Identification of the Reference Genes for Relative qRT-PCR Assay in Two Experimental Models of Rabbit and Horse Subcutaneous ASCs
Abstract
1. Introduction
2. Results
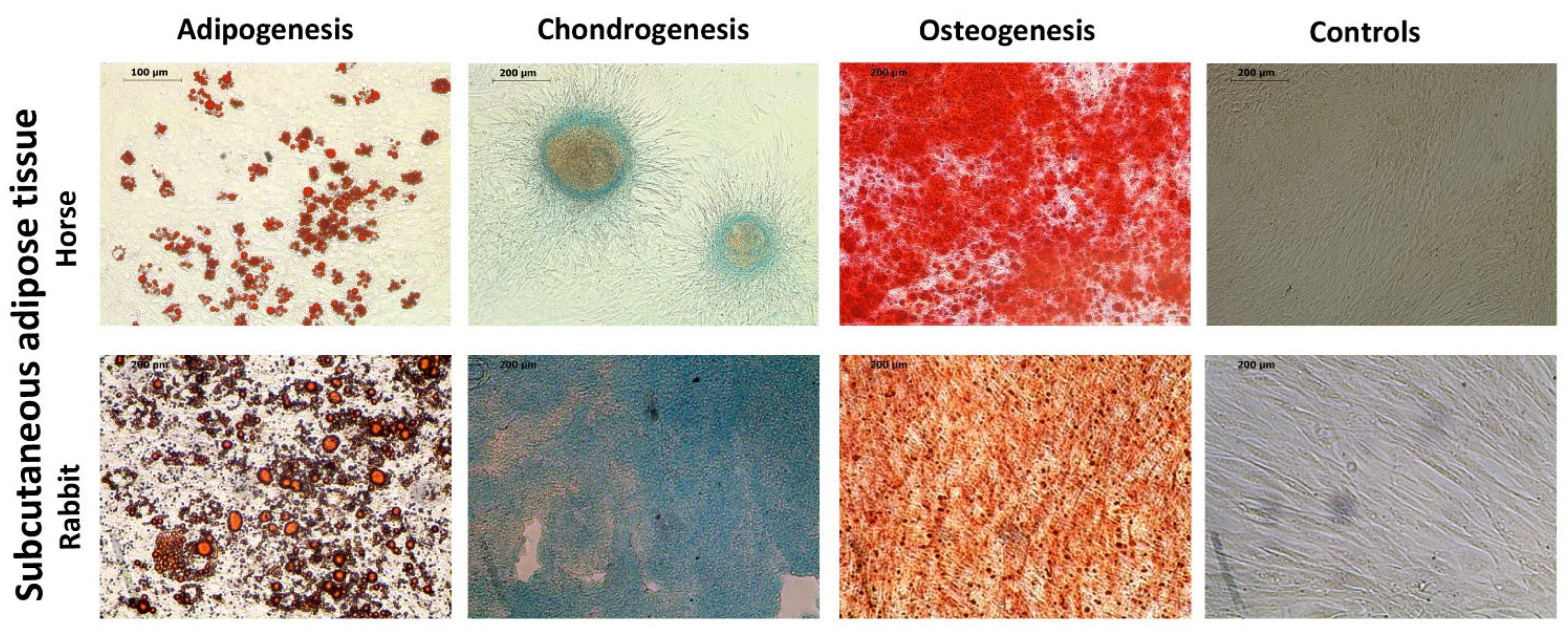
2.1. Identification and Characterization of ASCs
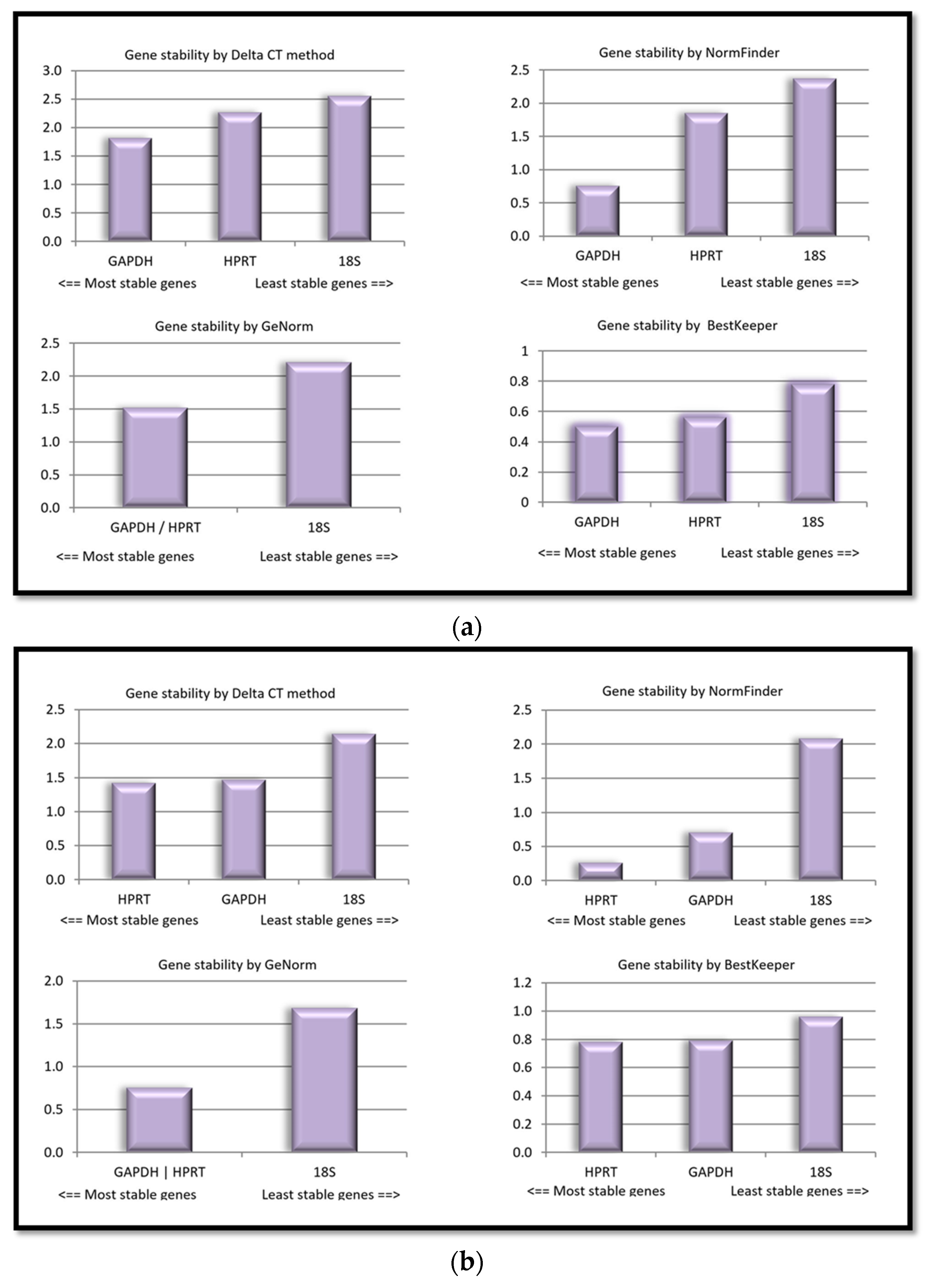
2.2. Expression Stability Evaluation of the Reference Genes Using Two Popular Algorithms
2.2.1. NormFinder
2.2.2. RefFinder
2.3. Expression Levels and Variation in Candidate Housekeeping Genes
2.4. Efficiency of the Reactions
3. Discussion
4. Materials and Methods
4.1. Experimental Design Rabbits
4.2. Experimental Design Horses
4.2.1. Adipogenic Differentiation
4.2.2. Osteogenic Differentiation
4.2.3. Chondrogenic Differentiation
4.2.4. Negative Controls
4.3. Gene Expression Analysis (RT-qPCR)
4.3.1. Rabbit Protocol
4.3.2. Horse Protocol
4.3.3. Standard Curve Construction
4.4. Evaluation of Expression Stability of Housekeeping Genes
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Nestorov, J.; Matić, G.; Elaković, I.; Tanić, N. Gene expression studies: How to obtain accurate and reliable data by quantitative real-time RT PCR. J. Med. Biochem. 2013, 32, 325–338. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. Biotechniques 2000, 29, 332–337. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and validation reference genes for qRT-PCR normalization in different cultivars during fruit ripening and softening of peach (Prunus persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Song, J.; Cho, J.; Park, J.; Hwang, J.H. Identification and validation of stable reference genes for quantitative real time PCR in different minipig tissues at developmental stages. BMC Genom. 2022, 23, 585. [Google Scholar] [CrossRef]
- Tsotetsi, T.N.; Collins, N.E.; Oosthuizen, M.C.; Sibeko-Matjila, K.P. Selection and evaluation of housekeeping genes as endogenous controls for quantification of mRNA transcripts in Theileria parva using quantitative real-time polymerase chain reaction (qPCR). PLoS ONE 2018, 13, 0196715. [Google Scholar] [CrossRef]
- De Jonge, H.J.M.; Fehrmann, R.S.N.; de Bont, E.S.J.M.; Hofstra, R.M.W.; Gerbens, F.; Kamps, W.A.; de Vries, E.G.E.; van der Zee, A.G.J.; te Merrman, G.J.; ter Elst, A. Evidence based selection of housekeeping genes. PLoS ONE 2007, 2, e898. [Google Scholar] [CrossRef]
