Visfatin Affects the Transcriptome of Porcine Luteal Cells during Early Pregnancy
Abstract
1. Introduction
2. Results
2.1. RNA Sequencing Data
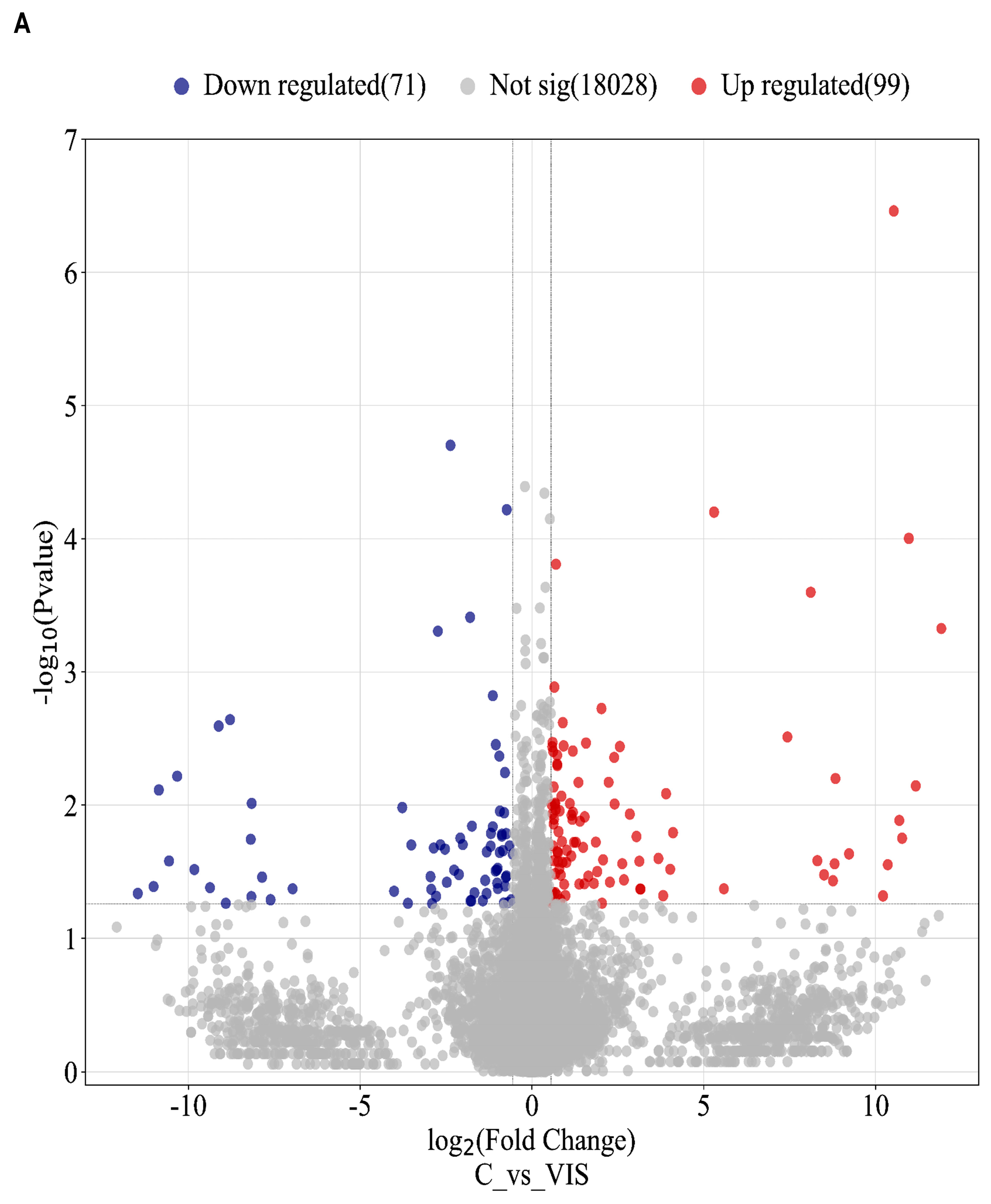
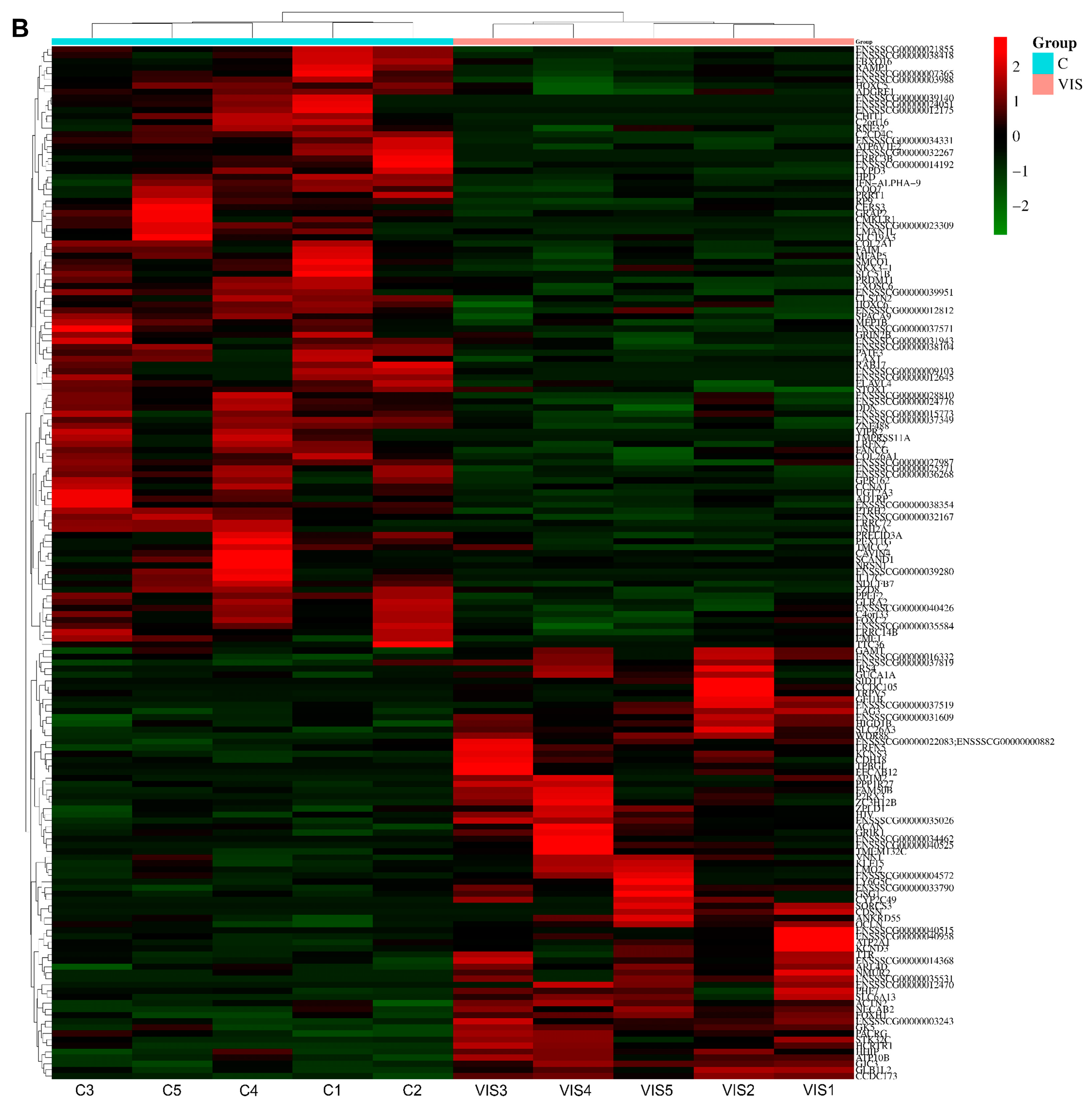
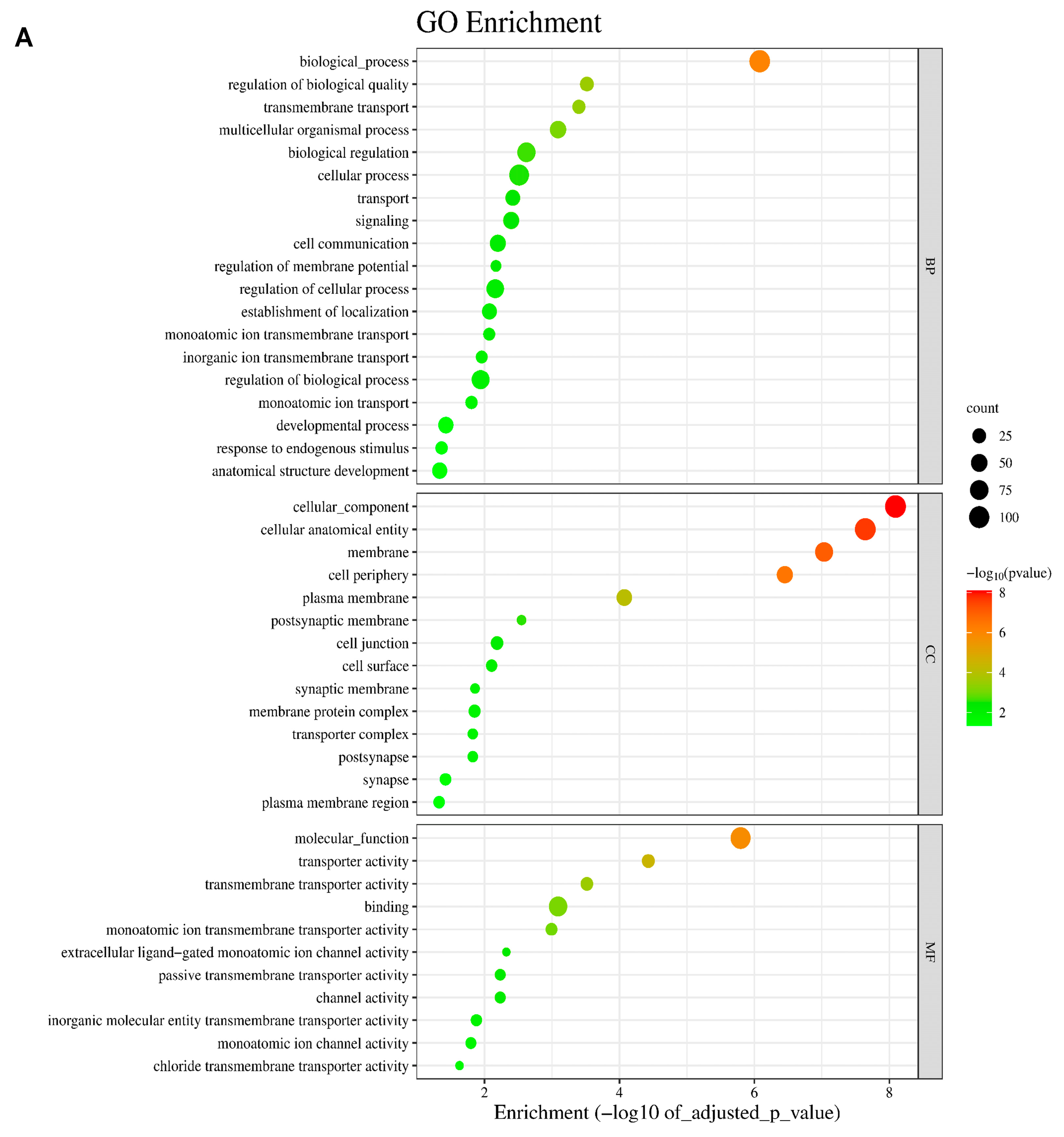
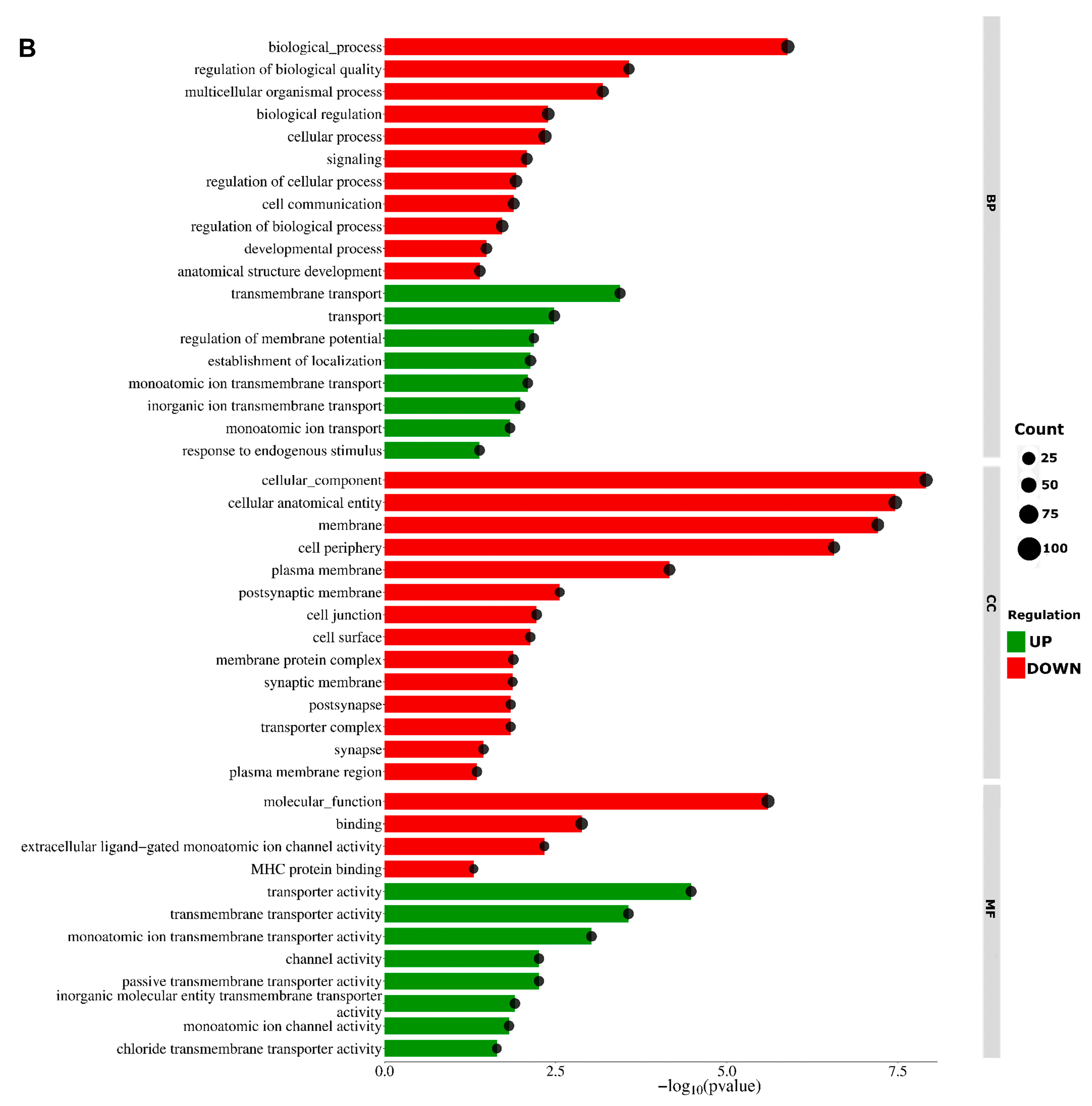
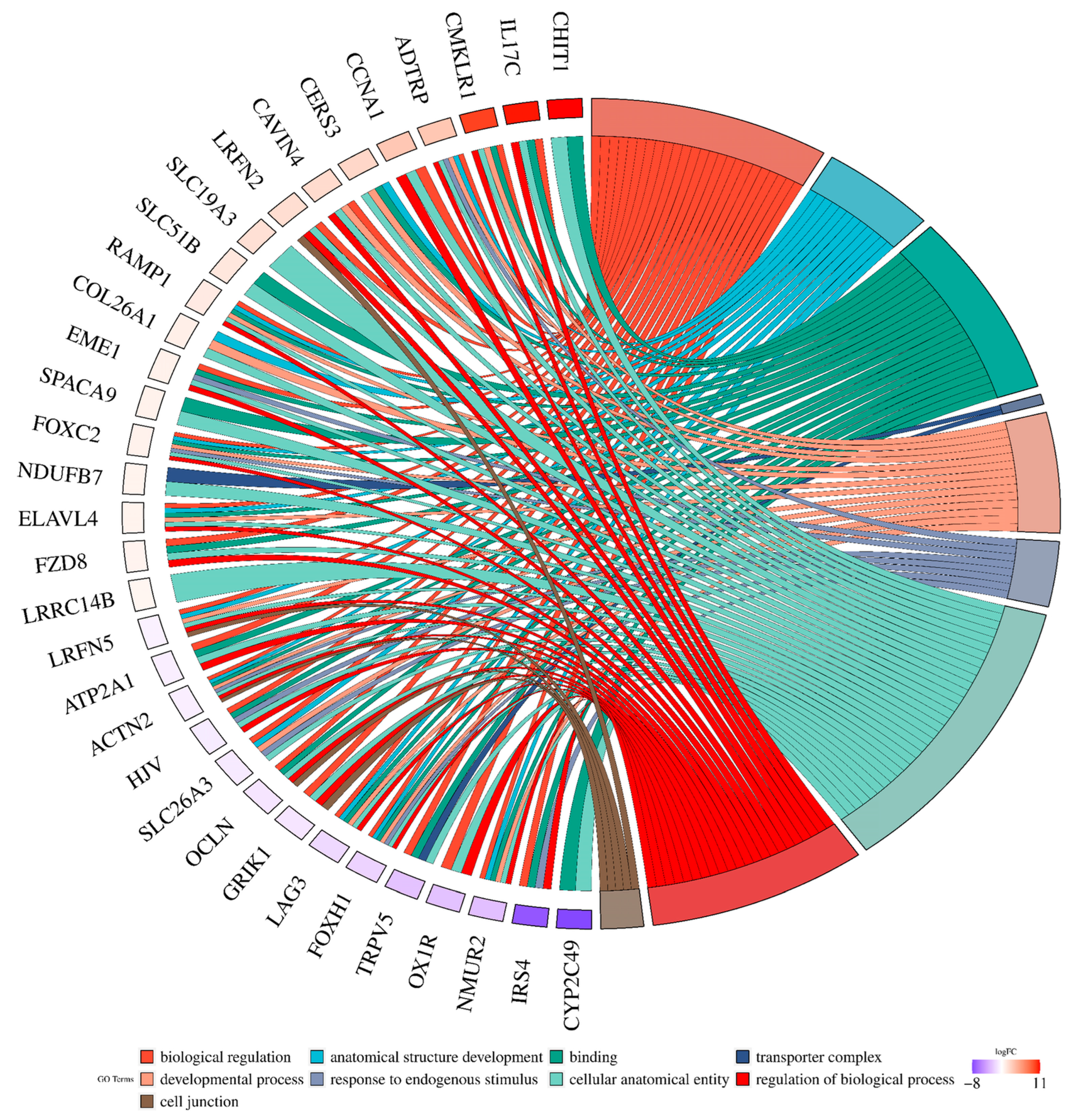
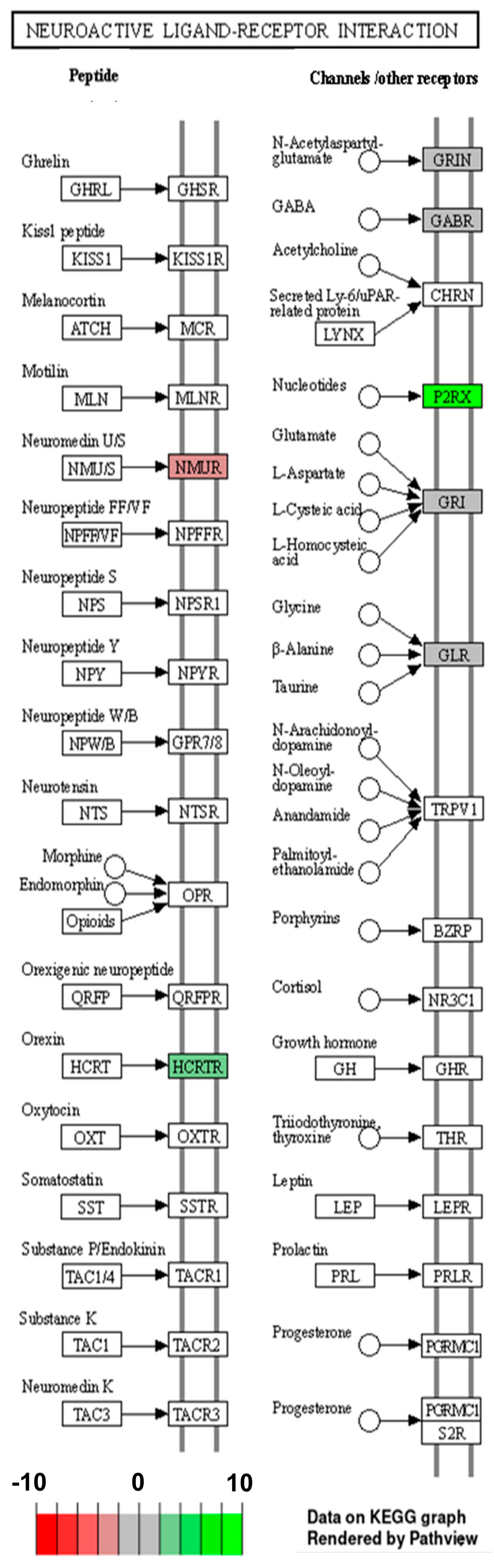
2.2. Analysis of Differentially Expressed Genes
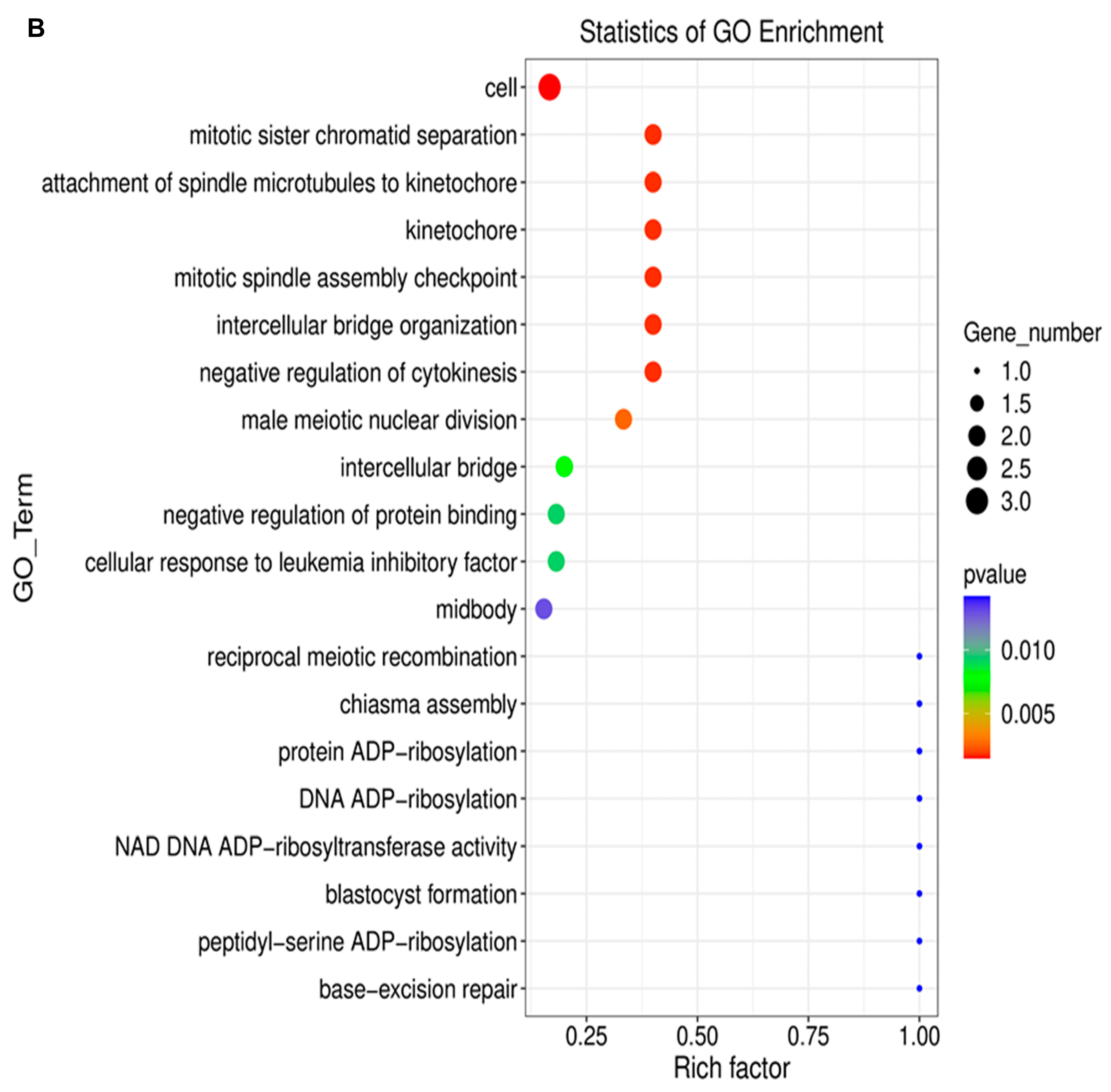
2.3. Long Noncoding RNA Identification and Functional Annotations
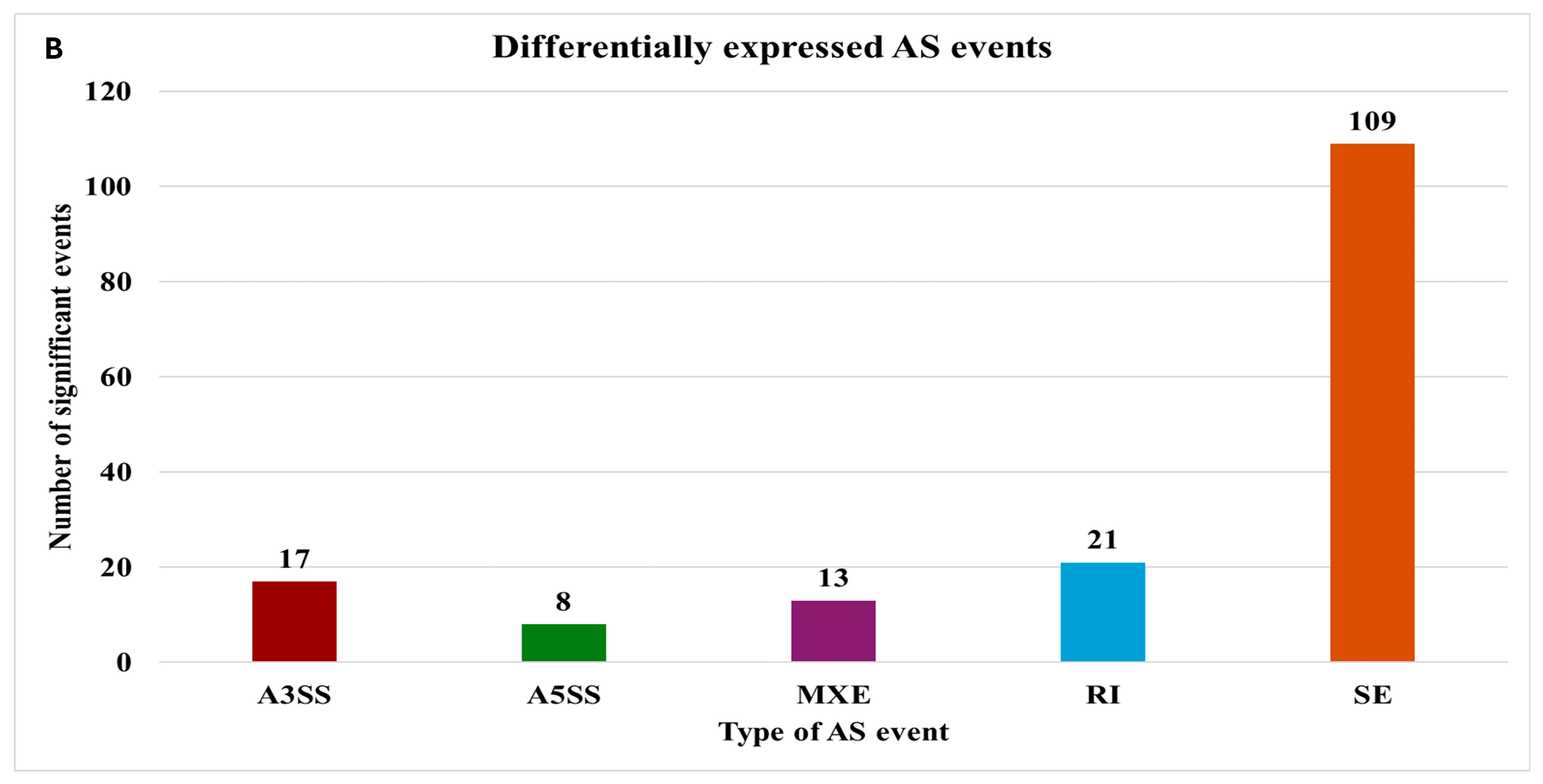
2.4. Alternative Splicing Event Analysis
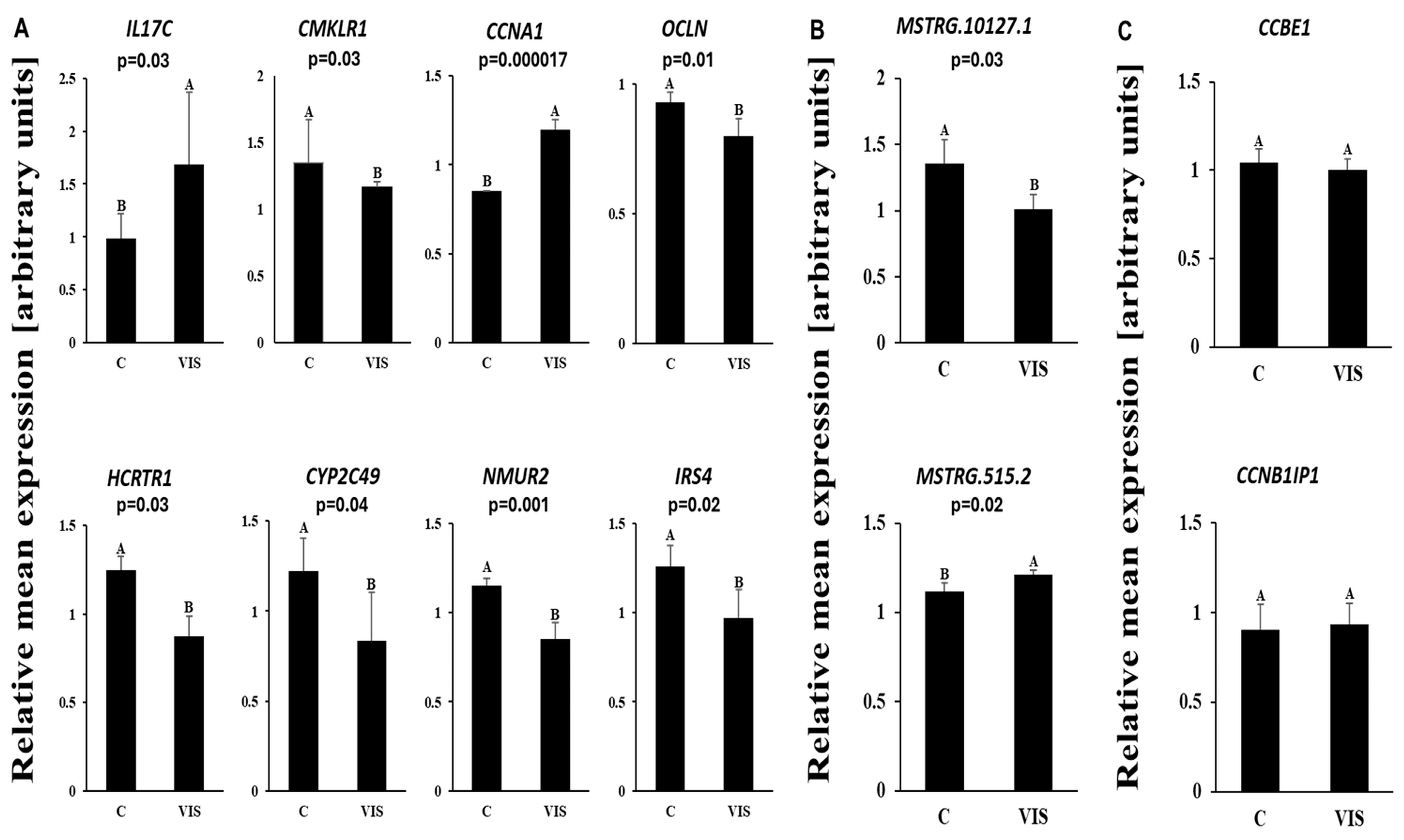
2.5. Validation of RNA-Seq Results
3. Discussion
4. Materials and Methods
4.1. Collection of Samples
4.2. In Vitro Culture of Luteal Cells and Total RNA Isolation
4.3. Library Construction and RNA Sequencing
4.4. Bioinformatic Analysis
4.4.1. Assembly of Transcripts and Processing of Differentially Expressed Transcripts
4.4.2. Functional Annotation of Target Genes
4.4.3. Long Noncoding RNA Identification, Target Gene Prediction, and Functional Analysis
4.4.4. Differential Alternative Splicing Event Analysis
4.5. Validation of DEGs and DELs by Quantitative Real-Time PCR (qPCR)
4.6. Validation of DASs by PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bazer, F.W.; Thatcher, W.W. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F2α by the uterine endometrium. Prostaglandins 1977, 14, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W. Pregnancy recognition signaling mechanisms in ruminants and pigs. J. Anim. Sci. Biotechnol. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Samborski, A.; Graf, A.; Krebs, S.; Kessler, B.; Reichenbach, M.; Reichenbach, H.-D.; Ulbrich, S.E.; Bauersachs, S. Transcriptome Changes in the Porcine Endometrium During the Preattachment Phase1. Biol. Reprod. 2013, 89, 134. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Seo, H.; Choi, Y.; Yoo, I.; Seo, M.; Lee, C.-K.; Kim, H.; Ka, H. Analysis of Stage-Specific Gene Expression Profiles in the Uterine Endometrium during Pregnancy in Pigs. PLoS ONE 2015, 10, e0143436. [Google Scholar] [CrossRef] [PubMed]
- Smolinska, N.; Szeszko, K.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Bors, K.; Wyrebek, J.; Kopij, G.; et al. Transcriptomic Analysis of Porcine Endometrium during Implantation after In Vitro Stimulation by Adiponectin. Int. J. Mol. Sci. 2019, 20, 1335. [Google Scholar] [CrossRef] [PubMed]
- Dobrzyn, K.; Kiezun, M.; Szeszko, K.; Gudelska, M.; Kisielewska, K.; Rytelewska, E.; Zaobidna, E.; Wyrebek, J.; Bors, K.; Kopij, G.; et al. The in vitro effect of orexin a on the porcine myometrial transcriptomic profile during the early-implantation period. Theriogenology 2019, 143, 157–167. [Google Scholar] [CrossRef]
- Orzechowska, K.; Kopij, G.; Paukszto, L.; Dobrzyn, K.; Kiezun, M.; Jastrzebski, J.; Kaminski, T.; Smolinska, N. Chemerin effect on transcriptome of the porcine endometrium during implantation determined by RNA-sequencing. Biol. Reprod. 2022, 107, 557–573. [Google Scholar] [CrossRef]
- Sugino, N. The Role of Oxygen Radical-mediated Signaling Pathways in Endometrial Function. Placenta 2007, 28, S133–S136. [Google Scholar] [CrossRef]
- Makowczenko, K.G.; Jastrzebski, J.P.; Szeszko, K.; Smolinska, N.; Paukszto, L.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kaminska, B.; Kaminski, T. Transcription Analysis of the Chemerin Impact on Gene Expression Profile in the Luteal Cells of Gilts. Genes 2020, 11, 651. [Google Scholar] [CrossRef] [PubMed]
- Makowczenko, K.G.; Jastrzebski, J.P.; Paukszto, L.; Dobrzyn, K.; Kiezun, M.; Smolinska, N.; Kaminski, T. Chemerin Im-pact on Alternative MRNA Transcription in the Porcine Luteal Cells. Cells 2022, 11, 715. [Google Scholar] [CrossRef] [PubMed]
