Studying the Effect of MBNL1 and MBNL2 Loss in Skeletal Muscle Regeneration
Abstract
1. Introduction
2. Results
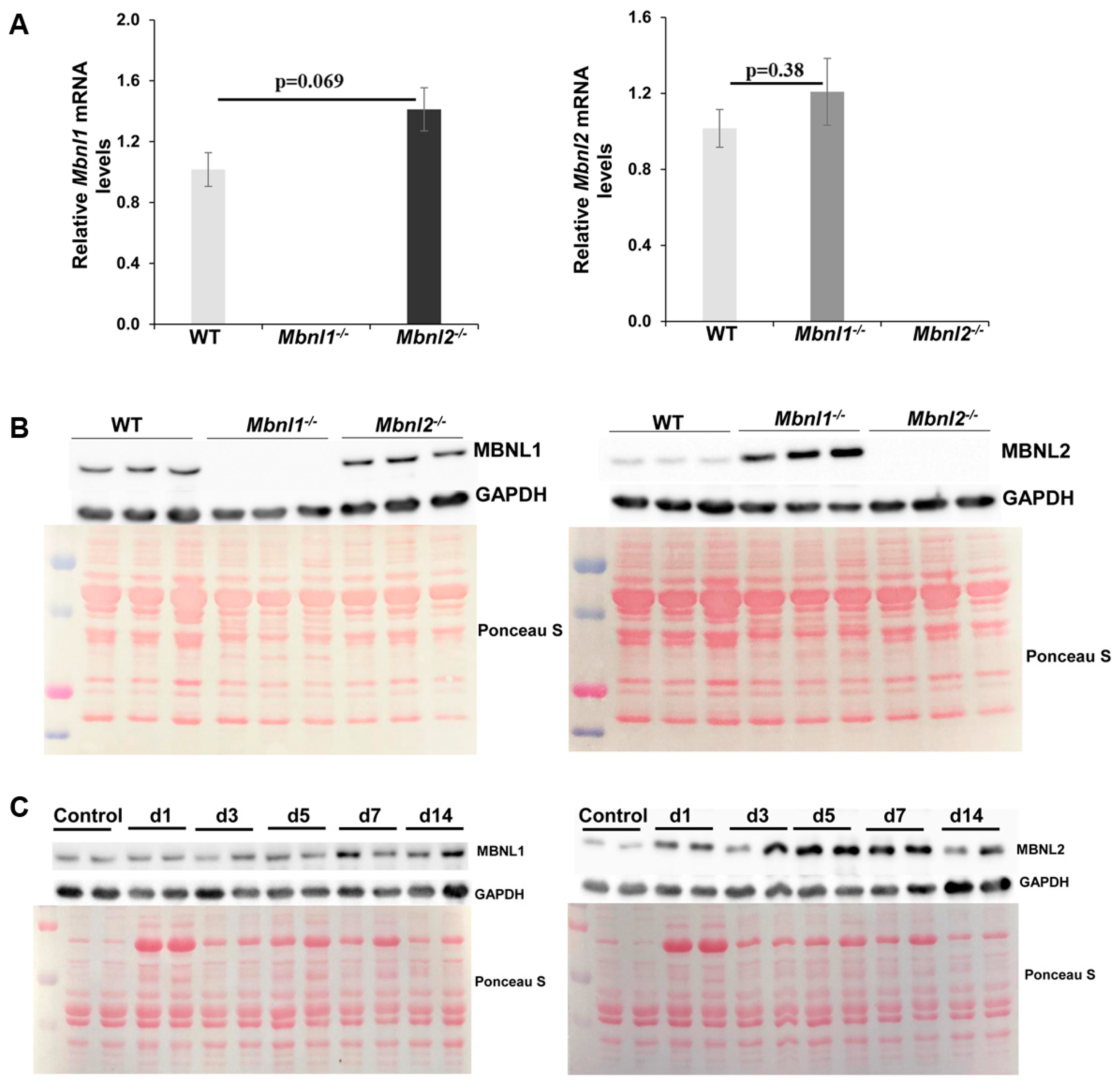
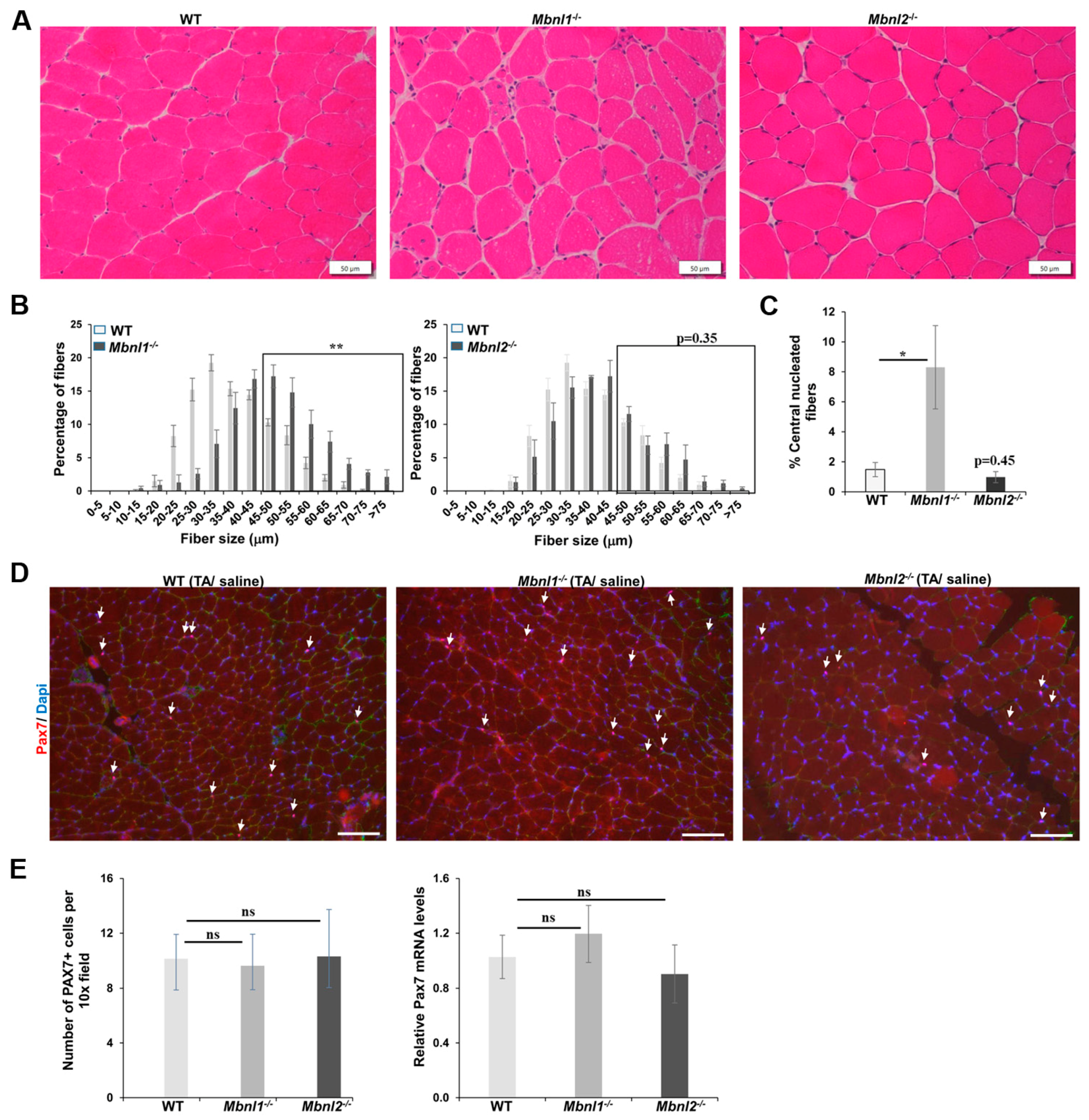
2.1. MBNLs, Muscle Histology, and Fiber Size Analysis
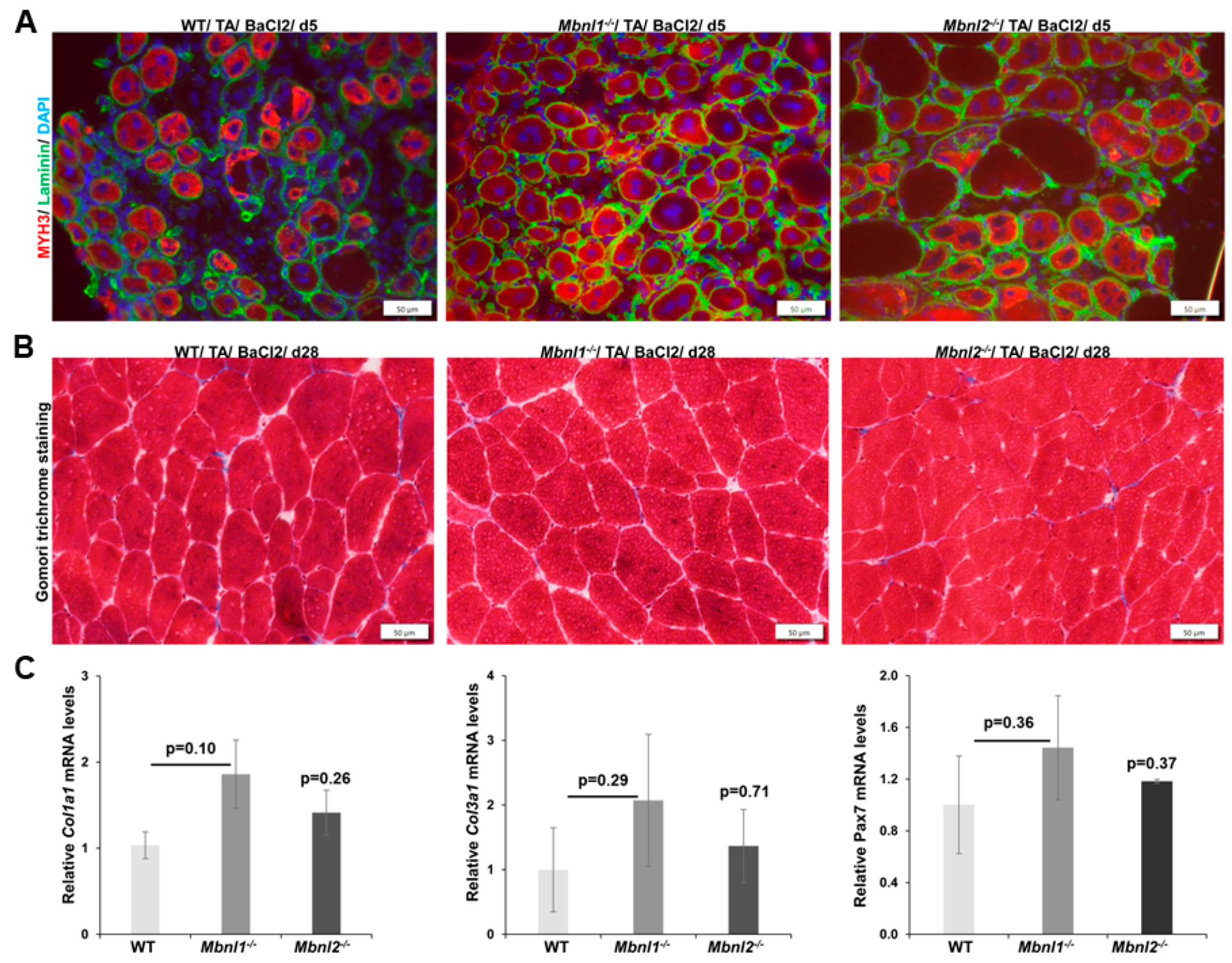
2.2. MBNLs and Skeletal Muscle Regeneration
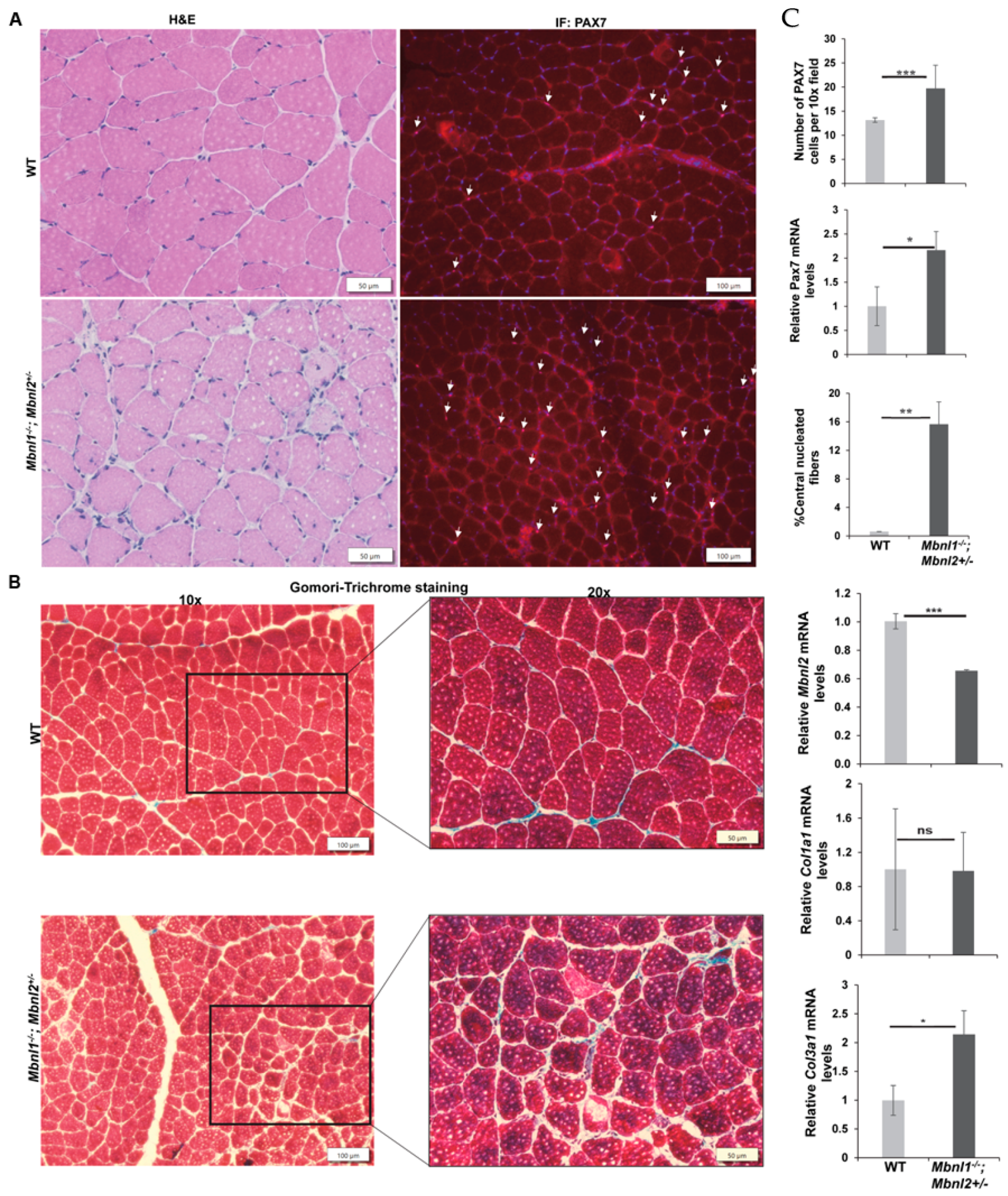
2.3. Combined MBNL1 Loss with Hemizygous Loss of MBNL2
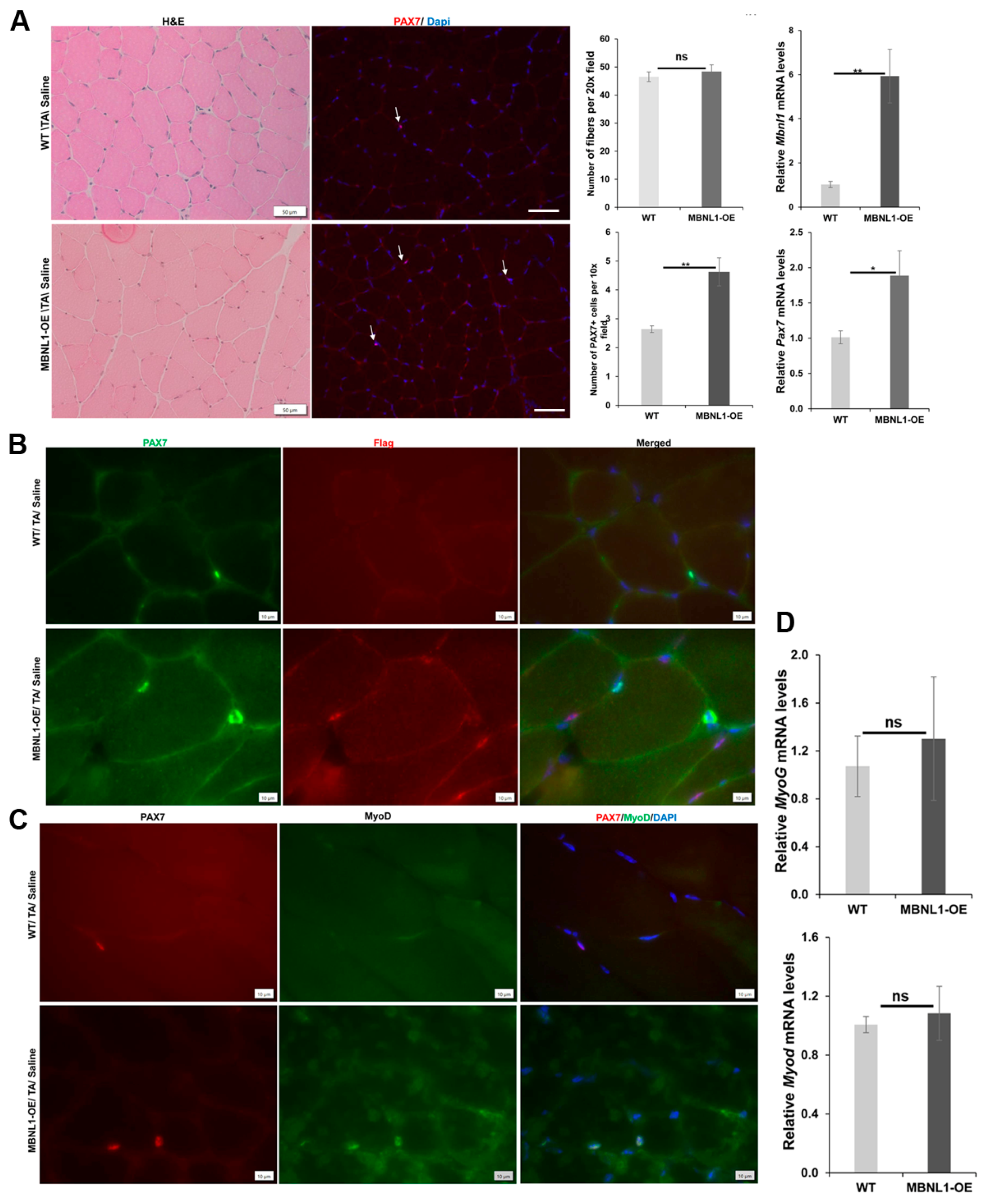
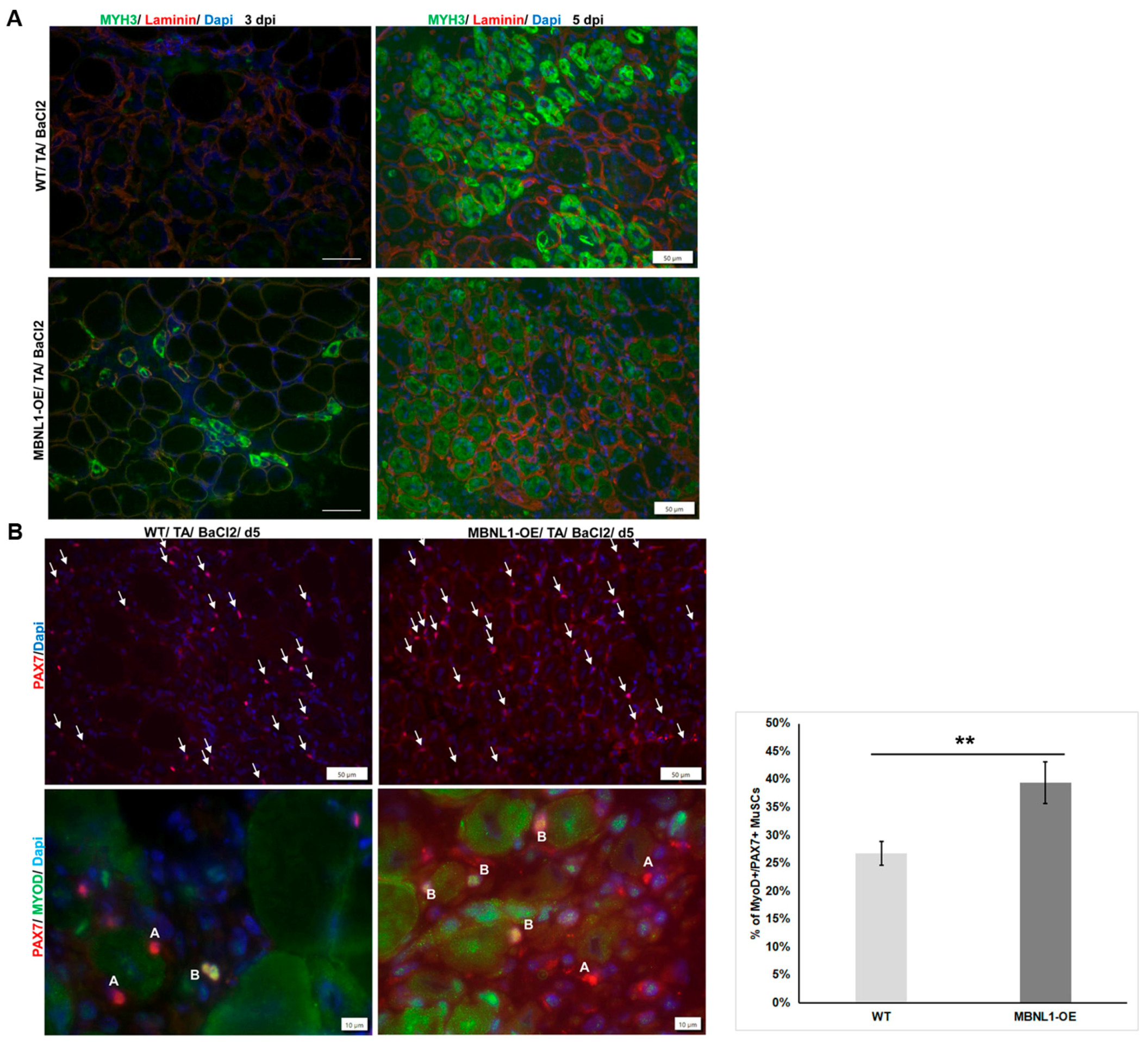
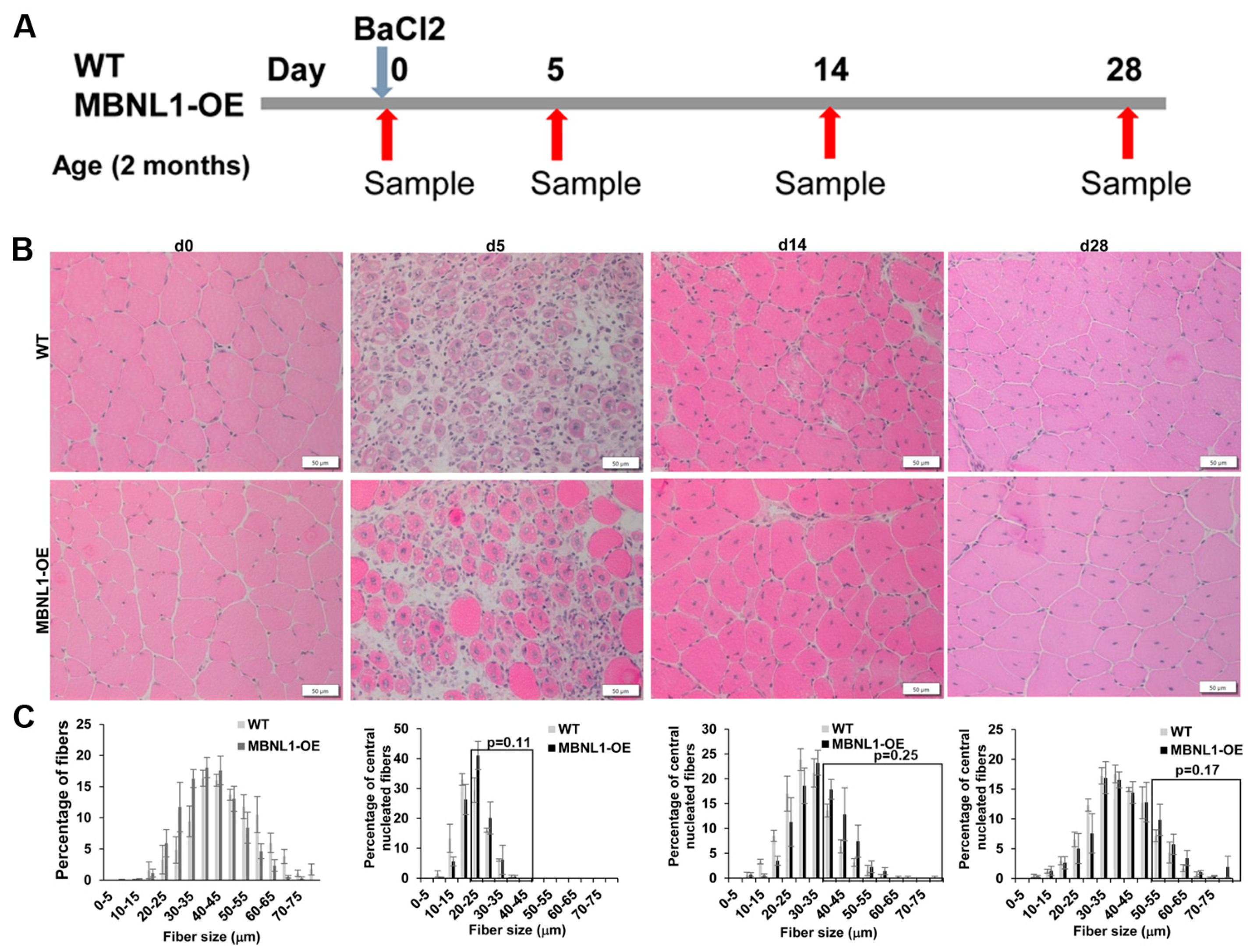
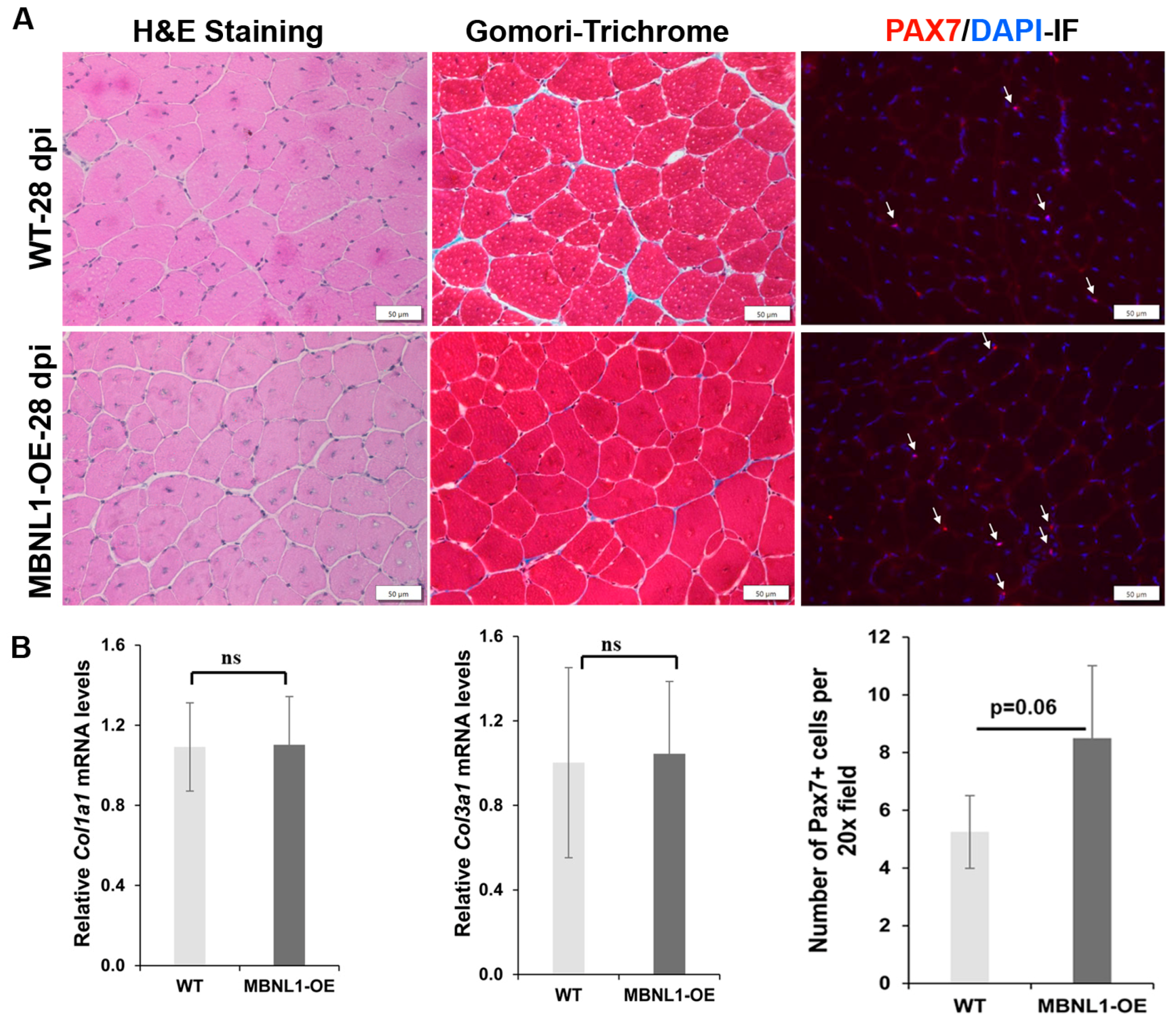
2.4. MBNL1 Overexpression and Skeletal Muscle Regeneration
3. Discussion
4. Materials and Methods
4.1. Experimental Mice
4.2. Muscle Regeneration
4.3. H&E and Gomori’s Trichrome Staining of Skeletal Muscles
4.4. Muscle Fiber Analysis
4.5. RNA Isolation and qRT-PCR Assays
4.6. Immunofluorescence and Western Blot
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brook, J.D.; McCurrach, M.E.; Harley, H.G.; Buckler, A.J.; Church, D.; Aburatani, H.; Hunter, K.; Stanton, V.P.; Thirion, J.P.; Hudson, T.; et al. Molecular basis of myotonic dystrophy: Expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell 1992, 69, 385. [Google Scholar] [CrossRef]
- Mahadevan, M.; Tsilfidis, C.; Sabourin, L.; Shutler, G.; Amemiya, C.; Jansen, G.; Neville, C.; Narang, M.; Barcelo, J.; O’Hoy, K.; et al. Myotonic dystrophy mutation: An unstable CTG repeat in the 3’ untranslated region of the gene. Science 1992, 255, 1253–1255. [Google Scholar] [CrossRef]
- Taneja, K.L.; McCurrach, M.; Schalling, M.; Housman, D.; Singer, R.H. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J. Cell Biol. 1995, 128, 995–1002. [Google Scholar] [CrossRef]
- Davis, B.M.; McCurrach, M.E.; Taneja, K.L.; Singer, R.H.; Housman, D.E. Expansion of a CUG trinucleotide repeat in the 3’ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc. Natl. Acad. Sci. USA 1997, 94, 7388–7393. [Google Scholar] [CrossRef]
- Mahadevan, M.S.A.J.D.; Paguio, A.P. A cell culture model of the myotonic dystrophy (DM) mutation. In Proceedings of the Annual Meeting of the American Society of Human Genetics, Denver, CO, USA, 27–31 October 1998. [Google Scholar]
- Amack, J.D.; Paguio, A.P.; Mahadevan, M.S. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum. Mol. Genet. 1999, 8, 1975–1984. [Google Scholar] [CrossRef]