- Guaita-Cespedes, M.; Grillo-Risco, R.; Hidalgo, M.R.; Fernández-Veledo, S.; Burks, D.J.; de la Iglesia-Vayá, M.; Galan, A.; Garcia-Garcia, F. Deciphering the sex bias in housekeeping gene expression in adipose tissue: A comprehensive meta-analysis of transcriptomic studies. Biol. Sex Differ. 2023, 14, 20. [Google Scholar] [CrossRef] [PubMed]
- Meyer-Siegler, K.; Mauro, D.J.; Seal, G.; Wurzer, J.; Deriel, J.K.; Sirover, M.A. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc. Natl. Acad. Sci. USA 1991, 88, 8460–8464. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Green, M.R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 1993, 259, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, E.J. Glyceraldehyde-3-phosphate dehydrogenase is required for vesicular transport in the early secretory pathway. J. Biol. Chem. 2001, 276, 2480–2486. [Google Scholar] [CrossRef] [PubMed]
- Tarze, A.; Deniaud, A.; Le Bras, M.; Maillier, E.; Mollé, D.; Larochette, N.; Zamzami, N.; Jan, G.; Kroemer, G.; Brenner, C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 2007, 26, 2606–2620. [Google Scholar] [CrossRef]
- Hara, M.R.; Agrawal, N.; Kim, S.F.; Cascio, M.B.; Fujimuro, M.; Ozeki, Y.; Takahashi, M.; Cheah, J.H.; Tankou, S.K.; Hester, L.D.; et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005, 7, 665–674. [Google Scholar] [CrossRef]
- Wilson, J.M.; Tarr, G.E.; Kelley, W.N. Human hypoxanthine (guanine) phosphoribosyltransferase: An amino acid substitution in a mutant form of the enzyme isolated from a patient with gout. Proc. Nat. Acad. Sci. USA 1983, 80, 870–873. [Google Scholar] [CrossRef]
- Stout, J.T.; Caskey, C.T. HPRT: Gene structure, expression, and mutation. Annu. Rev. Genet. 1985, 19, 127–148. [Google Scholar] [CrossRef]
- Becerra, A.; Lazcano, A. The role of gene duplication in the evolution of purine nucleotide salvage pathways. Orig. Life Evol. Biosphere. 1998, 28, 539–553. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Chen, D.; Monstein, H.J.; Hakanson, R. Effects of fasting on the expression of gastrin, cholecystokinin, and somatostatin genes and of various housekeeping genes in the pancreas and upper digestive tract of rats. Biochem. Biophys. Res. Commun. 1997, 231, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Cerca, S.; Pöll, G.; Gleizes, P.E.; Tschochner, H.; Milkereit, P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 2005, 20, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Nam, H.; Hopkins, R.G.; Morrison, R.F. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS ONE 2010, 5, e15208. [Google Scholar] [CrossRef]
