M.J. Hirschsprung disease, microcephaly, mental retardation, and characteristic facial features: Delineation of a new syndrome and identification of a locus at chromosome 2q22-q23. J. Med. Genet. 1998, 8, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, N.; Yamada, Y.; Yamada, K.; Ono, T.; Nomura, N.; Taniguchi, H.; Kitoh, H.; Mutoh, N.; Yamanaka, T.; Mushiake, K.; et al. Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease. Nat. Genet. 2001, 4, 369–370. [Google Scholar] [CrossRef]
- Cacheux, V.; Dastot-Le Moal, F.; Kääriäinen, H.; Bondurand, N.; Rintala, R.; Boissier, B.; Wilson, M.; Mowat, D.; Goossens, M. Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease. Hum. Mol. Genet. 2001, 14, 1503–1510. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.L.; Vivian, J.L.; Hagelstrom, R.T.; Hossain, W.A.; Golden, W.L.; Wassman, E.R.; Vanzo, R.J.; Butler, M.G. A Novel Partial Duplication of ZEB2 and Review of ZEB2 Involvement in Mowat-Wilson Syndrome. Mol. Syndromol. 2017, 4, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Mowat, D.R.; Wilson, M.J.; Goossens, M. Mowat-Wilson syndrome. J. Med. Genet. 2003, 5, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.P.; Feldman, J.; Mirzaa, F.M.; Pagon, R.A.; Wallace, S.E.; Bean, L.J.H.; Gripp, K.W.; Amemiya, A. (Eds.) GeneReviews [Internet]; University of Washington, Seattle: Seattle, WA, USA, 2023. [Google Scholar]
- Hegarty, S.V.; Sullivan, A.M.; O’Keeffe, G.W. Zeb2: A multifunctional regulator of nervous system development. Prog. Neurobiol. 2015, 132, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Zweier, C.; Thiel, C.T.; Dufke, A.; Crow, Y.J.; Meinecke, P.; Suri, M.; Ala-Mello, S.; Beemer, F.; Bernasconi, S.; Bianchi, P.; et al. Clinical and mutational spectrum of Mowat–Wilson syndrome. Eur. J. Med. Genet. 2005, 2, 97–111. [Google Scholar] [CrossRef]
- Saunders, C.J.; Zhao, W.; Ardinger, H.H. Comprehensive ZEB2 gene analysis for Mowat-Wilson syndrome in a North American cohort: A suggested approach to molecular diagnostics. Am. J. Med. Genet. A 2009, 11, 2527–2531. [Google Scholar] [CrossRef]
- Ghoumid, J.; Drevillon, L.; Alavi-Naini, S.M.; Bondurand, N.; Rio, M.; Briand-Suleau, A.; Nasser, M.; Goodwin, L.; Raymond, P.; Yanicostas, C.; et al. ZEB2 zinc-finger missense mutations lead to hypomorphic alleles and a mild Mowat–Wilson syndrome. Hum. Mol. Genet. 2013, 22, 2652–2661. [Google Scholar] [CrossRef]
- Zou, D.; Wang, L.; Wen, F.; Xiao, H.; Duan, J.; Zhang, T.; Yin, Z.; Dong, Q.; Guo, J.; Liao, J. Genotype-phenotype analysis in Mowat-Wilson syndrome associated with two novel and two recurrent ZEB2 variants. Exp. Ther. Med. 2020, 6, 263. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Peng, Q.; Ma, K.; Li, S.; Rao, C.; Zhong, B.; Lu, X. A novel nonsense mutation of ZEB2 gene in a Chinese patient with Mowat-Wilson syndrome. J. Clin. Lab. Anal. 2020, 9, e23413. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.; Luk, H.M.; Chung, B.H.; Fung, J.L.; Mak, H.H.; Lo, I.F.M. Mowat-Wilson syndrome in a Chinese population: A case series. Am. J. Med. Genet. A 2020, 182, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Wenger, T.L.; Harr, M.; Ricciardi, S.; Bhoj, E.; Santani, A.; Adam, M.P.; Barnett, S.S.; Ganetzky, R.; McDonald-McGinn, D.M.; Battaglia, D. CHARGE-like presentation, craniosynostosis and mild Mowat-Wilson Syndrome diagnosed by recognition of the distinctive facial gestalt in a cohort of 28 new cases. Am. J. Med. Genet. A. 2014, 164A, 2557–2566, Erratum in Am. J. Med. Genet. A 2015, 167, 1682–1683. [Google Scholar] [CrossRef] [PubMed]
- Mundhofir, F.E.; Yntema, H.G.; van der Burgt, I.; Hame, B.C.; Faradz, S.M.; van Bon, B.W. Mowat-Wilson syndrome: The first clinical and molecular report of an indonesian patient. Case Rep. Genet. 2012, 2012, 949507. [Google Scholar] [CrossRef]
- Murray, S.B.; Spangler, B.B.; Helm, B.M.; Vergano, S.S. Polymicrogyria in a 10-month-old boy with Mowat-Wilson syndrome. Am. J. Med. Genet. A 2015, 167A, 2402–2405. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yan, Y.C.; Li, Q.; Zhang, Z.; Xiao, P.; Yuan, X.Y.; Li, L.; Jiang, Q. Clinical and genetic features of Mowat-Wilson syndrome: An analysis of 3 cases. Zhongguo Dang Dai Er Ke Za Zhi 2019, 21, 468–473. (In Chinese) [Google Scholar] [CrossRef]
- Yamada, Y.; Nomura, N.; Yamada, K.; Matsuo, M.; Suzuki, Y.; Sameshima, K.; Kimura, R.; Yamamoto, Y.; Fukushi, D.; Fukuhara, Y.; et al. The spectrum of ZEB2 mutations causing the Mowat-Wilson syndrome in Japanese populations. Am. J. Med. Genet. A 2014, 164A, 1899–1908, Erratum in Am. J. Med. Genet. A 2015, 167, 1428. [Google Scholar] [CrossRef] [PubMed]
- Tronina, A.; Świerczyńska, M.; Filipek, E. First Case Report of Developmental Bilateral Cataract with a Novel Mutation in the ZEB2 Gene Observed in Mowat-Wilson Syndrome. Medicina 2023, 59, 101. [Google Scholar] [CrossRef]
- Jakubiak, A.; Szczałuba, K.; Badura-Stronka, M.; Kutkowska-Kaźmierczak, A.; Jakubiuk-Tomaszuk, A.; Chilarska, T.; Pilch, J.; Braun-Walicka, N.; Castaneda, J.; Wołyńska, K.; et al. Clinical characteristics of Polish patients with molecularly confirmed Mowat-Wilson syndrome. J. Appl. Genet. 2021, 62, 477–485. [Google Scholar] [CrossRef]
- Refaat, K.; Helmy, N.; Elawady, M.; El Ruby, M.; Kamel, A.; Mekkawy, M.; Ashaat, E.; Eid, O.; Mohamed, A.; Rady, M. Interstitial Deletion of 2q22.2q22.3 Involving the Entire ZEB2 Gene in a Case of Mowat-Wilson Syndrome. Mol. Syndromol. 2021, 12, 87–95. [Google Scholar] [CrossRef]