- Mahadevan, M.S.; Yadava, R.S.; Yu, Q.; Balijepalli, S.; Frenzel-McCardell, C.D.; Bourne, T.D.; Phillips, L.H. Reversible model of RNA toxicity and cardiac conduction defects in myotonic dystrophy. Nat. Genet. 2006, 38, 1066–1070. [Google Scholar] [CrossRef] [PubMed]
- Mankodi, A.; Logigian, E.; Callahan, L.; McClain, C.; White, R.; Henderson, D.; Krym, M.; Thornton, C.A. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science 2000, 289, 1769–1773. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.M.; Leger, A.J.; Pandey, S.K.; MacLeod, A.R.; Nakamori, M.; Cheng, S.H.; Wentworth, B.M.; Bennett, C.F.; Thornton, C.A. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature 2012, 488, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Echeverria, G.V.; Cooper, T.A. RNA-binding proteins in microsatellite expansion disorders: Mediators of RNA toxicity. Brain Res. 2012, 1462, 100–111. [Google Scholar] [CrossRef]
- Braz, S.O.; Acquaire, J.; Gourdon, G.; Gomes-Pereira, M. Of Mice and Men: Advances in the Understanding of Neuromuscular Aspects of Myotonic Dystrophy. Front. Neurol. 2018, 9, 519. [Google Scholar] [CrossRef]
- Gladman, J.T.; Yadava, R.S.; Mandal, M.; Yu, Q.; Kim, Y.K.; Mahadevan, M.S. NKX2-5, a modifier of skeletal muscle pathology due to RNA toxicity. Hum. Mol. Genet. 2015, 24, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Wei, C.; Iakova, P.; Bugiardini, E.; Schneider-Gold, C.; Meola, G.; Woodgett, J.; Killian, J.; Timchenko, N.A.; Timchenko, L.T. GSK3beta mediates muscle pathology in myotonic dystrophy. J. Clin. Investig. 2012, 122, 4461–4472. [Google Scholar] [CrossRef] [PubMed]
- Kuyumcu-Martinez, N.M.; Wang, G.S.; Cooper, T.A. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell 2007, 28, 68–78. [Google Scholar] [CrossRef]
- Morriss, G.R.; Rajapakshe, K.; Huang, S.; Coarfa, C.; Cooper, T.A. Mechanisms of skeletal muscle wasting in a mouse model for myotonic dystrophy type 1. Hum. Mol. Genet. 2018, 27, 2789–2804. [Google Scholar] [CrossRef]
- Yadava, R.S.; Foff, E.P.; Yu, Q.; Gladman, J.T.; Zheng, T.S.; Mahadevan, M.S. TWEAK Regulates Muscle Functions in a Mouse Model of RNA Toxicity. PLoS ONE 2016, 11, e0150192. [Google Scholar] [CrossRef] [PubMed]