- Ziecik, A.J.; Przygrodzka, E.; Jalali, B.M.; Kaczmarek, M.M. Regulation of the porcine corpus luteum during pregnancy. Reproduction 2018, 156, R57–R67. [Google Scholar] [CrossRef]
- Fukuhara, A.; Matsuda, M.; Nishizawa, M.; Segawa, K.; Tanaka, M.; Kishimoto, K.; Matsuki, Y.; Murakami, M.; Ichisaka, T.; Murakami, H.; et al. Visfatin: A Protein Secreted by Visceral Fat That Mimics the Effects of Insulin. Science 2005, 307, 426–430. [Google Scholar] [CrossRef]
- Samal, B.; Sun, Y.; Stearns, G.; Xie, C.; Suggs, S.; McNiece, I. Cloning and Characterization of the cDNA Encoding a Novel Human pre-B-Cell Colony-enhancing Factor. Mol. Cell. Biol. 1994, 14, 1432–1437. [Google Scholar] [CrossRef]
- Grolla, A.A.; Travelli, C.; Genazzani, A.A.; Sethi, J.K. Extracellular nicotinamide phosphoribosyltransferase, a new cancer metabokine. Br. J. Pharmacol. 2016, 173, 2182–2194. [Google Scholar] [CrossRef]
- Kim, M.-K.; Lee, J.H.; Kim, H.; Park, S.J.; Kim, S.H.; Kang, G.B.; Lee, Y.S.; Kim, J.B.; Kim, K.K.; Suh, S.W.; et al. Crystal Structure of Visfatin/Pre-B Cell Colony-enhancing Factor 1/Nicotinamide Phosphoribosyltransferase, Free and in Complex with the Anti-cancer Agent FK-866. J. Mol. Biol. 2006, 362, 66–77. [Google Scholar] [CrossRef]
- Maillard, V.; Elis, S.; Desmarchais, A.; Hivelin, C.; Lardic, L.; Lomet, D.; Uzbekova, S.; Monget, P.; Dupont, J. Visfatin and resistin in gonadotroph cells: Expression, regulation of LH secretion and signalling pathways. Reprod. Fertil. Dev. 2017, 29, 2479–2495. [Google Scholar] [CrossRef][Green Version]
- Reverchon, M.; Cornuau, M.; Cloix, L.; Ramé, C.; Guerif, F.; Royère, D.; Dupont, J. Visfatin is expressed in human granulosa cells: Regulation by metformin through AMPK/SIRT1 pathways and its role in steroidogenesis. Mol. Hum. Reprod. 2013, 19, 313–326. [Google Scholar] [CrossRef]
- Dupré, S.M.; Burt, D.W.; Talbot, R.; Downing, A.; Mouzaki, D.; Waddington, D.; Malpaux, B.; Davis, J.R.E.; Lincoln, G.A.; Loudon, A.S.I. Identification of Melatonin-Regulated Genes in the Ovine Pituitary Pars Tuberalis, a Target Site for Seasonal Hormone Control. Endocrinology 2008, 149, 5527–5539. [Google Scholar] [CrossRef] [PubMed]
- Reverchon, M.; Rame, C.; Bunel, A.; Chen, W.; Froment, P.; Dupont, J. VISFATIN (NAMPT) Improves in Vitro IGF1-Induced Steroidogenesis and IGF1 Receptor Signaling Through SIRT1 in Bovine Granulosa Cells1. Biol. Reprod. 2016, 94, 54. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.-J.; Tsai, E.-M.; Lee, J.-N.; Chen, Y.-L.; Lee, C.-H.; Chan, T.-F. The concentrations of visfatin in the follicular fluids of women undergoing controlled ovarian stimulation are correlated to the number of oocytes retrieved. Fertil. Steril. 2010, 93, 1844–1850. [Google Scholar] [CrossRef] [PubMed]
- Palin, M.-F.; Labrecque, B.; Beaudry, D.; Mayhue, M.; Bordignon, V.; Murphy, B.D. Visfatin expression is not associated with adipose tissue abundance in the porcine model. Domest. Anim. Endocrinol. 2008, 35, 58–73. [Google Scholar] [CrossRef]
- Mlyczyńska, E.; Zaobidna, E.; Rytelewska, E.; Dobrzyń, K.; Kieżun, M.; Kopij, G.; Szymańska, K.; Kurowska, P.; Dall’aglio, C.; Smolińska, N.; et al. Expression and regulation of visfatin/NAMPT in the porcine corpus luteum during the estrous cycle and early pregnancy. Anim. Reprod. Sci. 2023, 250, 107212. [Google Scholar] [CrossRef]
- Revollo, J.R.; Körner, A.; Mills, K.F.; Satoh, A.; Wang, T.; Garten, A.; Dasgupta, B.; Sasaki, Y.; Wolberger, C.; Townsend, R.R.; et al. Nampt/PBEF/Visfatin Regulates Insulin Secretion in β Cells as a Systemic NAD Biosynthetic Enzyme. Cell Metab. 2007, 6, 363–375. [Google Scholar] [CrossRef]
- Berndt, J.; Klöting, N.; Kralisch, S.; Kovacs, P.; Fasshauer, M.; Schön, M.R.; Stumvoll, M.; Blüher, M. Plasma Visfatin Concentrations and Fat Depot-Specific mRNA Expression in Humans. Diabetes 2005, 54, 2911–2916. [Google Scholar] [CrossRef] [PubMed]
