CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3
Abstract
1. Introduction
2. Results
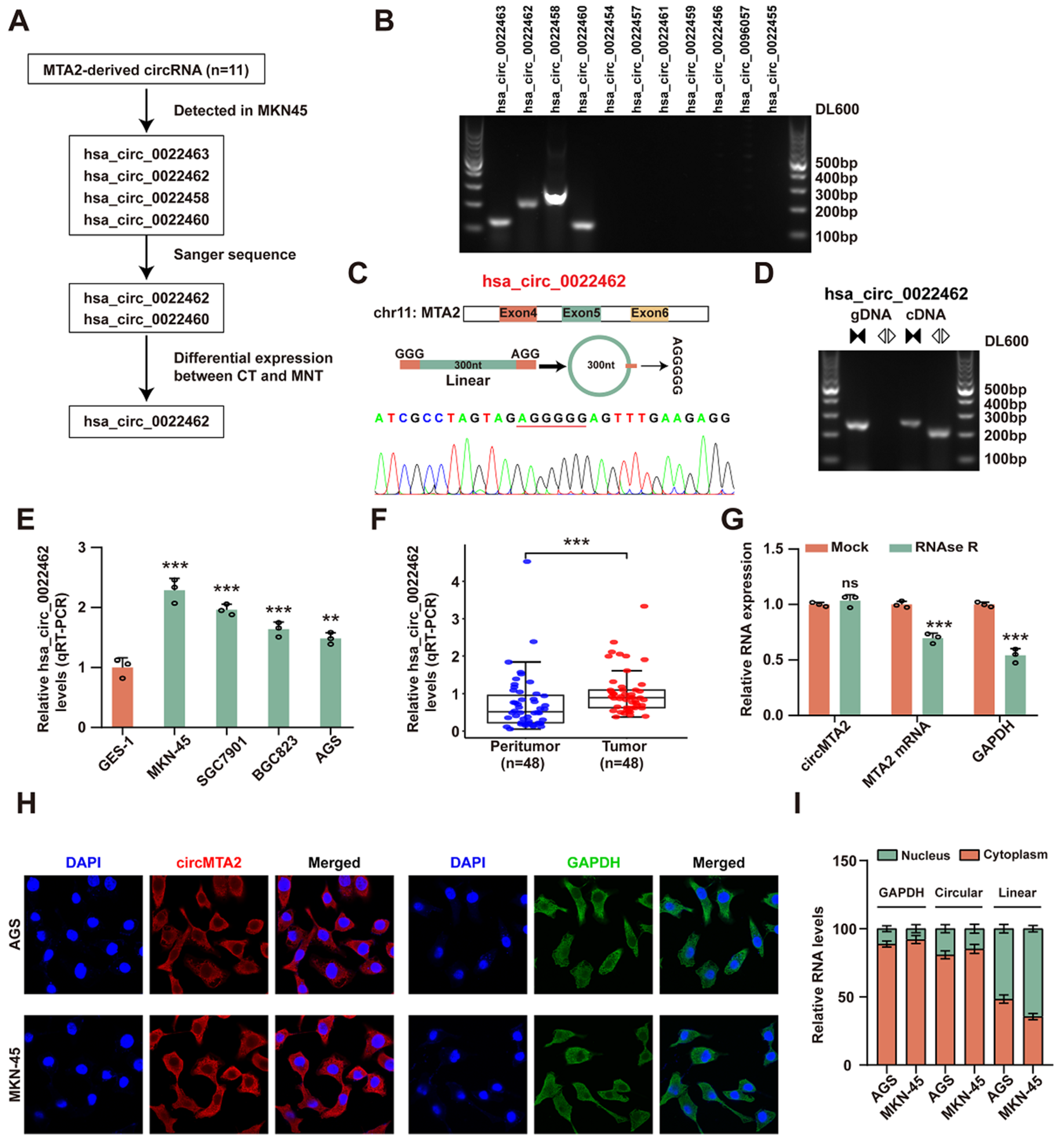
2.1. CircMTA2 Is Upregulated in the GC Tissue and Cells
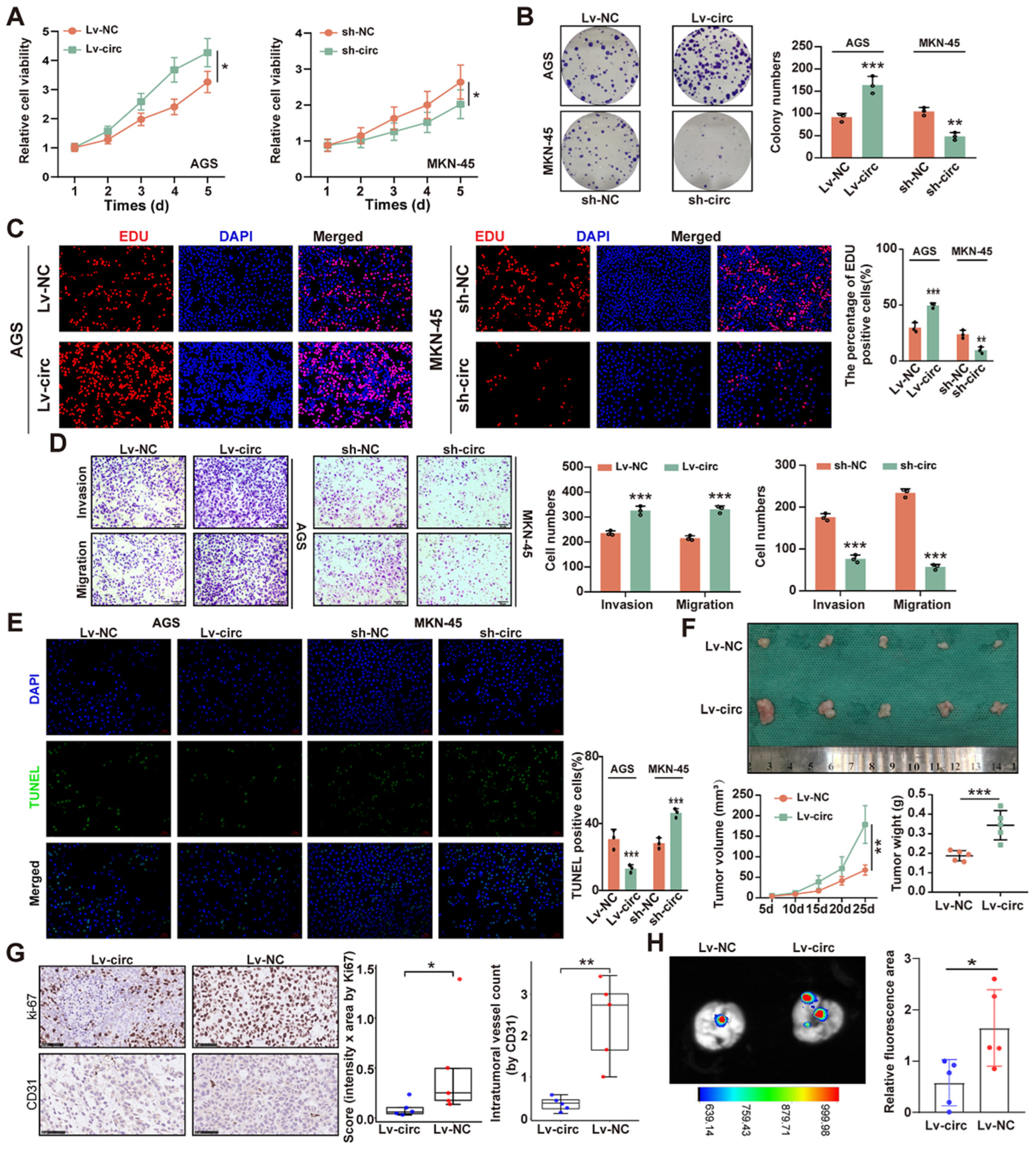
2.2. CircMTA2 Promotes the Growth and Aggressiveness of GC
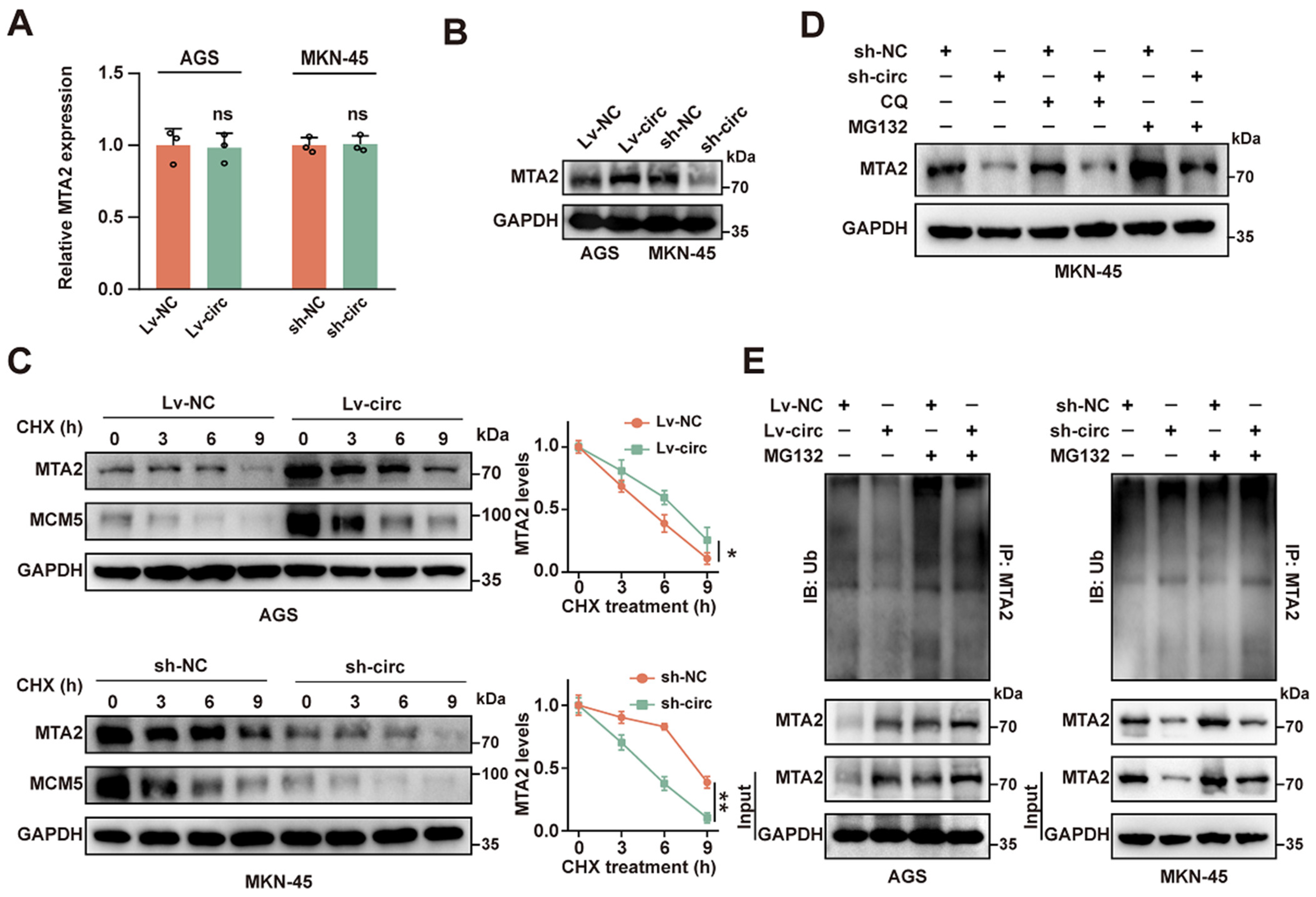
2.3. CircMTA2 Stabilities MTA2 Protein by Inhibiting Its Ubiquitination
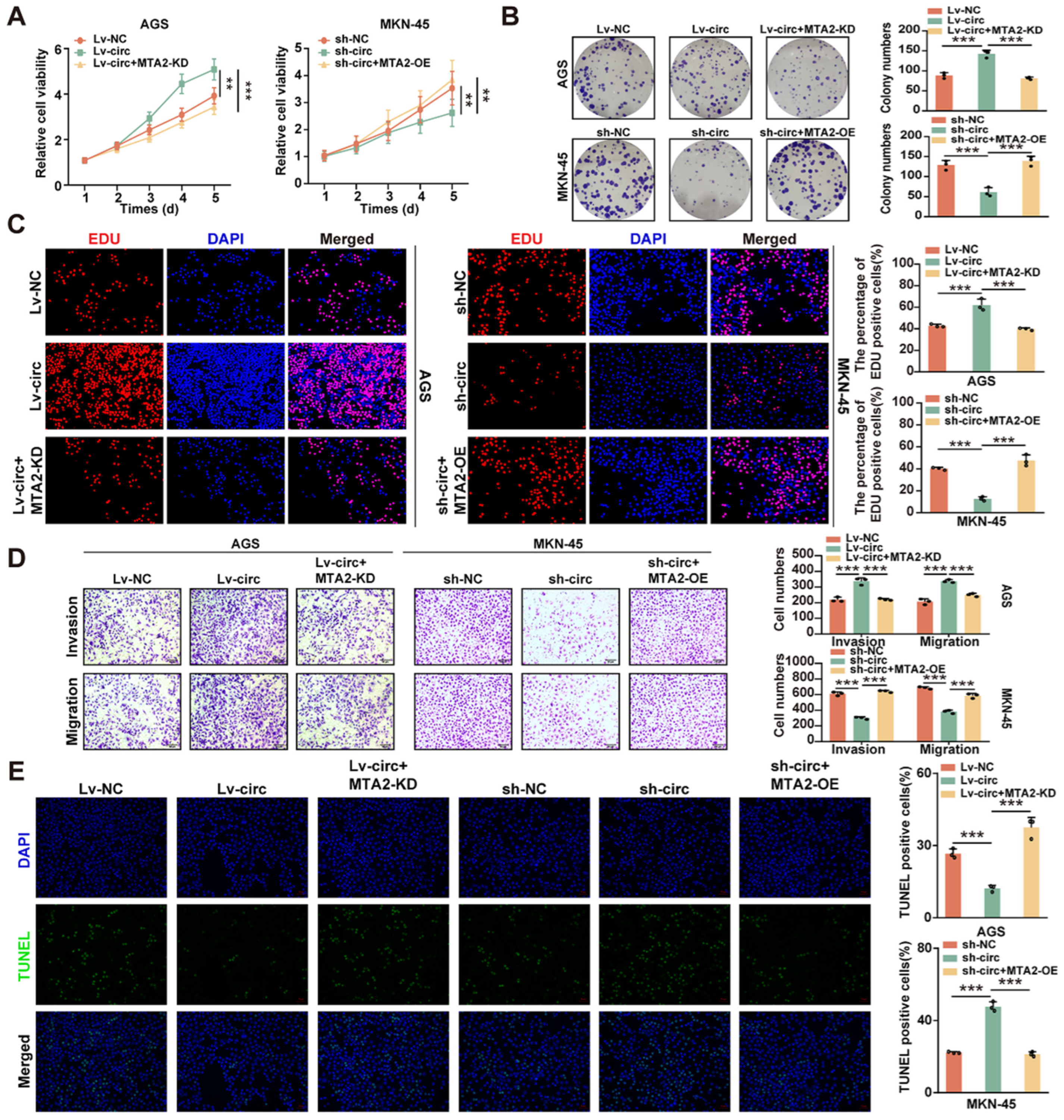
2.4. CircMTA2 Promotes Growth and Aggressiveness of GC by Upregulating MTA2 Expression
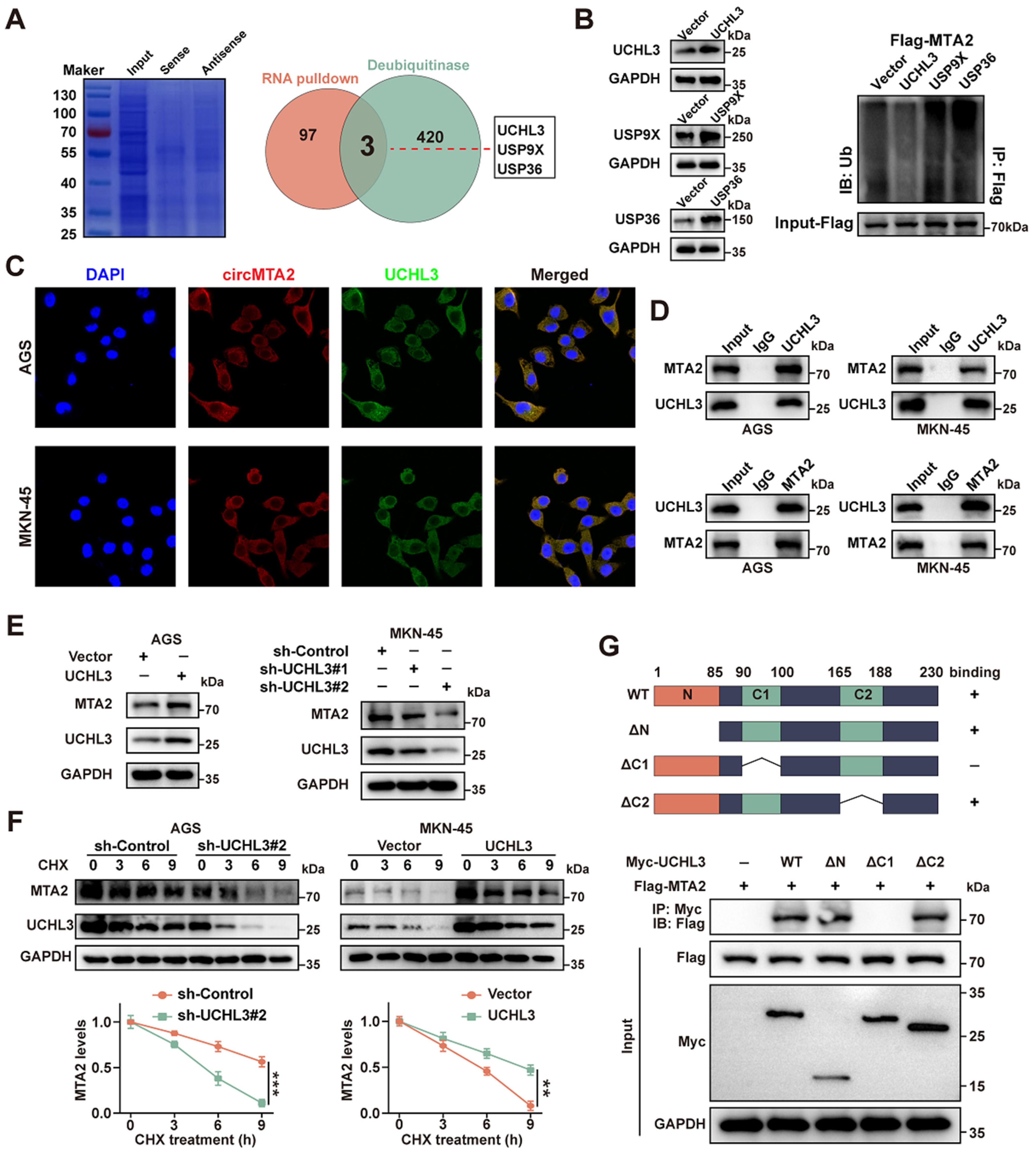
2.5. CircMTA2 Promotes MTA2 Expression via Interacting with UCHL3 in GC Cells
2.6. Exosomal circMTA2 Derived from GC Cells Enhances Cancer Cell Progression
3. Materials and Methods
3.1. Cell Lines and Treatment
3.2. Patient Tissue Samples
3.3. Fluorescence In Situ Hybridization (FISH)
3.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
3.5. RNase R and Actinomycin D Assays
3.6. RNA-Protein Immunoprecipitation (RIP) Assay
3.7. RNA Pulldown Assay
3.8. Western Blotting Analysis
3.9. CCK-8 Assay
3.10. Colony Formation Assay