- Yadava, R.S.; Frenzel-McCardell, C.D.; Yu, Q.; Srinivasan, V.; Tucker, A.L.; Puymirat, J.; Thornton, C.A.; Prall, O.W.; Harvey, R.P.; Mahadevan, M.S. RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression. Nat. Genet. 2008, 40, 61–68. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bigot, A.; Klein, A.F.; Gasnier, E.; Jacquemin, V.; Ravassard, P.; Butler-Browne, G.; Mouly, V.; Furling, D. Large CTG repeats trigger p16-dependent premature senescence in myotonic dystrophy type 1 muscle precursor cells. Am. J. Pathol. 2009, 174, 1435–1442. [Google Scholar] [CrossRef]
- Cisco, L.A.; Sipple, M.T.; Edwards, K.M.; Thornton, C.A.; Lueck, J.D. Verapamil mitigates chloride and calcium bi-channelopathy in a myotonic dystrophy mouse model. J. Clin. Investig. 2024, 134, e173576. [Google Scholar] [CrossRef]
- Conte, T.C.; Duran-Bishop, G.; Orfi, Z.; Mokhtari, I.; Deprez, A.; Cote, I.; Molina, T.; Kim, T.Y.; Tellier, L.; Roussel, M.P.; et al. Clearance of defective muscle stem cells by senolytics restores myogenesis in myotonic dystrophy type 1. Nat. Commun. 2023, 14, 4033. [Google Scholar] [CrossRef]
- Czubak, K.; Sedehizadeh, S.; Kozlowski, P.; Wojciechowska, M. An Overview of Circular RNAs and Their Implications in Myotonic Dystrophy. Int. J. Mol. Sci. 2019, 20, 4385. [Google Scholar] [CrossRef]
- Czubak, K.; Taylor, K.; Piasecka, A.; Sobczak, K.; Kozlowska, K.; Philips, A.; Sedehizadeh, S.; Brook, J.D.; Wojciechowska, M.; Kozlowski, P. Global Increase in Circular RNA Levels in Myotonic Dystrophy. Front. Genet. 2019, 10, 649. [Google Scholar] [CrossRef]
- Di Leo, V.; Lawless, C.; Roussel, M.P.; Gomes, T.B.; Gorman, G.S.; Russell, O.M.; Tuppen, H.A.L.; Duchesne, E.; Vincent, A.E. Resistance Exercise Training Rescues Mitochondrial Dysfunction in Skeletal Muscle of Patients with Myotonic Dystrophy Type 1. J. Neuromuscul. Dis. 2023, 10, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Hasuike, Y.; Mochizuki, H.; Nakamori, M. Cellular Senescence and Aging in Myotonic Dystrophy. Int. J. Mol. Sci. 2022, 23, 2339. [Google Scholar] [CrossRef]
- Li, M.; Zhuang, Y.; Batra, R.; Thomas, J.D.; Li, M.; Nutter, C.A.; Scotti, M.M.; Carter, H.A.; Wang, Z.J.; Huang, X.S.; et al. HNRNPA1-induced spliceopathy in a transgenic mouse model of myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2020, 117, 5472–5477. [Google Scholar] [CrossRef]
- Mikhail, A.I.; Nagy, P.L.; Manta, K.; Rouse, N.; Manta, A.; Ng, S.Y.; Nagy, M.F.; Smith, P.; Lu, J.Q.; Nederveen, J.P.; et al. Aerobic exercise elicits clinical adaptations in myotonic dystrophy type 1 patients independently of pathophysiological changes. J. Clin. Investig. 2022, 132, e156125. [Google Scholar] [CrossRef]