- Filippatos, T.D.; Derdemezis, C.S.; Gazi, I.F.; Lagos, K.; Kiortsis, D.N.; Tselepis, A.D.; Elisaf, M.S. Increased plasma visfatin levels in subjects with the metabolic syndrome. Eur. J. Clin. Investig. 2007, 38, 71–72. [Google Scholar] [CrossRef] [PubMed]
- Stofkova, A. Resistin and visfatin: Regulators of insulin sensitivity, inflammation and immunity. Endocr. Regul. 2010, 44, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Pavlová, T.; Novák, J.; Bienertová-Vašků, J. The role of visfatin (PBEF/Nampt) in pregnancy complications. J. Reprod. Immunol. 2015, 112, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Lovren, F.; Pan, Y.; Shukla, P.C.; Quan, A.; Teoh, H.; Szmitko, P.E.; Peterson, M.D.; Gupta, M.; Al-Omran, M.; Verma, S.; et al. Visfatin activates eNOS via Akt and MAP kinases and improves endothelial cell function and angiogenesis in vitro and in vivo: Translational implications for atherosclerosis. Am. J. Physiol. Metab. 2009, 296, E1440–E1449. [Google Scholar] [CrossRef] [PubMed]
- Adya, R.; Tan, B.K.; Punn, A.; Chen, J.; Randeva, H.S. Visfatin induces human endothelial VEGF and MMP-2/9 production via MAPK and PI3K/Akt signalling pathways: Novel insights into visfatin-induced angiogenesis. Cardiovasc. Res. 2007, 78, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-H.; Joo, B.-S.; Sun, S.-T.; Park, M.-J.; Son, J.-B.; Joo, J.-K.; Lee, K.-S. Administration of visfatin during superovulation improves developmental competency of oocytes and fertility potential in aged female mice. Fertil. Steril. 2012, 97, 1234–1241.e3. [Google Scholar] [CrossRef]
- Ognjanovic, S.; Bryant-Greenwood, G.D. Pre-B-cell colony-enhancing factor, a novel cytokine of human fetal membranes. Am. J. Obstet. Gynecol. 2002, 187, 1051–1058. [Google Scholar] [CrossRef]
- Reverchon, M.; Ramé, C.; Bertoldo, M.; Dupont, J. Adipokines and the Female Reproductive Tract. Int. J. Endocrinol. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Mazaki-Tovi, S.; Romero, R.; Kusanovic, J.P.; Vaisbuch, E.; Erez, O.; Than, N.G.; Chaiworapongsa, T.; Nhan-Chang, C.-L.; Pacora, P.; Gotsch, F.; et al. Visfatin in human pregnancy: Maternal gestational diabetes vis-à-vis neonatal birthweight. jpme 2008, 37, 218–231. [Google Scholar] [CrossRef] [PubMed]
- Mlyczynska, E.; Zaobidna, E.; Rytelweska, E.; Respekta, N.; Kopij, G.; Dobrzyń, K.; Kiezun, M.; Dall’aglio, C.; Smolinska, N.; Kaminski, T.; et al. Visfatin Effect on Progesterone Synthesis in the Porcine Corpus Luteum during the Estrous Cycle via Activation of MAPK and AMPK Signalling Pathways. In Proceedings of the 25th ESDAR Conference, Thessaloniki, Greece, 28 September 2022. Reproduction in Domestic Animals. [Google Scholar]
- Kang, Y.-J.; Yang, D.-C.; Kong, L.; Hou, M.; Meng, Y.-Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.-X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Constantin, S. Progress and Challenges in the Search for the Mechanisms of Pulsatile Gonadotropin-Releasing Hormone Secretion. Front. Endocrinol. 2017, 8, 180. [Google Scholar] [CrossRef] [PubMed]
- Berisha, B.; Schams, D. Ovarian function in ruminants. Domest. Anim. Endocrinol. 2005, 29, 305–317. [Google Scholar] [CrossRef]
- Jansova, D.; Tetkova, A.; Koncicka, M.; Kubelka, M.; Susor, A. Localization of RNA and translation in the mammalian oocyte and embryo. PLoS ONE 2018, 13, e0192544. [Google Scholar] [CrossRef]
- Yoshioka, S.; Abe, H.; Sakumoto, R.; Okuda, K. Proliferation of Luteal Steroidogenic Cells in Cattle. PLoS ONE 2013, 8, e84186. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.J.; Gao, F.; Yang, T.; Thakur, A.; Ren, H.; Li, Y.; Zhang, S.; Wang, T.; Chen, M.W. CDK-associated Cullin 1 promotes cell proliferation with activation of ERK1/2 in human lung cancer A549 cells. Biochem. Biophys. Res. Commun. 2013, 437, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Wang, F.; Qin, S.; Chen, Q.; Wang, Q.K. Coronary artery disease susceptibility gene ADTRP regulates cell cycle progression, proliferation, and apoptosis by global gene expression regulation. Physiol. Genom. 2016, 48, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, S.K.; Manterola, M.; Wolgemuth, D.J. Meiotic failure in cyclin A1-deficient mouse spermatocytes triggers apoptosis through intrinsic and extrinsic signaling pathways and 14-3-3 proteins. PLoS ONE 2017, 12, e0173926. [Google Scholar] [CrossRef] [PubMed]