- Musaad, W.; Lyons, A.; Allen, N.; Letshwiti, J. Mowat-Wilson syndrome presenting with Shone’s complex cardiac anomaly. BMJ Case Rep. 2022, 15, e246913. [Google Scholar] [CrossRef]
- Pachajoa, H.; Gomez-Pineda, E.; Giraldo-Ocampo, S.; Lores, J. Mowat-Wilson Syndrome as a Differential Diagnosis in Patients with Congenital Heart Defects and Dysmorphic Facies. Pharmgenom. Pers. Med. 2022, 15, 913–918. [Google Scholar] [CrossRef]
- Wu, L.; Wang, J.; Wang, L.; Xu, Q.; Zhou, B.; Zhang, Z.; Li, Q.; Wang, H.; Han, L.; Jiang, Q.; et al. Physical, language, neurodevelopment and phenotype-genotype correlation of Chinese patients with Mowat-Wilson syndrome. Front. Genet. 2022, 13, 1016677. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xu, W.; Wang, Q.; Lin, Y.; He, P.; Liu, Y.; Yuan, H. Three Novel De Novo ZEB2 Variants Identified in Three Unrelated Chinese Patients With Mowat-Wilson Syndrome and A Systematic Review. Front. Genet. 2022, 13, 853183. [Google Scholar] [CrossRef]
- Wei, L.; Han, X.; Li, X.; Han, B.; Nie, W. A Chinese Boy with Mowat-Wilson Syndrome Caused by a 10 bp Deletion in the ZEB2 Gene. Pharmgenom. Pers. Med. 2021, 14, 1041–1045. [Google Scholar] [CrossRef]
- Şenbil, N.; Arslan, Z.; Sayın Kocakap, D.B.; Bilgili, Y. A Case Report of a Prenatally Missed Mowat-Wilson Syndrome With Isolated Corpus Callosum Agenesis. Child. Neurol. Open. 2021, 8, 2329048X211006511. [Google Scholar] [CrossRef] [PubMed]
- Ivanovski, I.; Djuric, O.; Caraffi, S.G.; Santodirocco, D.; Pollazzon, M.; Rosato, S.; Cordelli, D.M.; Abdalla, E.; Accorsi, P.; Adam, M.P.; et al. Phenotype and genotype of 87 patients with Mowat-Wilson syndrome and recommendations for care. Genet. Med. 2018, 20, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://pathwaycommons.org/pc12/Pathway_751ce7c7ccec6e191682a33d7252aac8 (accessed on 1 December 2023).
- Long, J.; Zuo, D.; Park, M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 2005, 280, 35477–35489. [Google Scholar] [CrossRef]
- Chen, Y.; Banda, M.; Speyer, C.L.; Smith, J.S.; Rabson, A.B.; Gorski, D.H. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol. Cell. Biol. 2010, 15, 3902–3913. [Google Scholar] [CrossRef]
- Holland, W. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Jiang, R. Integrating human omics data to prioritize candidate genes. BMC Med. Genom. 2013, 6, 57. [Google Scholar] [CrossRef]
- Han, C.; Hong, K.-H.; Kim, Y.H.; Kim, M.-J.; Song, C.; Kim, M.J.; Kim, S.-J.; Raizada, M.K.; Oh, S.P. SMAD1 deficiency in either endothelial or smooth muscle cells can predispose mice to pulmonary hypertension. Hypertension 2013, 61, 1044–1052. [Google Scholar] [CrossRef]
- Calligaris, R.; Bottardi, S.; Cogoi, S.; Apezteguia, I.; Santoro, C. Alternative translation initiation site usage results in two functionally distinct forms of the GATA-1 transcription factor. Proc. Nat. Acad. Sci. USA 1995, 92, 11598–11602. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, L.A.; Lin, F.R.; Cuesta, I.; Friedman, D.; Jarnik, M.; Zaret, K.S. Opening of compacted chromatin by early developmental transcription factors HNF3(FoxA) and GATA-4. Mol. Cell 2002, 2, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, S.; Wesley, R.A.; Danner, R.L. Adjacent sequence controls the response polarity of nitric oxide-sensitive Sp factor binding sites. J. Biol. Chem. 2003, 278, 29192–29200. [Google Scholar] [CrossRef]
- Holtschke, T.; Löhler, J.; Kanno, Y.; Fehr, T.; Giese, N.; Rosenbauer, F.; Lou, J.; Knobeloch, K.P.; Gabriele, L.; Waring, J.F.; et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 1996, 87, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Rigby, P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef]
- Mansouri, A.; St-Onge, L.; Gruss, P. Role of Pax genes in endoderm-derived organs. Trends Endocr. Metab. 1999, 10, 164–167. [Google Scholar] [CrossRef]
- Kawamata, N.; Pennella, M.A.; Woo, J.L.; Berk, A.J.; Koffler, H.P. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene 2012, 31, 966–977. [Google Scholar] [CrossRef]
- Schreiber, E.; Tobler, A.; Malipiero, U.; Schaffner, W.; Fontana, A. cDNA cloning of human N-Oct 3, a nervous-system specific POU domain transcription factor binding to the octamer DNA motif. Nucleic Acids Res. 1993, 21, 253–258. [Google Scholar] [CrossRef][Green Version]
- Atanasoski, S.; Toldo, S.S.; Malipiero, U.; Schreiber, E.; Fries, R.; Fontana, A. Isolation of the human genomic brain-2/N-Oct 3 gene (POUF3) and assignment to chromosome 6q16. Genomics 1995, 26, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Wang, J.; Lee, S.; The, C.E.; Bunting, K.; Ma, L.; Shannon, M.F. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int. Immunol. 2009, 3, 227–235. [Google Scholar] [CrossRef]
- Wang, H.; Xiao, Z.; Zheng, J.; Wu, J.; Hu, X.L.; Yang, X.; Shen, Q. ZEB1 Represses Neural Differentiation and Cooperates with CTBP2 to Dynamically Regulate Cell Migration during Neocortex Development. Cell Rep. 2019, 27, 2335–2353.e6. [Google Scholar] [CrossRef]
- Güleray Lafcı, N.; Karaosmanoglu, B.; Taskiran, E.Z.; Simsek-Kiper, P.O.; Utine, G.E. Mutated Transcripts of ZEB2 Do Not Undergo Nonsense-Mediated Decay in Mowat-Wilson Syndrome. Mol. Syndromol. 2023, 14, 258–265. [Google Scholar] [CrossRef]
- Chinnadurai, G. CtBP Family Proteins: Unique Transcriptional Regulators in the Nucleus with Diverse Cytosolic Functions. In Madame Curie Bioscience Database [Internet]; Landes Bioscience: Austin, TX, USA, 2007. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6557/ (accessed on 1 November 2023).