- Gong, H.; Sun, L.; Chen, B.; Han, Y.; Pang, J.; Wu, W.; Qi, R.; Zhang, T.M. Evaluation of candidate reference genes for RT-qPCR studies in three metabolism related tissues of mice after caloric restriction. Sci. Rep. 2016, 6, 38513. [Google Scholar] [CrossRef]
- Almeida-Oliveira, F.; Leandro, J.G.; Ausina, P.; Sola-Penna, M.; Majerowicz, D. Reference genes for quantitative PCR in the adipose tissue of mice with metabolic disease. Biomed. Pharmacother. 2017, 88, 948–955. [Google Scholar] [CrossRef]
- Baer, P.C.; Geiger, H. Adipose-derived mesenchymal stromal/stem cells: Tissue localization, characterization, and heterogeneity. Stem Cells Int. 2012, 2012, 812693. [Google Scholar] [CrossRef]
- Vidal, M.A.; Kilroy, G.E.; Lopez, M.J.; Johnson, J.R.; Moore, R.M.; Gimble, J.M. Characterization of equine adipose tissue-derived stromal cells: Adipogenic and osteogenic capacity and comparison with bone marrow-derived mesenchymal stromal cells. Vet. Surg. 2007, 36, 613–622. [Google Scholar] [CrossRef]
- Corsini, N.S.; Knoblich, J.A. Human organoids: New strategies and methods for analyzing human development and disease. Cell 2022, 185, 2756–2769. [Google Scholar] [CrossRef]
- Tirpáková, M.; Vašíček, J.; Svoradová, A.; Baláži, A.; Tomka, M.; Bauer, M.; Makarevich, A.; Chrenek, P. Phenotypical characterization and neurogenic differentiation of rabbit adipose tissue-derived mesenchymal stem cells. Genes 2021, 12, 431. [Google Scholar] [CrossRef]
- Bukowska, J.; Szóstek-Mioduchowska, A.Z.; Kopcewicz, M.; Walendzik, K.; Machcińska, S.; Gawrońska-Kozak, B. Adipose-derived stromal/stem cells from large animal models: From basic to applied science. Stem Cell Rev. Rep. 2021, 17, 719–738. [Google Scholar] [CrossRef]
- Al Naem, M.; Bourebaba, L.; Kucharczyk, K.; Röcken, M.; Marycz, K. Therapeutic mesenchymal stromal stem cells: Isolation, characterization and role in equine regenerative medicine and metabolic disorders. Stem Cell Rev. Rep. 2020, 16, 301–322. [Google Scholar] [CrossRef]
- Ribitsch, I.; Baptista, P.M.; Lange-Consiglio, A.; Melotti, L.; Patruno, M.; Jenner, F.; Schnabl-Feichter, E.; Dutton, L.; Connolly, D.J.; van Steenbeek, F.; et al. Large animal models in regenerative medicine and tissue engineering: To do or not to do. Front. Bioeng. Biotechnol. 2020, 8, 972. [Google Scholar] [CrossRef] [PubMed]