3.11. 5-Ethynyl-20-Deoxyuridine (EdU) Analysis
3.12. Terminal Deoxynucleotidyl Transferase (TdT)-Mediated dUTP Biotin Nick End Labeling (TUNEL) Staining
3.13. Transwell Assay
3.14. Xenograft Assay
3.15. Immunohistochemical (IHC) Assay
3.16. Exosome Isolation and Characterization
4. Statistical Analysis
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 167–192. [Google Scholar] [CrossRef]
- Puliga, E.; Corso, S.; Pietrantonio, F.; Giordano, S. Microsatellite instability in Gastric Cancer: Between lights and shadows. Cancer Treat. Rev. 2021, 95, 102175. [Google Scholar] [CrossRef]
- Kakeji, Y.; Ishikawa, T.; Suzuki, S.; Akazawa, K.; Irino, T.; Miyashiro, I.; Ono, H.; Suzuki, H.; Tanabe, S.; Kadowaki, S.; et al. A retrospective 5-year survival analysis of surgically resected gastric cancer cases from the Japanese Gastric Cancer Association nationwide registry (2001–2013). Gastric Cancer 2022, 25, 1082–1093. [Google Scholar] [CrossRef]
- Chung, H.; Ko, Y.; Lee, I.S.; Hur, H.; Huh, J.; Han, S.U.; Kim, K.W.; Lee, J. Prognostic artificial intelligence model to predict 5 year survival at 1 year after gastric cancer surgery based on nutrition and body morphometry. J. Cachexia Sarcopenia Muscle 2023, 14, 847–859. [Google Scholar] [CrossRef]