- Kfir, S.; Basavaraja, R.; Wigoda, N.; Ben-Dor, S.; Orr, I.; Meidan, R. Genomic profiling of bovine corpus luteum maturation. PLoS ONE 2018, 13, e0194456. [Google Scholar] [CrossRef]
- Singh, M.K.; Nicolas, E.; Gherraby, W.; Dadke, D.; Lessin, S.; A Golemis, E. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene 2007, 26, 4825–4832. [Google Scholar] [CrossRef]
- Mine, N.; Kurose, K.; Konishi, H.; Araki, T.; Nagai, H.; Emi, M. Fusion of a Sequence from HEI10 (14q11) to the HMGIC Gene at 12ql5 in a Uterine Leiomyoma. Jpn. J. Cancer Res. 2001, 92, 135–139. [Google Scholar] [CrossRef]
- Toby, G.G.; Gherraby, W.; Coleman, T.R.; Golemis, E.A. A Novel RING Finger Protein, Human Enhancer of Invasion 10, Alters Mitotic Progression through Regulation of Cyclin B Levels. Mol. Cell. Biol. 2003, 23, 2109–2122. [Google Scholar] [CrossRef]
- Luo, C.; Pook, E.; Tang, B.; Zhang, W.; Li, S.; Leineweber, K.; Cheung, S.-H.; Chen, Q.; Bechem, M.; Hu, J.-S.; et al. Androgen inhibits key atherosclerotic processes by directly activating ADTRP transcription. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2017, 1863, 2319–2332. [Google Scholar] [CrossRef]
- Reynolds, L.P.; A Redmer, D. Expression of the angiogenic factors, basic fibroblast growth factor and vascular endothelial growth factor, in the ovary. J. Anim. Sci. 1998, 76, 1671–1681. [Google Scholar] [CrossRef]
- Lupu, C.; Zhu, H.; Popescu, N.I.; Wren, J.D.; Lupu, F. Novel protein ADTRP regulates TFPI expression and function in human endothelial cells in normal conditions and in response to androgen. Blood 2011, 118, 4463–4471. [Google Scholar] [CrossRef]
- Bauer, A.; Mylroie, H.; Thornton, C.C.; Calay, D.; Birdsey, G.M.; Kiprianos, A.P.; Wilson, G.K.; Soares, M.P.; Yin, X.; Mayr, M.; et al. Identification of cyclins A1, E1 and vimentin as downstream targets of heme oxygenase-1 in vascular endothelial growth factor-mediated angiogenesis. Sci. Rep. 2016, 6, 29417. [Google Scholar] [CrossRef]
- Patel, M.M.; Behar, A.R.; Silasi, R.; Regmi, G.; Sansam, C.L.; Keshari, R.S.; Lupu, F.; Lupu, C. Role of ADTRP (Androgen-Dependent Tissue Factor Pathway Inhibitor Regulating Protein) in Vascular Development and Function. J. Am. Hear. Assoc. 2018, 7, e010690. [Google Scholar] [CrossRef]
- Dakroub, A.; Nasser, S.A.; Kobeissy, F.; Yassine, H.M.; Orekhov, A.; Sharifi-Rad, J.; Iratni, R.; El-Yazbi, A.F.; Eid, A.H. Visfatin: An emerging adipocytokine bridging the gap in the evolution of cardiovascular diseases. J. Cell. Physiol. 2021, 236, 6282–6296. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-R.; Bae, S.-K.; Choi, K.-S.; Park, S.-Y.; Jun, H.O.; Lee, J.-Y.; Jang, H.-O.; Yun, I.; Yoon, K.-H.; Kim, Y.-J.; et al. Visfatin promotes angiogenesis by activation of extracellular signal-regulated kinase 1/2. Biochem. Biophys. Res. Commun. 2007, 357, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.C.; Best, L.; Lorenzo, L.; Casanova, J.-L.; Wacker, J.; Bertz, S.; Agaimy, A.; Harrer, T. A Multiplex Kindred with Hennekam Syndrome due to Homozygosity for a CCBE1 Mutation that does not Prevent Protein Expression. J. Clin. Immunol. 2015, 36, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, M.; Jha, S.K.; Tvorogov, D.; Anisimov, A.; Leppänen, V.M.; Holopainen, T.; Kivelä, R.; Ortega, S.; Kärpanen, T.; Alitalo, K. CCBE1 Enhances Lymphangiogenesis via a Disintegrin and Metalloprotease with Thrombospondin Mo-tifs-3-Mediated Vascular Endothelial Growth Factor-C Activation. Circulation 2014, 129, 1962–1971. [Google Scholar] [CrossRef]