- Ravel-Chapuis, A.; Duchesne, E.; Jasmin, B.J. Pharmacological and exercise-induced activation of AMPK as emerging therapies for myotonic dystrophy type 1 patients. J. Physiol. 2022, 600, 3249–3264. [Google Scholar] [CrossRef] [PubMed]
- Sabater-Arcis, M.; Moreno, N.; Sevilla, T.; Perez Alonso, M.; Bargiela, A.; Artero, R. Msi2 enhances muscle dysfunction in a myotonic dystrophy type 1 mouse model. Biomed. J. 2023, 100667. [Google Scholar] [CrossRef]
- Voellenkle, C.; Perfetti, A.; Carrara, M.; Fuschi, P.; Renna, L.V.; Longo, M.; Sain, S.B.; Cardani, R.; Valaperta, R.; Silvestri, G.; et al. Dysregulation of Circular RNAs in Myotonic Dystrophy Type 1. Int. J. Mol. Sci. 2019, 20, 1938. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Stock, L.; Valanejad, L.; Zalewski, Z.A.; Karns, R.; Puymirat, J.; Nelson, D.; Witte, D.; Woodgett, J.; Timchenko, N.A.; et al. Correction of GSK3beta at young age prevents muscle pathology in mice with myotonic dystrophy type 1. FASEB J. 2018, 32, 2073–2085. [Google Scholar] [CrossRef]
- Gambardella, S.; Rinaldi, F.; Lepore, S.M.; Viola, A.; Loro, E.; Angelini, C.; Vergani, L.; Novelli, G.; Botta, A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J. Transl. Med. 2010, 8, 48. [Google Scholar] [CrossRef]
- Perbellini, R.; Greco, S.; Sarra-Ferraris, G.; Cardani, R.; Capogrossi, M.C.; Meola, G.; Martelli, F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul. Disord. NMD 2011, 21, 81–88. [Google Scholar] [CrossRef]
- Perfetti, A.; Greco, S.; Cardani, R.; Fossati, B.; Cuomo, G.; Valaperta, R.; Ambrogi, F.; Cortese, A.; Botta, A.; Mignarri, A.; et al. Validation of plasma microRNAs as biomarkers for myotonic dystrophy type 1. Sci. Rep. 2016, 6, 38174. [Google Scholar] [CrossRef]
- Cerro-Herreros, E.; Fernandez-Costa, J.M.; Sabater-Arcis, M.; Llamusi, B.; Artero, R. Derepressing muscleblind expression by miRNA sponges ameliorates myotonic dystrophy-like phenotypes in Drosophila. Sci. Rep. 2016, 6, 36230. [Google Scholar] [CrossRef]
- Miller, J.W.; Urbinati, C.R.; Teng-Umnuay, P.; Stenberg, M.G.; Byrne, B.J.; Thornton, C.A.; Swanson, M.S. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000, 19, 4439–4448. [Google Scholar] [CrossRef] [PubMed]