- Patop, I.L.; Wust, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, W.; Zhou, Q.; Chen, C.; Yuan, W.; Liu, J.; Li, X.; Sun, Z. Roles of circRNAs in the tumour microenvironment. Mol. Cancer 2020, 19, 14. [Google Scholar] [CrossRef]
- Yu, T.; Wang, Y.; Fan, Y.; Fang, N.; Wang, T.; Xu, T.; Shu, Y. CircRNAs in cancer metabolism: A review. J. Hematol. Oncol. 2019, 12, 90. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, K.; Cui, J.; Xiong, J.; Wu, H.; Peng, T.; Guo, Y. Circ-MBOAT2 knockdown represses tumor progression and glutamine catabolism by miR-433-3p/GOT1 axis in pancreatic cancer. J. Exp. Clin. Cancer Res. 2021, 40, 124. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Wang, M.; Dong, Y.; Xu, B.; Chen, J.; Ding, Y.; Qiu, S.; Li, L.; Karamfilova Zaharieva, E.; Zhou, X.; et al. Circular RNA circRNF20 promotes breast cancer tumorigenesis and Warburg effect through miR-487a/HIF-1alpha/HK2. Cell Death Dis. 2020, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Hu, A.; Li, D.; Wang, J.; Guo, Y.; Liu, Y.; Li, H.; Chen, Y.; Wang, X.; Huang, K.; et al. Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer 2019, 18, 158. [Google Scholar] [CrossRef]