- Gugjoo, M.B.; Sharma, G.T. Equine mesenchymal stem cells: Properties, sources, characterization, and potential therapeutic applications. J. Equine Vet. Sci. 2019, 72, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Matsuhisa, F.; Kitajima, S.; Nishijima, K.; Akiyoshi, T.; Morimoto, M.; Fan, J. Transgenic rabbit models: Now and the future. Appl. Sci. 2020, 10, 7416. [Google Scholar] [CrossRef]
- Bacakova, L.; Zarubova, J.; Travnickova, M.; Musilkova, J.; Pajorova, J.; Slepicka, P.; Kasalkova, N.S.; Svorcik, V.; Kolska, Z.; Motarjemi, H.; et al. Stem cells: Their source, potency and use in regenerative therapies with focus on adipose-derived stem cells—A review. Biotechnol. Adv. 2018, 36, 1111–1126. [Google Scholar] [CrossRef]
- Fraser, J.K.; Wulur, I.; Alfonso, Z.; Hedrick, M.H. Fat tissue: An underappreciated source of stem cells for biotechnology. Trends Biotechnol. 2006, 24, 150–154. [Google Scholar] [CrossRef]
- Farré-Guasch, E.; Bravenboer, N.; Helder, M.N.; Schulten, E.A.; Ten Bruggenkate, C.M.; Klein-Nulend, J. Blood vessel formation and bone regeneration potential of the stromal vascular fraction seeded on a calcium phosphate scaffold in the human maxillary sinus floor elevation model. Materials 2018, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Scioli, M.G.; Storti, G.; D’Amico, F.; Gentile, P.; Kim, B.S.; Cervelli, V.; Orlandi, A. Adipose-derived stem cells in cancer progression: New perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 3296. [Google Scholar] [CrossRef]
- Zuk, P.A.; Zhu, M.; Mizuno, H.; Huang, J.J.; Futrell, W.; Katz, A.J.; Benhaim, P.; Lorenz, H.P.; Hedrick, M.H. Multilineage cells from human adipose tissue: Implications for cell-based therapies. Tissue Eng. 2001, 7, 211–228. [Google Scholar] [CrossRef]
- Quiroz, F.G.; Posada, O.M.; Gallego-Perez, D.; Higuita-Castro, N.; Sarassa, C.; Hansford, D.J.; Agudelo-Florez, P.; López, L.E. Housekeeping gene stability influences the quantification of osteogenic markers during stem cell differentiation to the osteogenic lineage. Cytotechnology 2010, 62, 109–120. [Google Scholar] [CrossRef]
- Wiraja, C.; Yeo, D.C.; Chong, M.S.; Xu, C. Nanosensors for continuous and noninvasive monitoring of mesenchymal stem cell osteogenic differentiation. Small 2016, 12, 1342–1350. [Google Scholar] [CrossRef]