- Kanadia, R.N.; Johnstone, K.A.; Mankodi, A.; Lungu, C.; Thornton, C.A.; Esson, D.; Timmers, A.M.; Hauswirth, W.W.; Swanson, M.S. A muscleblind knockout model for myotonic dystrophy. Science 2003, 302, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y.; Li, M.; Manchanda, M.; Batra, R.; Charizanis, K.; Mohan, A.; Warren, S.A.; Chamberlain, C.M.; Finn, D.; Hong, H.; et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol. Med. 2013, 5, 1887–1900. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.D.; Sznajder, L.J.; Bardhi, O.; Aslam, F.N.; Anastasiadis, Z.P.; Scotti, M.M.; Nishino, I.; Nakamori, M.; Wang, E.T.; Swanson, M.S. Disrupted prenatal RNA processing and myogenesis in congenital myotonic dystrophy. Genes. Dev. 2017, 31, 1122–1133. [Google Scholar] [CrossRef]
- Grimby, G.; Hedberg, M.; Henriksson, K.G.; Johansson, G.; Wigerstad-Lossing, I.; Sellden, U.; Orndahl, G. Muscle function and morphology in myotonic dystrophy. Acta Medica Scand. 1988, 224, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Andre, L.M.; Ausems, C.R.M.; Wansink, D.G.; Wieringa, B. Abnormalities in Skeletal Muscle Myogenesis, Growth, and Regeneration in Myotonic Dystrophy. Front. Neurol. 2018, 9, 368. [Google Scholar] [CrossRef]
- Mateos-Aierdi, A.J.; Goicoechea, M.; Aiastui, A.; Fernandez-Torron, R.; Garcia-Puga, M.; Matheu, A.; Lopez de Munain, A. Muscle wasting in myotonic dystrophies: A model of premature aging. Front. Aging Neurosci. 2015, 7, 125. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, L. Molecular mechanisms of muscle atrophy in myotonic dystrophies. Int. J. Biochem. Cell Biol. 2013, 45, 2280–2287. [Google Scholar] [CrossRef]
- Blau, H.M.; Cosgrove, B.D.; Ho, A.T. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015, 21, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Yadava, R.S.; Mandal, M.; Giese, J.M.; Rigo, F.; Bennett, C.F.; Mahadevan, M.S. Modeling muscle regeneration in RNA toxicity mice. Hum. Mol. Genet. 2021, 30, 1111–1130. [Google Scholar] [CrossRef]
- Yadava, R.S.; Yu, Q.; Mandal, M.; Rigo, F.; Bennett, C.F.; Mahadevan, M.S. Systemic therapy in an RNA toxicity mouse model with an antisense oligonucleotide therapy targeting a non-CUG sequence within the DMPK 3’UTR RNA. Hum. Mol. Genet. 2020, 29, 1440–1453. [Google Scholar] [CrossRef]