- Chen, Y.S.; Hung, T.W.; Su, S.C.; Lin, C.L.; Yang, S.F.; Lee, C.C.; Yeh, C.F.; Hsieh, Y.H.; Tsai, J.P. MTA2 as a Potential Biomarker and Its Involvement in Metastatic Progression of Human Renal Cancer by miR-133b Targeting MMP-9. Cancers 2019, 11, 1851. [Google Scholar] [CrossRef]
- Covington, K.R.; Fuqua, S.A. Role of MTA2 in human cancer. Cancer Metastasis Rev. 2014, 33, 921–928. [Google Scholar] [CrossRef]
- Zhang, H.; Chang, Z.; Qin, L.N.; Liang, B.; Han, J.X.; Qiao, K.L.; Yang, C.; Liu, Y.R.; Zhou, H.G.; Sun, T. MTA2 triggered R-loop trans-regulates BDH1-mediated beta-hydroxybutyrylation and potentiates propagation of hepatocellular carcinoma stem cells. Signal Transduct. Target. Ther. 2021, 6, 135. [Google Scholar] [CrossRef]
- Li, A.; Guo, Y.; Yin, Z.; Liu, X.; Xie, G. MTA2 is one of 14 Transcription factors predicting recurrence free survival in gastric cancer and promotes cancer progression by targeting MCM5. J. Cancer 2023, 14, 262–274. [Google Scholar] [CrossRef]