- Li, X.; Han, M.; Zhang, H.; Liu, F.; Pan, Y.; Zhu, J.; Liao, Z.; Chen, X.; Zhang, B. Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma. Biomark. Res. 2022, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Birkhoff, J.C.; Huylebroeck, D.; Conidi, A. ZEB2, the Mowat-Wilson Syndrome Transcription Factor: Confirmations, Novel Functions, and Continuing Surprises. Genes 2021, 12, 1037. [Google Scholar] [CrossRef] [PubMed]
- Macias, M.J.; Martin-Malpartida, P.; Massagué, J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 2015, 40, 296–308. [Google Scholar] [CrossRef]
- Bürglin, T.R. Homeodomain subtypes and functional diversity. Subcell. Biochem. 2011, 52, 95–122. [Google Scholar] [CrossRef] [PubMed]
| Our Study ID | Publication ID/ (Patient Number) | ZEB2 Exon | ZEB2 Gene Variant or Defect | Protein Defect | Type of Genetic Defect | ZEB2 Protein Domain |
|---|---|---|---|---|---|---|
| 1 | Mowat–Wilson Syndrome Foundation [MWSF]/(P1) | - | c.2180del | p.Leu727Tyrfs*7 | Frameshift | - |
| 2 | MWSF/(P2) | 8 | c.2083C>T | p.Arg695Ter | Nonsense | HD |
| 3 | MWSF/(P3) | 9 | c.3002del | p.Cys1001LeufsX74 | Frameshift | C-ZFa |
| 4 | MWSF/(P4) | 8 | c.2761C>T | p.Arg921* | Nonsense | - |
| 5 | MWSF/(P5) | 9 | c.3095G>A | p.Cys1032Tyr | Missense | C-ZFb |
| 6 | MWSF/(P6) | 10 | c.3213dup | p.Q1072AfsX52 | Frameshift | C-ZFb |
| 7 | MWSF/(P7) | 6 | c.696C>G | p.Tyr232* | Nonsense | N-ZFa |
| 8 | MWSF/(P8) | 6 | c.805C>T | p.Q269X | Nonsense | N-ZFb |
| 9 | MWSF/(P9) | 8 | c.2061del | p.Phe687Leufs*2 | Frameshift | HD |
| 10 | MWSF/(P10) | 10 | - | p.Tyr999* | Nonsense | C-ZFa |
| 11 | MWSF/(P11) | 3 | c.108del | p.E37fsX74 | Frameshift | - |
| 12 | MWSF/(P12) | 8 | c.2721del | p.Thr908LeufsTer22 | Frameshift | - |
| 13 | MWSF/(P13) | 2 | c.81_84dup | p.Asp29Leufs*2 | Frameshift | - |
| 14 | MWSF/(P14) | 8 | c.2094C>A | p.Y698X | Nonsense | HD |
| 15 | MWSF/(P15) | 6 | c.763C>T | p.Q255X | Nonsense | N-ZFa |
| 16 | MWSF/(P16) | intron 3 | c.331+1_331+2dup | - | Splicing | - |
| 17 | MWSF/(P17) | 10 | c.3533del | - | Deletion | - |
| 18 | MWSF/(P18) | 10 | c.3196C>T | p.His1066Tyr | Missense | C-ZFb |
| 19 | MWSF/(P19) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 20 | MWSF/(P20) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 21 | MWSF/(P21) | 8 | c.2367del | - | Frameshift | CID |
| 22 | MWSF/(P22) | 6 | c.674C>A | p.S225X | Nonsense | N-ZFa |
| 23 | MWSF/(P23) | 8 | c.909_910ins | p.H304Ffs*5 | Frameshift | N-ZFc |
| 24 | MWSF/(P24) | 8 | c.1795del | p.His599MetfsX8 | Frameshift | - |
| 25 | MWSF/(P25) | 2 | c.31del | p.Arg11Glyfs*16 | Frameshift | - |
| 26 | MWSF/(P26) | 8 | c.1571_1572ins | p.Ser524Argfs*4 | Frameshift | - |
| 27 | MWSF/(P27) | 1–10 | 6.2 Mb deletion Ch2q22.1-q22.3 | - | Chromosome deletion | - |
| 28 | MWSF/(P28) | 8 | c.1956C>A | p.Y652X | Nonsense | HD |
| 29 | MWSF/(P29) | 1–10 | 6.9 Mb deletion Ch2q22.1-q22.3 | - | Chromosome deletion | - |
| 30 | MWSF/(P30) | 4 | c.357_358del | p.Met120GlyfsX11 | Frameshift | - |
| 31 | MWSF/(P31) | - | 144 kb deletion Ch2q22.3 | - | Chromosome deletion | - |
| 32 | MWSF/(P32) | 10 | c.3212_3215dup | p.Gln1072HisfsTer53 | Frameshift | C-ZFb |
| 33 | [13] Zou et al., 2020/(P1) | 3 | c.290G>A | p.Trp97X | Nonsense | - |
| 34 | Zou et al., 2020/(P2) | 8 | c.1067_1068ins | p.Val357Aspfs*15 | Frameshift | - |
| 35 | Zou et al., 2020/(P3) | 8 | c.2761C>T | p.Arg921X | Nonsense | - |
| 36 | Zou et al., 2020/(P4) | 8 | c.3214C>T | p.Gln1072X | Nonsense | C-ZFb |
| 37 | [14] Hu et al., 2020/(P1) | 3 | c.250G>T | p.E84* | Nonsense | - |
| 38 | [15] Ho et al., 2020/(P1) | 8&9 | ZEB2 gene Exons 8 and 9 deletion | - | Deletion | - |
| 39 | Ho et al., 2020/(P2) | 8 | c.1472_c.1473ins | p.Met491llefs*9 | Small insertion, Frameshift | - |
| 40 | Ho et al., 2020/(P3) | 8 | c.2083C>T | p.Arg695* | Nonsense | HD |
| 41 | Ho et al., 2020/(P4) | 8 | c.1387del | p.Val463Phefs*24 | Frameshift | SMD |
| 42 | Ho et al., 2020/(P5) | 8 | c.2646del | p.Val883Cysfs*4 | Small deletion, Frameshift | - |
| 43 | Ho et al., 2020/(P6) | 3 | c.189del | p.Ser64Valfs*11 | Small deletion, Frameshift | - |
| 44 | Ho et al., 2020/(P7) | 3 | c.189del | p.Ser64Valfs*11 | Small deletion, Frameshift | - |
| 45 | Ho et al., 2020/(P8) | 1–10 | ZEB2 gene Exons 1-10 deletion | - | Deletion | - |
| 46 | Ho et al., 2020/(P9) | 9 | c1297C>T | p.Gln433* | Nonsense | - |
| 47 | Ho et al., 2020/(P10) | 10 | c.3335del | p.Tyr1112Cysfs*128 | Small deletion, Frameshift | - |
| 48 | Ho et al., 2020/(P11) | 1–10 | ZEB2 gene Exons 1-10 deletion | - | Deletion | - |