- Poulos, M.G.; Batra, R.; Li, M.; Yuan, Y.; Zhang, C.; Darnell, R.B.; Swanson, M.S. Progressive impairment of muscle regeneration in muscleblind-like 3 isoform knockout mice. Hum. Mol. Genet. 2013, 22, 3547–3558. [Google Scholar] [CrossRef]
- Charizanis, K.; Lee, K.Y.; Batra, R.; Goodwin, M.; Zhang, C.; Yuan, Y.; Shiue, L.; Cline, M.; Scotti, M.M.; Xia, G.; et al. Muscleblind-like 2-mediated alternative splicing in the developing brain and dysregulation in myotonic dystrophy. Neuron 2012, 75, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Hardy, D.; Besnard, A.; Latil, M.; Jouvion, G.; Briand, D.; Thepenier, C.; Pascal, Q.; Guguin, A.; Gayraud-Morel, B.; Cavaillon, J.M.; et al. Comparative Study of Injury Models for Studying Muscle Regeneration in Mice. PLoS ONE 2016, 11, e0147198. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; Dumont, N.A.; Rudnicki, M.A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 2013, 14, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Charge, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Cooper, R.N.; Tajbakhsh, S.; Mouly, V.; Cossu, G.; Buckingham, M.; Butler-Browne, G.S. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J. Cell Sci. 1999, 112 Pt 17, 2895–2901. [Google Scholar] [CrossRef]
- Wang, Y.X.; Rudnicki, M.A. Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 2011, 13, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Chamberlain, C.M.; Ranum, L.P. Mouse model of muscleblind-like 1 overexpression: Skeletal muscle effects and therapeutic promise. Hum. Mol. Genet. 2012, 21, 4645–4654. [Google Scholar] [CrossRef] [PubMed]
- Yadava, R.S.; Kim, Y.K.; Mandal, M.; Mahadevan, K.; Gladman, J.T.; Yu, Q.; Mahadevan, M.S. MBNL1 overexpression is not sufficient to rescue the phenotypes in a mouse model of RNA toxicity. Hum. Mol. Genet. 2019, 28, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, L.; Hu, R.C.; Miller, A.N.; Lucas, L.; Cooper, T.A. Alternative splicing mediates the compensatory upregulation of MBNL2 upon MBNL1 loss-of-function. Nucleic Acids Res. 2023, 51, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Tohgi, H.; Kawamorita, A.; Utsugisawa, K.; Yamagata, M.; Sano, M. Muscle histopathology in myotonic dystrophy in relation to age and muscular weakness. Muscle Nerve 1994, 17, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Sahgal, V.; Bernes, S.; Sahgal, S.; Lischwey, C.; Subramani, V. Skeletal muscle in preterm infants with congenital myotonic dystrophy. Morphologic and histochemical study. J. Neurol. Sci. 1983, 59, 47–55. [Google Scholar] [CrossRef]