- Jha, S.K.; Rauniyar, K.; Karpanen, T.; Leppänen, V.-M.; Brouillard, P.; Vikkula, M.; Alitalo, K.; Jeltsch, M. Efficient activation of the lymphangiogenic growth factor VEGF-C requires the C-terminal domain of VEGF-C and the N-terminal domain of CCBE1. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Hwang, G.; Sun, F.; O’Brien, M.; Eppig, J.J.; Handel, M.A.; Jordan, P.W. SMC5/6 Is Required for the Formation of Segre-gation-Competent Bivalent Chromosomes during Meiosis i in Mouse Oocytes. Development 2017, 144, 1648–1660. [Google Scholar] [CrossRef]
- Ward, J.; Reinholdt, L.G.; Motley, W.W.; Niswander, L.M.; Deacon, D.C.; Griffin, L.B.; Langlais, K.K.; Backus, V.L.; Schimenti, K.J.; O’Brien, M.J.; et al. Mutation in Mouse Hei10, an E3 Ubiquitin Ligase, Disrupts Meiotic Crossing Over. PLOS Genet. 2007, 3, e139. [Google Scholar] [CrossRef]
- Hogan, B.M.; Bos, F.L.; Bussmann, J.; Witte, M.; Chi, N.C.; Duckers, H.J.; Schulte-Merker, S. ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat. Genet. 2009, 41, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Smolinska, N.; Kiezun, M.; Dobrzyn, K.; Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Zaobidna, E.; Bogus-Nowakowska, K.; Wyrebek, J.; Bors, K.; et al. Expression of Chemerin and Its Receptors in the Porcine Hypothalamus and Plasma Chemerin Levels during the Oestrous Cycle and Early Pregnancy. Int. J. Mol. Sci. 2019, 20, 3887. [Google Scholar] [CrossRef] [PubMed]
- Kisielewska, K.; Rytelewska, E.; Gudelska, M.; Kiezun, M.; Dobrzyn, K.; Bogus-Nowakowska, K.; Kaminska, B.; Smolinska, N.; Kaminski, T. Expression of chemerin receptors CMKLR1, GPR1 and CCRL2 in the porcine pituitary during the oestrous cycle and early pregnancy and the effect of chemerin on MAPK/Erk1/2, Akt and AMPK signalling pathways. Theriogenology 2020, 157, 181–198. [Google Scholar] [CrossRef] [PubMed]
- Rytelewska, E.; Kisielewska, K.; Kiezun, M.; Dobrzyn, K.; Gudelska, M.; Rak, A.; Dupont, J.; Kaminska, B.; Kaminski, T.; Smolinska, N. Expression of chemerin and its receptors in the ovaries of prepubertal and mature gilts. Mol. Reprod. Dev. 2020, 87, 739–762. [Google Scholar] [CrossRef] [PubMed]
- Gudelska, M.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kisielewska, K.; Kaminska, B.; Kaminski, T.; Smolinska, N. The expression of chemerin and its receptors (CMKLR1, GPR1, CCRL2) in the porcine uterus during the oestrous cycle and early pregnancy and in trophoblasts and conceptuses. Animal 2020, 14, 2116–2128. [Google Scholar] [CrossRef] [PubMed]
- Helfer, G.; Wu, Q.-F. Chemerin: A multifaceted adipokine involved in metabolic disorders. J. Endocrinol. 2018, 238, R79–R94. [Google Scholar] [CrossRef]
- Yoshimura, T.; Oppenheim, J.J. Chemokine-like receptor 1 (CMKLR1) and chemokine (C–C motif) receptor-like 2 (CCRL2); Two multifunctional receptors with unusual properties. Exp. Cell Res. 2011, 317, 674–684. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Kiezun, M.; Dobrzyn, K.; Bors, K.; Wyrebek, J.; Kaminska, B.; Kaminski, T.; Smolinska, N. The effect of orexin a on the StAR, CYP11A1 and HSD3B1 gene expression, as well as progesterone and androstenedione secretion in the porcine uterus during early pregnancy and the oestrous cycle. Theriogenology 2019, 143, 179–190. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kiezun, M.; Kisielewska, K.; Gudelska, M.; Dobrzyn, K.; Kaminska, B.; Kaminski, T.; Smolinska, N. Chemerin as a modulator of ovarian steroidogenesis in pigs: An in vitro study. Theriogenology 2020, 160, 95–101. [Google Scholar] [CrossRef]
- Nitkiewicz, A.; Smolinska, N.; Przala, J.; Kaminski, T. Expression of orexin receptors 1 (OX1R) and 2 (OX2R) in the porcine ovary during the oestrous cycle. Regul. Pept. 2010, 165, 186–190. [Google Scholar] [CrossRef]