| 49 | Ho et al., 2020/(P12) | 7 | c.857_858del | p.Glu286Valfs*8 | Small deletion, Frameshift | N-ZFb |
| 50 | Ho et al., 2020/(P13) | 3 | c.291G>A | p.Trp97* | Nonsense | - |
| 51 | Ho et al., 2020/(P14) | 8 | c.2865C>A | p.Tyr955* | Nonsense | - |
| 52 | Ho et al., 2020/(P15) | 9 | c.169delins | p.Leu565* | Small indel, Frameshift | - |
| 53 | [16] Wenger et al., 2014/(P1) | 8 | - | p.R695X | Nonsense | HD |
| 54 | Wenger et al., 2014/(P2) | 8 | - | p.Q384X | Nonsense | - |
| 55 | Wenger et al., 2014/(P3) | 8 | - | p.G1182KfsX59 | Frameshift | - |
| 56 | Wenger et al., 2014/(P4) | 5 | - | p.E181Rfs211X | Frameshift | - |
| 57 | Wenger et al., 2014/(P5) | 9 | - | p.F1008C | Missense | C-ZFa |
| 58 | Wenger et al., 2014/(P6) | 7 | c.808-1G>T | - | Splicing | N-ZF |
| 59 | Wenger et al., 2014/(P7) | 10 | - | p.Ser1071Pro | Missense | C-ZFb |
| 60 | Wenger et al., 2014/(P8) | 8 | - | p.L894FfsX53 | Frameshift | - |
| 61 | Wenger et al., 2014/(P9) | 8 | - | p.S434Vfs7X | Frameshift | - |
| 62 | Wenger et al., 2014/(P10) | 8 | - | p.R695X | Nonsense | HD |
| 63 | Wenger et al., 2014/(P11) | 8 | - | p.R695X | Nonsense | HD |
| 64 | Wenger et al., 2014/(P12) | 8 | - | p.R695X | Nonsense | HD |
| 65 | Wenger et al., 2014/(P13) | 8 | - | p.M476WfsX11 | Frameshift | - |
| 66 | Wenger et al., 2014/(P14) | 8 | - | p.P906LfsX24 | Frameshift | - |
| 67 | Wenger et al., 2014/(P15) | 6 | - | p.Q209X | Nonsense | - |
| 68 | Wenger et al., 2014/(P16) | 8 | - | p.V627SfsX4 | Frameshift | - |
| 69 | Wenger et al., 2014/(P17) | 9 | - | p.S1011AfsX53 | Frameshift | C-ZFa |
| 70 | Wenger et al., 2014/(P18) | 6 | - | p.R218RfsX21 | Frameshift | - |
| 71 | Wenger et al., 2014/(P19) | 8 | - | p.V621AfsX25 | Frameshift | - |
| 72 | Wenger et al., 2014/(P20) | 1–10 | - | - | Whole gene deletion | - |
| 73 | Wenger et al., 2014/(P21) | 8 | - | p.S359TfsX3 | Frameshift | - |
| 74 | Wenger et al., 2014/(P22) | 8 | - | p.R695X | Nonsense | HD |
| 75 | Wenger et al., 2014/(P23) | 8 | - | p.Q962X | Nonsense | - |
| 76 | Wenger et al., 2014/(P24) | 8 | - | p.A907LfsX23 | Frameshift | - |
| 77 | Wenger et al., 2014/(P25) | 8 | - | p.G351VfsX19 | Frameshift | - |
| 78 | Wenger et al., 2014/(P26) | 5 | - | p.E181RfsX211 | Frameshift | - |
| 79 | Wenger et al., 2014/(P27) | 8 | c.1224del | - | Deletion | - |
| 80 | Wenger et al., 2014/(P28) | 8 | - | p.I390LfsX6 & p.L388F | Frameshift | - |
| 81 | [6] Baxter et al., 2017/(P1) | 1&2 | - | - | Partial duplication | - |
| 82 | [17] Mundhofir et al., 2012/(P1) | 8 | c.1965C>G | p.Tyr652∗ | Nonsense | HD |
| 83 | [18] Murray et al., 2015/(P1) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 84 | [19] Wang et al., 2019/(P1) | 7 | c.904C>T | p.R302X | Nonsense | N-ZFc |
| 85 | Wang et al., 2019/(P2) | 6 | c.756C>A | p.Y252X | Nonsense | N-ZFa |
| 86 | Wang et al., 2019/(P3) | 8 | c.2761C>T | p.R921X | Nonsense | - |
| 87 | [20] Yamada et al., 2014/(P1) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 88 | Yamada et al., 2014/(P2) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 89 | Yamada et al., 2014/(P3) | 3 | c.259G>T | p.E87X | Nonsense | - |
| 90 | Yamada et al., 2014 (P4) | 7 | c.811C>T | p.Q271X | Nonsense | N-ZFb |
| 91 | Yamada et al., 2014/(P5) | 7 | c.904C>T | p.R302X | Nonsense | N-ZFc |
| 92 | Yamada et al., 2014/(P6) | 8 | c.936C>A | p.C312X | Nonsense | N-ZFc |
| 93 | Yamada et al., 2014/(P7) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 94 | Yamada et al., 2014/(P8) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 95 | Yamada et al., 2014/(P9) | 8 | c.1027C>T | p.R343X | Nonsense | - |
| 96 | Yamada et al., 2014/(P10) | 8 | c.1298C>T | p.Q433X | Nonsense | - |
| 97 | Yamada et al., 2014/(P11) | 8 | c.1489C>T | p.Q497X | Nonsense | - |
| 98 | Yamada et al., 2014/(P12) | 8 | c.1645A>T | p.R549X | Nonsense | - |
| 99 | Yamada et al.,2014/(P13) | 8 | c.1825G>T | p.E609X | Nonsense | - |
| 100 | Yamada et al., 2014/(P14) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 101 | Yamada et al., 2014/(P15) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 102 | Yamada et al., 2014/(P16) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 103 | Yamada et al., 2014/(P17) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 104 | Yamada et al., 2014/(P18) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 105 | Yamada et al., 2014/(P19) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 106 | Yamada et al., 2014/(P20) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 107 | Yamada et al., 2014/(P21) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 108 | Yamada et al., 2014/(P22) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 109 | Yamada et al., 2014/(P23) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 110 | Yamada et al., 2014/(P24) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 111 | Yamada et al., 2014/(P25) | 8 | c.2083C>T | p.R695X | Nonsense | HD |