- Sarnat, H.B.; Silbert, S.W. Maturational arrest of fetal muscle in neonatal myotonic dystrophy. A pathologic study of four cases. Arch. Neurol. 1976, 33, 466–474. [Google Scholar] [CrossRef]
- Ludatscher, R.M.; Kerner, H.; Amikam, S.; Gellei, B. Myotonia dystrophica with heart involvement: An electron microscopic study of skeletal, cardiac, and smooth muscle. J. Clin. Pathol. 1978, 31, 1057–1064. [Google Scholar] [CrossRef]
- Furling, D.; Coiffier, L.; Mouly, V.; Barbet, J.P.; St Guily, J.L.; Taneja, K.; Gourdon, G.; Junien, C.; Butler-Browne, G.S. Defective satellite cells in congenital myotonic dystrophy. Hum. Mol. Genet. 2001, 10, 2079–2087. [Google Scholar] [CrossRef]
- Ishimoto, S.; Goto, I.; Ohta, M.; Kuroiwa, Y. A quantitative study of the muscle satellite cells in various neuromuscular disorders. J. Neurol. Sci. 1983, 62, 303–314. [Google Scholar] [CrossRef]
- Thornell, L.E.; Lindstom, M.; Renault, V.; Klein, A.; Mouly, V.; Ansved, T.; Butler-Browne, G.; Furling, D. Satellite cell dysfunction contributes to the progressive muscle atrophy in myotonic dystrophy type 1. Neuropathol. Appl. Neurobiol. 2009, 35, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Ganassi, M.; Zammit, P.S. Involvement of muscle satellite cell dysfunction in neuromuscular disorders: Expanding the portfolio of satellite cell-opathies. Eur. J. Transl. Myol. 2022, 32, 10064. [Google Scholar] [CrossRef] [PubMed]
- Andre, L.M.; van Cruchten, R.T.P.; Willemse, M.; Bezstarosti, K.; Demmers, J.A.A.; van Agtmaal, E.L.; Wansink, D.G.; Wieringa, B. Recovery in the Myogenic Program of Congenital Myotonic Dystrophy Myoblasts after Excision of the Expanded (CTG)n Repeat. Int. J. Mol. Sci. 2019, 20, 5685. [Google Scholar] [CrossRef] [PubMed]
- Franck, S.; Couvreu De Deckersberg, E.; Bubenik, J.L.; Markouli, C.; Barbe, L.; Allemeersch, J.; Hilven, P.; Duque, G.; Swanson, M.S.; Gheldof, A.; et al. Myotonic dystrophy type 1 embryonic stem cells show decreased myogenic potential, increased CpG methylation at the DMPK locus and RNA mis-splicing. Biol. Open 2022, 11, bio058978. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Weng, W.C.; Stock, L.; Lindquist, D.; Martinez, A.; Gourdon, G.; Timchenko, N.; Snape, M.; Timchenko, L. Correction of Glycogen Synthase Kinase 3beta in Myotonic Dystrophy 1 Reduces the Mutant RNA and Improves Postnatal Survival of DMSXL Mice. Mol. Cell Biol. 2019, 39, e00155-19. [Google Scholar] [CrossRef] [PubMed]