- Li, J.; Hu, Z.; de Lecea, L. The hypocretins/orexins: Integrators of multiple physiological functions. Br. J. Pharmacol. 2013, 171, 332–350. [Google Scholar] [CrossRef]
- Li, T.; Li, T.; Li, T.; Xu, W.; Xu, W.; Ouyang, J.; Lu, X.; Sherchan, P.; Lenahan, C.; Lenahan, C.; et al. Orexin A Alleviates Neuroinflammation via OXR2/CaMKKβ/AMPK Signaling Pathway after ICH in Mice. J. Neuroinflam. 2020, 17, 1–15. [Google Scholar] [CrossRef]
- Kiezun, M.; Smolinska, N.; Dobrzyn, K.; Szeszko, K.; Rytelewska, E.; Kaminski, T. The effect of orexin A on CYP17A1 and CYP19A3 expression and on oestradiol, oestrone and testosterone secretion in the porcine uterus during early pregnancy and the oestrous cycle. Theriogenology 2017, 90, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Nitkiewicz, A.; Smolinska, N.; Maleszka, A.; Chojnowska, K.; Kaminski, T. Expression of orexins and their precursor in the porcine ovary and the influence of orexins on ovarian steroidogenesis in pigs. Anim. Reprod. Sci. 2014, 148, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Ciccimarra, R.; Bussolati, S.; Grasselli, F.; Grolli, S.; Ragionieri, L.; Ravanetti, F.; Botti, M.; Gazza, F.; Cacchioli, A.; Di Lecce, R.; et al. Orexin system in swine ovarian follicles. Domest. Anim. Endocrinol. 2018, 62, 49–59. [Google Scholar] [CrossRef]
- Basini, G.; Ciccimarra, R.; Bussolati, S.; Grolli, S.; Ragionieri, L.; Ravanetti, F.; Botti, M.; Gazza, F.; Cacchioli, A.; Di Lecce, R.; et al. Orexin A in swine corpus luteum. Domest. Anim. Endocrinol. 2018, 64, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Silveyra, P.; Lux-Lantos, V.; Libertun, C. Both Orexin Receptors Are Expressed in Rat Ovaries and Fluctuate with the Es-trous Cycle: Effects of Orexin Receptor Antagonists on Gonadotropins and Ovulation. Am. J. Physiol. Endocrinol. Metab. 2007, 293, 977–985. [Google Scholar] [CrossRef]
- Grafe, L.A.; Eacret, D.; Luz, S.; Gotter, A.L.; Renger, J.J.; Winrow, C.J.; Bhatnagar, S. Orexin 2 receptor regulation of the hypothalamic–pituitary–adrenal (HPA) response to acute and repeated stress. Neuroscience 2017, 348, 313–323. [Google Scholar] [CrossRef]
- Solano, M.E.; Arck, P.C. Steroids, Pregnancy and Fetal Development. Front. Immunol. 2020, 10, 3017. [Google Scholar] [CrossRef]
- Malendowicz, L.K.; Jedrzejczak, N.; Belloni, A.S.; Trejter, M.; Hochól, A.; Nussdorfer, G.G. Effects of orexins A and B on the secretory and proliferative activity of immature and regenerating rat adrenal glands. Histol. Histopathol. 2001, 16, 713–717. [Google Scholar] [CrossRef]
- Xu, S.; Wang, D.; Zhou, D.; Lin, Y.; Che, L.; Fang, Z.; Wu, D. Reproductive Hormone and Transcriptomic Responses of Pituitary Tissue in Anestrus Gilts Induced by Nutrient Restriction. PLoS ONE 2015, 10, e0143219. [Google Scholar] [CrossRef] [PubMed]
- Dejana, E. Endothelial cell–cell junctions: Happy together. Nat. Rev. Mol. Cell Biol. 2004, 5, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The Control of Vascular Integrity by Endothelial Cell Junctions: Molecular Basis and Pathological Implications. Dev. Cell 2009, 16, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Groten, T.; Fraser, H.; Duncan, W.; Konrad, R.; Kreienberg, R.; Wulff, C. Cell junctional proteins in the human corpus luteum: Changes during the normal cycle and after HCG treatment. Hum. Reprod. 2006, 21, 3096–3102. [Google Scholar] [CrossRef][Green Version]
- Wulff, C.; Wiegand, S.J.; Saunders, P.T.K.; Scobie, G.A.; Fraser, H.M. Angiogenesis During Follicular Development in the Primate and its Inhibition by Treatment with Truncated Flt-1-Fc (Vascular Endothelial Growth Factor TrapA40)*. Endocrinology 2001, 142, 3244–3254. [Google Scholar] [CrossRef] [PubMed]