| 112 | Yamada et al., 2014/(P26) | 8 | c.2399C>G | p.S800X | Nonsense | CID |
| 113 | Yamada et al., 2014/(P27) | 8 | c.2615C>G | p.S872X | Nonsense | - |
| 114 | Yamada et al., 2014/(P28) | 8 | c.2761C>T | p.R921X | Nonsense | - |
| 115 | Yamada et al., 2014/(P29) | 8 | c.2761C>T | p.R921X | Nonsense | - |
| 116 | Yamada et al., 2014/(P1) | 3 | c.162_164del | p.P55Lfs*20 | Frameshift | - |
| 117 | Yamada et al., 2014/(P2) | 3 | c.175_182del | p.T60Sfs*3 | Frameshift | - |
| 118 | Yamada et al., 2014/(P3) | 3 | c270_272del | p.G91Vfs*17 | Frameshift | - |
| 119 | Yamada et al., 2014/(P4) | 3 | c.311_312dup | p.A105Sfs*16 | Frameshift | - |
| 120 | Yamada et al., 2014/(P5) | 5 | c.459_460del | p.E154Rfs*58 | Frameshift | - |
| 121 | Yamada et al., 2014/(P6) | 6 | c.635_638dup | p.P214Lfs*26 | Frameshift | - |
| 122 | Yamada et al., 2014/(P7) | 6 | c.647del | p.C216Sfs*8 | Frameshift | - |
| 123 | Yamada et al., 2014/(P8) | 6 | c.759_760dup | p.Q255Pfs*8 | Frameshift | N-ZFa |
| 124 | Yamada et al., 2014/(P9) | 7 | c.852_855del | p.T285Rfs*9 | Frameshift | N-ZFb |
| 125 | Yamada et al., 2014/(P10) | 7 | c.855_858del | p.E286Vfs*8 | Frameshift | N-ZFb |
| 126 | Yamada et al., 2014/(P11) | 7 | c.862_863del | p.G288Afs*10 | Frameshift | N-ZFb |
| 127 | Yamada et al., 2014/(P12) | 8 | c.1169ins | p.I390Tfs*41 | Frameshift | - |
| 128 | Yamada et al., 2014/(P13) | 8 | c.1169_1170del | p.T392Qfs*4 | Frameshift | - |
| 129 | Yamada et al., 2014/(P14) | 8 | c.1174_1178del | p.T392Nfs*3 | Frameshift | - |
| 130 | Yamada et al., 2014/(P15) | 8 | c.1176del | p.E393Nfs*3 | Frameshift | - |
| 131 | Yamada et al., 2014/(P16) | 8 | c.1212_1213del | p.A405Lfs*12 | Frameshift | - |
| 132 | Yamada et al., 2014/(P17) | 8 | c.1268_1273del | p.S424Lfs*2 | Frameshift | - |
| 133 | Yamada et al., 2014/(P18) | 8 | c.1280_1286del7ins | p.G427Dfs*2 | Frameshift | - |
| 134 | Yamada et al., 2014/(P19) | 8 | c.1334_1337dup | p.L447Ffs*9 | Frameshift | SMD |
| 135 | Yamada et al., 2014/(P20) | 8 | c.1395_1408del14ins | p.Q465Hfs*9 | Frameshift | SMD |
| 136 | Yamada et al., 2014/(P21) | 8 | c.1417del | p.R473Gfs*14 | Frameshift | SMD |
| 137 | Yamada et al., 2014/(P22) | 8 | c.1417del | p.R473Gfs*14 | Frameshift | SMD |
| 138 | Yamada et al., 2014/(P23) | 8 | c.1421_1426dup | p.M476Nfs*6 | Frameshift | SMD |
| 139 | Yamada et al., 2014/(P24) | 8 | c.1492_1493del | p.P498Lfs*18 | Frameshift | - |
| 140 | Yamada et al., 2014/(P25) | 8 | c.1534_1535del | p.G512Vfs*4 | Frameshift | - |
| 141 | Yamada et al., 2014/(P26) | 8 | c.1822del | p.E608Kfs*13 | Frameshift | - |
| 142 | Yamada et al., 2014/(P27) | 8 | c.1966_1967del | p.M656Vfs*17 | Frameshift | HD |
| 143 | Yamada et al., 2014/(P28) | 8 | c.2178_2180del | p.L727Ifs*28 | Frameshift | - |
| 144 | Yamada et al., 2014/(P29) | 8 | c.2254dup | p.T752Nfs*4 | Frameshift | - |
| 145 | Yamada et al., 2014/(P30) | 8 | c. 2282del | p.T761Kfs*26 | Frameshift | CID |
| 146 | Yamada et al., 2014/(P31) | 8 | c.2349_2351dup | p.S784Ffs*11 | Frameshift | CID |
| 147 | Yamada et al., 2014/(P32) | 8 | c.2579del | p.L860Rfs*3 | Frameshift | CID |
| 148 | Yamada et al., 2014/(P33) | 8 | c.2740_2743dup | p.S916Dfs*34 | Frameshift | - |
| 149 | Yamada et al., 2014/(P34) | 10 | c.3608_3614del | p.D1204Rfs*29 | Frameshift | - |
| 150 | [21] Tronina et al., 2023/(P1) | 6 | - | p.Gln694Ter | Missense | HD |
| 151 | [22] Jakubiak et al., 2021/(P1) | 8 | c.1027C > T | p.R343* | Nonsense | - |
| 152 | Jakubiak et al., 2021/(P2) | 1–10 | - | - | Deletion | - |
| 153 | Jakubiak et al., 2021/(P3) | 6 | c.648C > A | p.C216* | Nonsense | - |
| 154 | Jakubiak et al., 2021/(P4) | 10 | ZEB2 gene Exon 10 deletion | - | Deletion | - |
| 155 | Jakubiak et al., 2021/(P5) | 8 | c.1946del | p.I649Tfs*17 | Frameshift | HD |
| 156 | Jakubiak et al., 2021/(P6) | 6 | c.607ins | p.Thr203IlefsTer37 | Frameshift | - |
| 157 | Jakubiak et al., 2021/(P7) | 4 | c.399_400dup | p.Thr134IlefsTer3 | Frameshift | - |
| 158 | Jakubiak et al., 2021/(P8) | 8 | c.1276T > A | p.Leu426Ile | Missense | - |
| 159 | Jakubiak et al., 2021/(P9) | 6 | c.696C > G | p.Y232* | Nonsense | N-ZFa |
| 160 | Jakubiak et al., 2021/(P10) | - | 8 Mb deletion 2q22.3q23.3 | - | Chromosome deletion | - |