- Kanadia, R.N.; Shin, J.; Yuan, Y.; Beattie, S.G.; Wheeler, T.M.; Thornton, C.A.; Swanson, M.S. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc. Natl. Acad. Sci. USA 2006, 103, 11748–11753. [Google Scholar] [CrossRef] [PubMed]
- Nakamori, M.; Sobczak, K.; Puwanant, A.; Welle, S.; Eichinger, K.; Pandya, S.; Dekdebrun, J.; Heatwole, C.R.; McDermott, M.P.; Chen, T.; et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann. Neurol. 2013, 74, 862–872. [Google Scholar] [CrossRef]
- Cardani, R.; Baldassa, S.; Botta, A.; Rinaldi, F.; Novelli, G.; Mancinelli, E.; Meola, G. Ribonuclear inclusions and MBNL1 nuclear sequestration do not affect myoblast differentiation but alter gene splicing in myotonic dystrophy type 2. Neuromuscul. Disord. NMD 2009, 19, 335–343. [Google Scholar] [CrossRef]
- Pelletier, R.; Hamel, F.; Beaulieu, D.; Patry, L.; Haineault, C.; Tarnopolsky, M.; Schoser, B.; Puymirat, J. Absence of a differentiation defect in muscle satellite cells from DM2 patients. Neurobiol. Dis. 2009, 36, 181–190. [Google Scholar] [CrossRef]
- Langlois, M.A.; Lee, N.S.; Rossi, J.J.; Puymirat, J. Hammerhead ribozyme-mediated destruction of nuclear foci in myotonic dystrophy myoblasts. Mol. Ther. J. Am. Soc. Gene Ther. 2003, 7, 670–680. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadava, R.S.; Mandal, M.; Mahadevan, M.S. Studying the Effect of MBNL1 and MBNL2 Loss in Skeletal Muscle Regeneration. Int. J. Mol. Sci. 2024, 25, 2687. https://doi.org/10.3390/ijms25052687
Yadava RS, Mandal M, Mahadevan MS. Studying the Effect of MBNL1 and MBNL2 Loss in Skeletal Muscle Regeneration. International Journal of Molecular Sciences. 2024; 25(5):2687. https://doi.org/10.3390/ijms25052687
Chicago/Turabian StyleYadava, Ramesh S., Mahua Mandal, and Mani S. Mahadevan. 2024. "Studying the Effect of MBNL1 and MBNL2 Loss in Skeletal Muscle Regeneration" International Journal of Molecular Sciences 25, no. 5: 2687. https://doi.org/10.3390/ijms25052687
APA StyleYadava, R. S., Mandal, M., & Mahadevan, M. S. (2024). Studying the Effect of MBNL1 and MBNL2 Loss in Skeletal Muscle Regeneration. International Journal of Molecular Sciences, 25(5), 2687. https://doi.org/10.3390/ijms25052687