- Watanabe, Y.; Taguchi, K.; Tanaka, M. Ubiquitin, Autophagy and Neurodegenerative Diseases. Cells 2020, 9, 2022. [Google Scholar] [CrossRef]
- Yan, B.; Zhang, Y.; Liang, C.; Liu, B.; Ding, F.; Wang, Y.; Zhu, B.; Zhao, R.; Yu, X.Y.; Li, Y. Stem cell-derived exosomes prevent pyroptosis and repair ischemic muscle injury through a novel exosome/circHIPK3/ FOXO3a pathway. Theranostics 2020, 10, 6728–6742. [Google Scholar] [CrossRef]
- Wang, X.; Chen, T.; Li, C.; Li, W.; Zhou, X.; Li, Y.; Luo, D.; Zhang, N.; Chen, B.; Wang, L.; et al. CircRNA-CREIT inhibits stress granule assembly and overcomes doxorubicin resistance in TNBC by destabilizing PKR. J. Hematol. Oncol. 2022, 15, 122. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, F.; Fang, E.; Xiao, W.; Mei, H.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019, 26, 1346–1364. [Google Scholar] [CrossRef]
- Lu, X.; Chu, C.S.; Fang, T.; Rayon-Estrada, V.; Fang, F.; Patke, A.; Qian, Y.; Clarke, S.H.; Melnick, A.M.; Zhang, Y.; et al. MTA2/NuRD Regulates B Cell Development and Cooperates with OCA-B in Controlling the Pre-B to Immature B Cell Transition. Cell Rep. 2019, 28, 472–485.e5. [Google Scholar] [CrossRef]