- Zainuddin, A.; Chua, K.H.; Abdul Rahim, N.; Makpol, S. Effect of experimental treatment on GAPDH mRNA expression as a housekeeping gene in human diploid fibroblasts. BMC Mol. Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tang, H.; Zhang, Y.; Deng, R.; Shao, L.; Liu, Y.; Li, F.; Wang, X.; Zhou, L. Identification of suitable reference genes for quantitative RT-PCR during 3T3-L1 adipocyte differentiation. Int. J. Mol. Med. 2014, 33, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Salmerón, C.; Riera-Heredia, N.; Gutiérrez, J.; Navarro, I.; Capilla, E. Adipogenic gene expression in gilthead sea bream mesenchymal stem cells from different origin. Front. Endocrinol. 2016, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Krautgasser, C.; Mandl, M.; Hatzmann, F.M.; Waldegger, P.; Mattesich, M.; Zwerschke, W. Reliable reference genes for expression analysis of proliferating and adipogenically differentiating human adipose stromal cells. Cell. Mol. Biol. Lett. 2019, 24, 14. [Google Scholar] [CrossRef] [PubMed]
- Gorzelniak, K.; Janke, J.; Engeli, S.; Sharma, A.M. Validation of endogenous controls for gene expression studies in human adipocytes and preadipocytes. Horm. Metab. Res. 2001, 33, 625–627. [Google Scholar] [CrossRef]
- Fink, T.; Lund, P.; Pilgaard, L.; Rasmussen, J.G.; Duroux, M.; Zachar, V. Instability of standard PCR reference genes in adipose-derived stem cells during propagation, differentiation and hypoxic exposure. BMC Mol. Biol. 2008, 9, 98. [Google Scholar] [CrossRef]
- Ebrahimi, R.; Toolabi, K.; Jannat Ali Pour, N.; Mohassel Azadi, S.; Bahiraee, A.; Zamani-Garmsiri, F.; Emamgholipour, S. Adipose tissue gene expression of long non-coding RNAs; MALAT1, TUG1 in obesity: Is it associated with metabolic profile and lipid homeostasis-related genes expression? Diabetol. Metab. Syndr. 2020, 12, 36. [Google Scholar] [CrossRef]
- Bourebaba, L.; Kornicka-Garbowska, K.; Al Naem, M.; Röcken, M.; Łyczko, J.; Marycz, K. MSI-1436 improves EMS adipose derived progenitor stem cells in the course of adipogenic differentiation through modulation of ER stress, apoptosis, and oxidative stress. Stem Cell Res. Ther. 2021, 12, 97. [Google Scholar] [CrossRef]
- El-Gindy, Y.M.; Hafsa, S.H.A.; El-Deeb, N.M. The expression of liver TNF-α gene, liver and small intestine histology of thermal stressed growing rabbits affected by allicin and lycopene. J. Therm. Biol. 2023, 113, 103521. [Google Scholar] [CrossRef]