| 161 | Jakubiak et al., 2021/(P11) | 1–10 | - | - | Deletion | - |
| 162 | Jakubiak et al., 2021/(P12) | 3–10 | - | - | Deletion | - |
| 163 | Jakubiak et al., 2021/(P13) | 7 | c.857-858del | p.Glu286ValfsTer8 | Frameshift | N-ZFb |
| 164 | Jakubiak et al., 2021/(P14) | 6 | c.607ins | p.Thr203IlefsTer37 | Frameshift | - |
| 165 | Jakubiak et al., 2021/(P15) | 8 | c.1445T > G | p.Leu482* | Nonsense | SMD |
| 166 | Jakubiak et al., 2021/(P16) | 3 | c.84T > G | p.Tyr28* | Nonsense | - |
| 167 | Jakubiak et al., 2021/(P17) | 8 | c.1421-1426del | p.Gln474_Met476delins | Insertion, deletion | SMD |
| 168 | Jakubiak et al., 2021/(P18) | 8 | c.2230A > G | p.Ile744Val | Missense | - |
| 169 | Jakubiak et al., 2021/(P19) | 10 | c.3202G > T | p.Gly1068Cys | Missense | C-ZFb |
| 170 | Jakubiak et al., 2021/(P20) | 8 | c.2087_2088del | Lys696Serfs*24 | Frameshift | HD |
| 171 | Jakubiak et al., 2021/(P21) | 8 | c.2073G > A | p.Trp691Ter | Nonsense | HD |
| 172 | Jakubiak et al.,2021/(P22) | - | - | - | Deletion | - |
| 173 | Jakubiak et al., 2021/(P23) | 8 | c.2562_2564del | p.N855Lfs*3 | Frameshift | CID |
| 174 | Jakubiak et al., 2021/(P24) | 8 | c.1177dup | p.E393Gfs*7 | Frameshift | - |
| 175 | Jakubiak et al., 2021/(P25) | 8 | c.1437_1440del | p.H304Qfs*3 | Frameshift | N-ZFc |
| 176 | Jakubiak et al., 2021/(P26) | 8 | c.2083C > T | p.Arg695Ter | Nonsense | HD |
| 177 | Jakubiak et al., 2021/(P27) | 8 | c.2083C > T | p.Arg695Ter | Nonsense | HD |
| 178 | Jakubiak et al., 2021/(P28) | 3–10 | 258.2 kb deletion | - | Chromosome deletion | - |
| 179 | [23] Refaat et al., 2021/(P1) | - | 2.27 Mb deletion | - | Chromosome deletion | - |
| 180 | [24] Musaad et al., 2022/(P1) | - | Chr2:145161574del | - | Frameshift | - |
| 181 | [25] Pachajoa et al., 2022/(P1) | 8 | c.2761C>T | p.Arg921Ter | Nonsense | - |
| 182 | [26] Wu et al., 2022/(P1) | 8 | c.2417del | p.Phe807Serfs*11 | Frameshift | CID |
| 183 | Wu et al., 2022/(P2) | 8 | c.1200T>A | p.Tyr400X | Nonsense | - |
| 184 | Wu et al., 2022/(P3) | 8 | c.1027C>T | p.Arg343X | Nonsense | - |
| 185 | Wu et al., 2022/(P4) | 8 | c.2621del | p.Asn874Ilefs*12 | Frameshift | - |
| 186 | Wu et al., 2022/(P5) | 8 | c.2456C>G | p.Ser819X | Nonsense | CID |
| 187 | Wu et al., 2022/(P6) | 8 | c.2002del | p.Glu668Serfs*8 | Frameshift | HD |
| 188 | Wu et al., 2022/(P7) | 2–10 | Chr2:145147017-145274917del | - | Large deletion | - |
| 189 | Wu et al., 2022/(P8) | 5 | c.492_517del | p.Glu164Aspfs*9 | Frameshift | - |
| 190 | Wu et al., 2022/(P9) | 6 | c.779dup | p.Met260Ilefs*19 | Frameshift | N-ZFa |
| 191 | Wu et al., 2022/(P10) | 1–10 | chr2:141213978-148010654del | - | Large deletion | - |
| 192 | Wu et al., 2022/(P11) | 8 | c.2083C>T | p.Arg695X | Nonsense | HD |
| 193 | Wu et al., 2022/(P12) | 1–10 | chr2:145000000-145351228 del | - | Large deletion | - |
| 194 | Wu et al., 2022/(P13) | 7 | c.904C>T | p.Arg302X | Nonsense | N-ZFc |
| 195 | Wu et al., 2022/(P14) | 8 | c.2712del | p.Pro906Leufs*24 | Frameshift | - |
| 196 | Wu et al., 2022/(P15) | 8 | c.2670_2677del | p.Ala891Phefs*55 | Frameshift | - |
| 197 | Wu et al., 2022/(P16) | 8 | c.2177_2180del | p.Ser726Tyrfs*7 | Frameshift | - |
| 198 | Wu et al., 2022/(P17) | 1–10 | chr2:138434153-145285163 del | - | Large Deletion | - |
| 199 | Wu et al., 2022/(P18) | 8 | c.2851C>T | p.Gln951X | Nonsense | - |
| 200 | Wu et al., 2022/(P19) | 8 | c.1426dup | p.Met476Asnfs*5 | Frameshift | SMD |
| 201 | Wu et al., 2022/(P20) | 8 | c.2761C>T | p.Arg921X | Nonsense | - |
| 202 | Wu et al., 2022/(P21) | 8 | c.1027C>T | p.Arg343X | Nonsense | - |
| 203 | Wu et al., 2022/(P22) | 8 | c.1106_1115delins | p.Leu369X | Nonsense | - |
| 204 | [27] Fu et al., 2022/(P1) | 8 | c.2136del | p.Lys713Serfs*3 | Frameshift | - |
| 205 | Fu et al., 2022/(P2) | 8 | c.2740del | p. Gln914Argfs*16 | Frameshift | - |
| 206 | Fu et al., 2022/(P3) | 7 | c.808-2del | - | Splicing | N-ZFb |
| 207 | [28] Wei et al., 2021/(P1) | 8 | c.1137_1146del | p.S380Nfs*13 | Deletion | - |
| 208 | [29] Şenbil et al., 2021/(P1) | 6 | c.646dup | p.Cys216LeufsTer23 | Frameshift | - |
| 209 | [30] Ivanoski et al., 2018/(P1) | 6 | c.805C>T | p.Q269* | Nonsense | N-ZFb |
| 210 | Ivanoski et al., 2018/(P2) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 211 | Ivanoski et al., 2018/(P3) | 1–10 | 7.3 Mb deletion Ch2q22.1q22.3 | - | Large deletion | - |
| 212 | Ivanoski et al., 2018/(P4) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 213 | Ivanoski et al., 2018/(P5) | 8 | c.1052_1057delins | p.G351Vfs*19 | Small deletion, Frameshift | - |