- Rodewald, M.; Herr, D.; Fraser, H.; Hack, G.; Kreienberg, R.; Wulff, C. Regulation of tight junction proteins occludin and claudin 5 in the primate ovary during the ovulatory cycle and after inhibition of vascular endothelial growth factor. Mol. Hum. Reprod. 2007, 13, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Sundfeldt, K.; Piontkewitz, Y.; Billig, H.; Hedin, L. E-cadherin-catenin complex in the rat ovary: Cell-specific expression during folliculogenesis and luteal formation. Reproduction 2000, 118, 375–385. [Google Scholar] [CrossRef]
- Kawagishi, R.; Tahara, M.; Morishige, K.; Sakata, M.; Tasaka, K.; Ikeda, W.; Morimoto, K.; Takai, Y.; Murata, Y. Expression of nectin-2 in mouse granulosa cells. Eur. J. Obstet. Gynecol. Reprod. Biol. 2005, 121, 71–76. [Google Scholar] [CrossRef]
- Jiao, X.; Xue, J.; Gong, P.; Bao, X.; Wu, Y.; Zhang, Y.; Jiang, Y.; Yang, Z. Analyzing clinical and genetic characteristics of a cohort with multiple congenital anomalies-hypotonia-seizures syndrome (MCAHS). Orphanet J. Rare Dis. 2020, 15, 1–12. [Google Scholar] [CrossRef]
- Lee, H.N.; Mitra, M.; Bosompra, O.; Corney, D.C.; Johnson, E.L.; Rashed, N.; Ho, L.D.; Coller, H.A.; Yao, X.; Chaponnier, C.; et al. RECK isoforms have opposing effects on cell migration. Mol. Biol. Cell 2018, 29, 1825–1838. [Google Scholar] [CrossRef]
- Lee, H.N.; Bosompra, O.A.; Coller, H.A. RECK isoforms differentially regulate fibroblast migration by modulating tubulin post-translational modifications. Biochem. Biophys. Res. Commun. 2019, 510, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Levin, G.; Coelho, T.M.; Nóbrega, N.G.; Trombetta-Lima, M.; Sogayar, M.C.; Carreira, A.C.O. Spatio-temporal expression profile of matrix metalloproteinase (Mmp) modulators Reck and Sparc during the rat ovarian dynamics. Reprod. Biol. Endocrinol. 2018, 16, 116. [Google Scholar] [CrossRef] [PubMed]
- Labrador, V.; Brun, C.; König, S.; Roatti, A.; Baertschi, A.J. Peptidyl-Glycine α-Amidating Monooxygenase Targeting and Shaping of Atrial Secretory Vesicles. Circ. Res. 2004, 95, e98–e109. [Google Scholar] [CrossRef] [PubMed]
- Czyzyk, T.A.; Ning, Y.; Hsu, M.-S.; Peng, B.; Mains, R.E.; Eipper, B.A.; Pintar, J.E. Deletion of peptide amidation enzymatic activity leads to edema and embryonic lethality in the mouse. Dev. Biol. 2005, 287, 301–313. [Google Scholar] [CrossRef]
- Petojevic, T.; Pesavento, J.J.; Costa, A.; Liang, J.; Wang, Z.; Berger, J.M.; Botchan, M.R. Cdc45 (cell division cycle protein 45) guards the gate of the Eukaryote Replisome helicase stabilizing leading strand engagement. Proc. Natl. Acad. Sci. USA 2015, 112, E249–E258. [Google Scholar] [CrossRef]
- Fu, Y.; Lv, Z.; Kong, D.; Fan, Y.; Dong, B. High abundance of CDC45 inhibits cell proliferation through elevation of HSPA6. Cell Prolif. 2022, 55, e13257. [Google Scholar] [CrossRef]
- Akins, E.L.; Morrissette, M.C. Gross ovarian changes during estrous cycle of swine. Am. J. Vet. Res. 1968, 29, 1953–1957. [Google Scholar]
- Anderson, L.L. Growth, protein content and distribution of early pig embryos. Anat. Rec. 1978, 190, 143–153. [Google Scholar] [CrossRef]
- Makowczenko, K.G.; Jastrzebski, J.P.; Kiezun, M.; Paukszto, L.; Dobrzyn, K.; Smolinska, N.; Kaminski, T. Adaptation of the Porcine Pituitary Transcriptome, Spliceosome and Editome during Early Pregnancy. Int. J. Mol. Sci. 2023, 24, 5946. [Google Scholar] [CrossRef] [PubMed]
- Paukszto, L.; Mikolajczyk, A.; Jastrzebski, J.P.; Majewska, M.; Dobrzyn, K.; Kiezun, M.; Smolinska, N.; Kaminski, T. Transcriptome, Spliceosome and Editome Expression Patterns of the Porcine Endometrium in Response to a Single Subclinical Dose of Salmonella Enteritidis Lipopolysaccharide. Int. J. Mol. Sci. 2020, 21, 4217. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence; Babraham Institute: Cambridge, UK, 2023. [Google Scholar]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.M.; Satterstrom, F.K.; Peng, M.; Brand, H.; Collins, R.L.; Dong, S.; Wamsley, B.; Klei, L.; Wang, L.; Hao, S.P.; et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022, 54, 1320–1331. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Ge-nome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vella, G.; Ritzmann, F.; Wolf, L.; Kamyschnikov, A.; Stodden, H.; Herr, C.; Slevogt, H.; Bals, R.; Beisswenger, C. IL-17C contributes to NTHi-induced inflammation and lung damage in experimental COPD and is present in sputum during acute exacerbations. PLoS ONE 2021, 16, e0243484. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Nambu, A.; Numata, T.; Yoshizaki, T.; Narushima, S.; Shimura, E.; Hiraishi, Y.; Arae, K.; Morita, H.; Matsumoto, K.; et al. The roles of IL-17C in T cell-dependent and -independent inflammatory diseases. Sci. Rep. 2018, 8, 15750. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Mach, N.; Esteve-Codina, A.; Corominas, J.; Castelló, A.; Ballester, M.; Estellé, J.; Ibáñez-Escriche, N.; Fernández, A.I.; Pérez-Enciso, M.; et al. Liver Transcriptome Profile in Pigs with Extreme Phenotypes of Intramuscular Fatty Acid Composition. BMC Genom. 2012, 13, 547. [Google Scholar] [CrossRef]
- Cantarini, M.C.; de la Monte, S.M.; Pang, M.; Tong, M.; D’Errico, A.; Trevisani, F.; Wands, J.R. Aspartyl-asparagyl β hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology 2006, 44, 446–457. [Google Scholar] [CrossRef]
- Zhou, X.; He, Y.; Li, N.; Bai, G.; Pan, X.; Zhang, Z.; Zhang, H.; Li, J.; Yuan, X. DNA methylation mediated RSPO2 to promote follicular development in mammals. Cell Death Dis. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Li, X.; Niu, M.; Su, J.; Ma, Z.; Jin, M.; Qiao, W.; Zhang, Y.; Feng, Y.; An, N.; Hou, Y.; et al. Cloning and expression patterns of neuromedin U and its receptors in pigs. Neuropeptides 2017, 64, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-H.; Park, J.-K.; Son, D.; Hwang, J.Y.; Lee, D.-K.; Ka, H.; Park, J.; Lee, C.-K. Reactivation of Endogenous Genes and Epigenetic Remodeling Are Barriers for Generating Transgene-Free Induced Pluripotent Stem Cells in Pig. PLoS ONE 2016, 11, e0158046. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo-Weaver, M.; Fuerst, R.; Campbell, S.; Meehan, B.; McNeilly, F.; Adair, B.; Allan, G. A fluorimeter-based RT-PCR method for the detection and quantitation of porcine cytokines. J. Immunol. Methods 1999, 230, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Bogacka, I.; Przała, J.; Siawrys, G.; Kaminski, T.; Smolinska, N. The Expression of Short Form of Leptin Receptor Gene during Early Pregnancy in the Pig Examined by Quantitative Real Time RT-PCR. J. Physiol. Pharmacol. 2006, 57, 479–489. [Google Scholar] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kopij, G.; Kiezun, M.; Dobrzyn, K.; Zaobidna, E.; Zarzecka, B.; Rak, A.; Kaminski, T.; Kaminska, B.; Smolinska, N. Visfatin Affects the Transcriptome of Porcine Luteal Cells during Early Pregnancy. Int. J. Mol. Sci. 2024, 25, 2339. https://doi.org/10.3390/ijms25042339
Kopij G, Kiezun M, Dobrzyn K, Zaobidna E, Zarzecka B, Rak A, Kaminski T, Kaminska B, Smolinska N. Visfatin Affects the Transcriptome of Porcine Luteal Cells during Early Pregnancy. International Journal of Molecular Sciences. 2024; 25(4):2339. https://doi.org/10.3390/ijms25042339
Chicago/Turabian StyleKopij, Grzegorz, Marta Kiezun, Kamil Dobrzyn, Ewa Zaobidna, Barbara Zarzecka, Agnieszka Rak, Tadeusz Kaminski, Barbara Kaminska, and Nina Smolinska. 2024. "Visfatin Affects the Transcriptome of Porcine Luteal Cells during Early Pregnancy" International Journal of Molecular Sciences 25, no. 4: 2339. https://doi.org/10.3390/ijms25042339
APA StyleKopij, G., Kiezun, M., Dobrzyn, K., Zaobidna, E., Zarzecka, B., Rak, A., Kaminski, T., Kaminska, B., & Smolinska, N. (2024). Visfatin Affects the Transcriptome of Porcine Luteal Cells during Early Pregnancy. International Journal of Molecular Sciences, 25(4), 2339. https://doi.org/10.3390/ijms25042339