- Radtke, C.L.; Nino-Fong, R.; Gonzalez, B.P.E.; Stryhn, H.; McDuffee, L.A. Characterization and osteogenic potential of equine muscle tissue–and periosteal tissue–derived mesenchymal stem cells in comparison with bone marrow–and adipose tissue–derived mesenchymal stem cells. Am. J. Vet. Res. 2013, 74, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Azarpeykan, S.; Dittmer, K.E. Evaluation of housekeeping genes for quantitative gene expression analysis in the equine kidney. J. Equine Sci. 2016, 27, 165–168. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hotham, W.E.; Thompson, C.; Szu-Ting, L.; Henson, F.M.D. The anti-inflammatory effects of equine bone marrow stem cell-derived extracellular vesicles on autologous chondrocytes. Vet. Rec. Open 2021, 8, 22. [Google Scholar] [CrossRef]
- Jarazo, J.M. Effect of Tissue Source on Adult Equine Multipotent Stromal Cell Pluripotency Induction Treatment with Synthetic mRNA. Master’s Thesis, Louisiana State University and Agricultural & Mechanical College, Baton Rouge, LA, USA, 2014. [Google Scholar]
- Bruynsteen, L.; Erkens, T.; Peelman, L.J.; Ducatelle, R.; Janssens, G.P.J.; Harris, P.A.; Hesta, M. Expression of inflammation-related genes is associated with adipose tissue location in horses. BMC Vet. Res. 2013, 9, 240. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.W.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Nazari, F.; Parham, A.; Maleki, A.F. GAPDH, β-actin and β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose-and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015, 57, 18. [Google Scholar] [CrossRef] [PubMed]
- De Spiegelaere, W.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Raabe, O.; Shell, K.; Würtz, A.; Reich, C.M.; Wenisch, S.; Arnhold, S. Further insights into the characterization of equine adipose tissue-derived mesenchymal stem cells. Vet. Res. Commun. 2011, 35, 355–365. [Google Scholar] [CrossRef]
- Vachkova, E.; Bosnakovski, D.; Yonkova, P.; Grigorova, N.; Ivanova, Z.; Todorov, P.; Penchev, G.; Milanova, A.; Simeonova, G.; Stanilova, S.; et al. Adipogenic potential of stem cells derived from rabbit subcutaneous and visceral adipose tissue in vitro. Vitr. Cell. Dev. Biol.-Anim. 2016, 52, 829–837. [Google Scholar] [CrossRef]
- Arnhold, S.; Elashry, M.I.; Klymiuk, M.C.; Geburek, F. Investigation of Stemness and Multipotency of Equine Adipose-Derived Mesenchymal Stem Cells (ASCs) from Different Fat Sources in Comparison with Lipoma. Stem Cell Res. Ther. 2019, 10, 309. [Google Scholar] [CrossRef]
| Gene Name | Stability Value | |
|---|---|---|
| Rabbit | Horse | |
| 18S | 0.473 | 0.589 |
| GAPDH | 0.136 | 0.235 |
| HPRT | 0.263 | 0.071 |