- Si, W.; Liu, X.; Wei, R.; Zhang, Y.; Zhao, Y.; Cui, L.; Hong, T. MTA2-mediated inhibition of PTEN leads to pancreatic ductal adenocarcinoma carcinogenicity. Cell Death Dis. 2019, 10, 206. [Google Scholar] [CrossRef]
- Lin, C.L.; Ying, T.H.; Yang, S.F.; Chiou, H.L.; Chen, Y.S.; Kao, S.H.; Hsieh, Y.H. MTA2 silencing attenuates the metastatic potential of cervical cancer cells by inhibiting AP1-mediated MMP12 expression via the ASK1/MEK3/p38/YB1 axis. Cell Death Dis. 2021, 12, 451. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Q.; Zhou, Y.; Ouyang, Q.; Dai, W.; Chen, J.; Ding, P.; Li, L.; Zhang, X.; Zhang, W.; et al. Overexpression of MTA1 inhibits the metastatic ability of ZR-75-30 cells in vitro by promoting MTA2 degradation. Cell Commun. Signal. 2019, 17, 4. [Google Scholar] [CrossRef]
- Mofers, A.; Pellegrini, P.; Linder, S.; D’Arcy, P. Proteasome-associated deubiquitinases and cancer. Cancer Metastasis Rev. 2017, 36, 635–653. [Google Scholar] [CrossRef]