| 214 | Ivanoski et al., 2018/(P6) | 6 | c.696C>G | p.Y232* | Nonsense | N-ZFa |
| 215 | Ivanoski et al., 2018/(P7) | 8 | c.1073_1122delins | p.S359Tfs*3 | Small indel, Frameshift | - |
| 216 | Ivanoski et al., 2018/(P8) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 217 | Ivanoski et al., 2018/(P9) | 1–10 | 2.6 Mb deletion Ch2q22.2q22.3 | - | Large deletion | - |
| 218 | Ivanoski et al., 2018/(P10) | 8 | c.2718del | p.A907Lfs*23 | Small deletion, Frameshift | - |
| 219 | Ivanoski et al., 2018/(P11) | 8 | c.2180T>A | p.L727* | Nonsense | - |
| 220 | Ivanoski et al., 2018/(P12) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 221 | Ivanoski et al., 2018/(P13) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 222 | Ivanoski et al., 2018/(P14) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 223 | Ivanoski et al., 2018/(P15) | 5 | c.460G>T | p.E154* | Nonsense | - |
| 224 | Ivanoski et al., 2018/(P16) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 225 | Ivanoski et al., 2018/(P17) | 8 | c.1426dup | p.M476Nfs*6 | Small insertion, Frameshift | SMD |
| 226 | Ivanoski et al., 2018/(P18) | 1–10 | 4.6 Mb deletion Ch2q22q22.3 | - | Large deletion | - |
| 227 | Ivanoski et al., 2018/(P19) | 8 | c.2682_2687delins | p.L894Ffs*36 | Small indel, Frameshift | - |
| 228 | Ivanoski et al., 2018/(P20) | 3 | c.274G>T | p.G92* | Nonsense | - |
| 229 | Ivanoski et al., 2018/(P21) | 8 | c.2053C>T | p.Q685* | Nonsense | HD |
| 230 | Ivanoski et al., 2018/(P23) | 9 | c.3031del | p.S1011Afs*64 | Small deletion, Frameshift | C-ZFa |
| 231 | Ivanoski et al., 2018/(P24) | 8 | c.2227del | p.S743Lfs*2 | Small deletion, Frameshift | - |
| 232 | Ivanoski et al., 2018/(P25) | 5 | c.460del | p.E154Rfs*58 | Small deletion, Frameshift | - |
| 233 | Ivanoski et al., 2018/(P26) | 6 | c.625C>T | p.Q209* | Nonsense | - |
| 234 | Ivanoski et al., 2018/(P27) | 7 | c.817del | p.L273* | Nonsense | N-ZFb |
| 235 | Ivanoski et al., 2018/(P28) | 8 | c.1635_1636ins | p.D546Lfs*11 | Small insertion, Frameshift | - |
| 236 | Ivanoski et al., 2018/(P29) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 237 | Ivanoski et al., 2018/(P30) | 3 | c.310C>T | p.Q104* | Nonsense | - |
| 238 | Ivanoski et al., 2018/(P31) | 8 | c.2701C>T | p.Q901* | Nonsense | - |
| 239 | Ivanoski et al., 2018/(P32) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 240 | Ivanoski et al., 2018/(P34) | 8 | c.2718del | p.A907Lfs*23 | Small deletion, Frameshift | - |
| 241 | Ivanoski et al., 2018/(P35) | 8 | c.2317_2318del | p.E773Kfs*8 | Small deletion, Frameshift | CID |
| 242 | Ivanoski et al., 2018/(P36) | 1–10 | 0.6 Mb deletion Ch2q22.2 | - | Large deletion | - |
| 243 | Ivanoski et al., 2018/(P37) | 3 | c.264_267del | p.I88Mfs*19 | Small deletion, Frameshift | - |
| 244 | Ivanoski et al., 2018/(P38) | 8 | c.1541del | p.P414Rfs*2 | Small deletion, Frameshift | - |
| 245 | Ivanoski et al., 2018/(P39) | 8 | c.2856del | p.R953Efs*24 | Small deletion, Frameshift | - |
| 246 | Ivanoski et al., 2018/(P40) | 8 | c.2254dup | p.Y752Nfs*4 | Small insertion, Frameshift | - |
| 247 | Ivanoski et al., 2018/(P41) | 8 | c.930C>A | p.Y310* | Nonsense | N-ZFc |
| 248 | Ivanoski et al., 2018/(P42) | 8&9 | c.917_3067del | p.E307_G1023del | Deletion, in-frame | N-ZFc C-ZFa |
| 249 | Ivanoski et al., 2018/(P43) | 8 | c.975C>A | p.Y325* | Nonsense | N-ZFc |
| 250 | Ivanoski et al., 2018/(P44) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 251 | Ivanoski et al., 2018/(P45) | 6 | c.691dup | p.Y232Vfs*7 | Frameshift | N-ZFa |
| 252 | Ivanoski et al., 2018/(P46) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 253 | Ivanoski et al., 2018/(P47) | 7 | c.901del | p.L301Cfs*37 | Small deletion, Frameshift | N-ZFc |
| 254 | Ivanoski et al., 2018/(P48) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 255 | Ivanoski et al., 2018/(P49) | 3 | c.81_84dup | p.D29Lfs*2 | Small insertion, Frameshift | - |
| 256 | Ivanoski et al., 2018/(P50) | 8 | c.1202dup | p.K401Ifs*17 | Small insertion, Frameshift | - |
| 257 | Ivanoski et al., 2018/(P51) | 6 | c.648C>A | p.C216* | Nonsense | - |
| 258 | Ivanoski et al., 2018/(P52) | 8 | c.1910C>G | p.S637* | Nonsense | - |
| 259 | Ivanoski et al., 2018/(P53) | 8 | c.2713del | p.P906Lfs*24 | Small deletion, Frameshift | - |
| 260 | Ivanoski et al., 2018/(P54) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 261 | Ivanoski et al., 2018/(P55) | 5 | c.540del | p.E181Rfs*31 | Small deletion, Frameshift | - |
| 262 | Ivanoski et al., 2018/(P56) | 6 | c.609del | p.P204Qfs*9 | Small deletion, Frameshift | - |