| Gene Name | Mean Ct Values with Their SD | |
|---|---|---|
| Rabbit | Horse | |
| 18S | 17.01 ± 1.05 | 19.03 ± 1.15 |
| GAPDH | 23.28 ± 0.40 | 23.48 ± 1.26 |
| HPRT | 27.74 ± 0.88 | 26.91 ± 0.94 |
| Gene Name | Slopes/Efficiency (%) of PCR Reaction | |
|---|---|---|
| Rabbit | Horse | |
| 18S | −3.449/94.96 | −3.389/97.28 |
| GAPDH | −3.333/99.54 | −3.337/99.38 |
| HPRT | −3.331/99.62 | −3.345/99.04 |
| Abbreviation | Full Name | Forward | Reverse | Product Length | NCBI Accession № |
|---|---|---|---|---|---|
| HORSE | |||||
| HPRT | Hypoxanthine phosphoribosyltransferase | CCAGTCAACAGGGGACATAA | GCTTGCGACCTTGACCATCT | 163 | AY372182.1 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | TCCCTGCTTCTACTGGTGCT | CGTATTTGGCAGCTTTCTCC | 147 | NM_001163856.1 |
| 18S | 18S ribosomal RNA | ATGCGGCGGCGTTATTCC | GCTATCAATCTGTCAATCCTGTCC | 204 | NR_046271.1 |
| RABBITS | |||||
| HPRT | Hypoxanthine phosphoribosyltransferase | CCCCAGCGTTGTGATTAGTG | GCCTCCCATCTCCTTCATCA | 163 | ENSOCUT00000030995.2 |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | GAACGGGAAGCTGGTCATCA | GAAGACGCCAGTGGATTCCA | 118 | GCA_000003625.1 |
| 18S | 18S ribosomal RNA | ATCAGATACCGTCGTAGTTC | TTCCGTCAATTCCTTTAAG | 167 | NR_033238.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, Z.; Petrova, V.; Grigorova, N.; Vachkova, E. Identification of the Reference Genes for Relative qRT-PCR Assay in Two Experimental Models of Rabbit and Horse Subcutaneous ASCs. Int. J. Mol. Sci. 2024, 25, 2292. https://doi.org/10.3390/ijms25042292
Ivanova Z, Petrova V, Grigorova N, Vachkova E. Identification of the Reference Genes for Relative qRT-PCR Assay in Two Experimental Models of Rabbit and Horse Subcutaneous ASCs. International Journal of Molecular Sciences. 2024; 25(4):2292. https://doi.org/10.3390/ijms25042292
Chicago/Turabian StyleIvanova, Zhenya, Valeria Petrova, Natalia Grigorova, and Ekaterina Vachkova. 2024. "Identification of the Reference Genes for Relative qRT-PCR Assay in Two Experimental Models of Rabbit and Horse Subcutaneous ASCs" International Journal of Molecular Sciences 25, no. 4: 2292. https://doi.org/10.3390/ijms25042292
APA StyleIvanova, Z., Petrova, V., Grigorova, N., & Vachkova, E. (2024). Identification of the Reference Genes for Relative qRT-PCR Assay in Two Experimental Models of Rabbit and Horse Subcutaneous ASCs. International Journal of Molecular Sciences, 25(4), 2292. https://doi.org/10.3390/ijms25042292