- Bonacci, T.; Emanuele, M.J. Dissenting degradation: Deubiquitinases in cell cycle and cancer. Semin. Cancer Biol. 2020, 67, 145–158. [Google Scholar] [CrossRef]
- Tang, J.; Yang, Q.; Mao, C.; Xiao, D.; Liu, S.; Xiao, L.; Zhou, L.; Wu, G.; Tao, Y. The deubiquitinating enzyme UCHL3 promotes anaplastic thyroid cancer progression and metastasis through Hippo signaling pathway. Cell Death Differ. 2023, 30, 1247–1259. [Google Scholar] [CrossRef]
- Ouyang, L.; Yan, B.; Liu, Y.; Mao, C.; Wang, M.; Liu, N.; Wang, Z.; Liu, S.; Shi, Y.; Chen, L.; et al. The deubiquitylase UCHL3 maintains cancer stem-like properties by stabilizing the aryl hydrocarbon receptor. Signal Transduct. Target. Ther. 2020, 5, 78. [Google Scholar] [CrossRef]
- Yang, R.; Chen, H.; Xing, L.; Wang, B.; Hu, M.; Ou, X.; Chen, H.; Deng, Y.; Liu, D.; Jiang, R.; et al. Hypoxia-induced circWSB1 promotes breast cancer progression through destabilizing p53 by interacting with USP10. Mol. Cancer 2022, 21, 88. [Google Scholar] [CrossRef]
- Shang, A.; Gu, C.; Wang, W.; Wang, X.; Sun, J.; Zeng, B.; Chen, C.; Chang, W.; Ping, Y.; Ji, P.; et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol. Cancer 2020, 19, 117. [Google Scholar] [CrossRef]
- Pan, Z.; Zhao, R.; Li, B.; Qi, Y.; Qiu, W.; Guo, Q.; Zhang, S.; Zhao, S.; Xu, H.; Li, M.; et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol. Cancer 2022, 21, 16. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, G.; Lei, B.; Yin, Z.; Xu, F.; Liu, X. CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3. Int. J. Mol. Sci. 2024, 25, 2817. https://doi.org/10.3390/ijms25052817
Xie G, Lei B, Yin Z, Xu F, Liu X. CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3. International Journal of Molecular Sciences. 2024; 25(5):2817. https://doi.org/10.3390/ijms25052817
Chicago/Turabian StyleXie, Gengchen, Bo Lei, Zhijie Yin, Fei Xu, and Xinghua Liu. 2024. "CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3" International Journal of Molecular Sciences 25, no. 5: 2817. https://doi.org/10.3390/ijms25052817
APA StyleXie, G., Lei, B., Yin, Z., Xu, F., & Liu, X. (2024). CircMTA2 Drives Gastric Cancer Progression through Suppressing MTA2 Degradation via Interacting with UCHL3. International Journal of Molecular Sciences, 25(5), 2817. https://doi.org/10.3390/ijms25052817