| 263 | Ivanoski et al., 2018/(P57) | 5 | c.477_484del | p.H159Qfs*10 | Small deletion, Frameshift | - |
| 264 | Ivanoski et al., 2018/(P58) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 265 | Ivanoski et al., 2018/(P59) | 5–8 | c.403_2887del | p.V135Gfs*5 | Frameshift | - |
| 266 | Ivanoski et al., 2018/(P60) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 267 | Ivanoski et al., 2018/(P61) | 1–10 | 16.7 Mb deletion Ch2q21.1q22.3 | - | Large deletion | - |
| 268 | Ivanoski et al., 2018/(P62) | 6 | c.653_654ins | p.G219Pfs*21 | Small insertion, Frameshift | - |
| 269 | Ivanoski et al., 2018/(P63) | 8 | c.1851del | p.H617Qfs*4 | Small deletion, Frameshift | - |
| 270 | Ivanoski et al., 2018/(P64) | 4 | c.389_390dup | p.T134Lfs*3 | Small insertion, Frameshift | - |
| 271 | Ivanoski et al., 2018/(P65) | 8 | c.2076del | p.F692Lfs*24 | Small deletion, Frameshift | HD |
| 272 | Ivanoski et al., 2018/(P66) | 8 | c.2083C>T | p.R695* | Nonsense | HD |
| 273 | Ivanoski et al., 2018/(P67) | 7 | c.823C>T | p.Q275* | Nonsense | N-ZFb |
| 274 | Ivanoski et al., 2018/(P68) | 8 | c.1313del | p.H438Pfs*2 | Small deletion, Frameshift | SMD |
| 275 | Ivanoski et al., 2018/(P69) | 1&2 | ZEB2 gene Exons 1 and 2 deletion | - | Large deletion | - |
| 276 | Ivanoski et al., 2018/(P70) | 8 | c.1027C>T | p.R343* | Nonsense | - |
| 277 | Ivanoski et al., 2018/(P71) | 6 | c.648C>A | p.C216* | Nonsense | - |
| 278 | Ivanoski et al., 2018/(P72) | 1–10 | ZEB2 gene deletion | - | Large deletion | - |
| 279 | Ivanoski et al., 2018/(P73) | 8 | c.1381C>T | p.Q461* | Nonsense | SMD |
| 280 | Ivanoski et al., 2018/(P74) | 10 | ZEB2 gene Exon 10 deletion (3′UTR c.3642+750) | - | Large deletion | - |
| 281 | Ivanoski et al., 2018/(P75) | 8 | c.1946del | p.I649Tfs*17 | Small deletion, Frameshift | HD |
| 282 | Ivanoski et al., 2018/(P76) | 2 | c.32dup | p.R11Pfs*9 | Small insertion, Frameshift | - |
| 283 | Ivanoski et al., 2018/(P77) | 6 | c.761del | p.Q255Sfs*7 | Small deletion, Frameshift | N-ZFa |
| 284 | Ivanoski et al., 2018/(P78) | 8 | c.1553del | p.H518Lfs*26 | Small deletion, Frameshift | - |
| 285 | Ivanoski et al., 2018/(P79) | 8 | c.936C>A | p.C312* | Nonsense | N-ZFc |
| 286 | Ivanoski et al., 2018/(P80) | 3 | c.187_228dup | p.A77Lfs*13 | Small insertion, Frameshift | - |
| 287 | Ivanoski et al., 2018/(P81) | 8 | c.1150C>T | p.Q384* | Nonsense | - |
| 288 | Ivanoski et al., 2018/(P82) | 5 | c.553_554ins | p.R185Lfs*28 | Small insertion, Frameshift | - |
| 289 | Ivanoski et al., 2018/(P83) | 7 | c.857_858del | p.E286Vfs*8 | Small deletion, Frameshift | N-ZFb |
| 290 | Ivanoski et al., 2018/(P84) | 10 | c.3567_3568ins | p.M1190Pfs*52 | Small insertion, Frameshift | - |
| 291 | Ivanoski et al., 2018/(P85) | 8 | c.2677del | p.P893Lfs*37 | Small deletion, Frameshift | - |
| 292 | Ivanoski et al., 2018/(P86) | 8 | c.2372del | p.T791Nfs*26 | Small deletion, Frameshift | CID |
| 293 | Ivanoski et al., 2018/(P87) | 6 | c.715del | p.E239Rfs*23 | Small deletion, Frameshift | N-ZFa |
| 294 | Ivanoski et al., 2018/(P88) | IVS1 | c.-69-2A>C | p.M1_N24delins | Splicing | - |
| 295 | Ivanoski et al., 2018/(P89) | 8 | c.1578_1579delins | p.D527Tfs*17 | Small indel, Frameshift | - |
| 296 | [12] Ghoumid et al., 2013/(P1) | 10 | c.3134A>G | p.His1045Arg | Missense | C-ZFb |
| 297 | Ghoumid et al., 2013/(P2) | 10 | c.3164A>G | p.Tyr1055Cys | Missense | C-ZFb |
| 298 | Ghoumid et al., 2013/(P3) | 10 | c.3211T>C | p.Ser1071Pro | Missense | C-ZFb |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
St. Peter, C.; Hossain, W.A.; Lovell, S.; Rafi, S.K.; Butler, M.G. Mowat–Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions. Int. J. Mol. Sci. 2024, 25, 2838. https://doi.org/10.3390/ijms25052838
St. Peter C, Hossain WA, Lovell S, Rafi SK, Butler MG. Mowat–Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions. International Journal of Molecular Sciences. 2024; 25(5):2838. https://doi.org/10.3390/ijms25052838
Chicago/Turabian StyleSt. Peter, Caroline, Waheeda A. Hossain, Scott Lovell, Syed K. Rafi, and Merlin G. Butler. 2024. "Mowat–Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions" International Journal of Molecular Sciences 25, no. 5: 2838. https://doi.org/10.3390/ijms25052838
APA StyleSt. Peter, C., Hossain, W. A., Lovell, S., Rafi, S. K., & Butler, M. G. (2024). Mowat–Wilson Syndrome: Case Report and Review of ZEB2 Gene Variant Types, Protein Defects and Molecular Interactions. International Journal of Molecular Sciences, 25(5), 2838. https://doi.org/10.3390/ijms25052838






