Ectopic Expression of PtrLBD39 Retarded Primary and Secondary Growth in Populus trichocarpa
Abstract
1. Introduction
2. Results
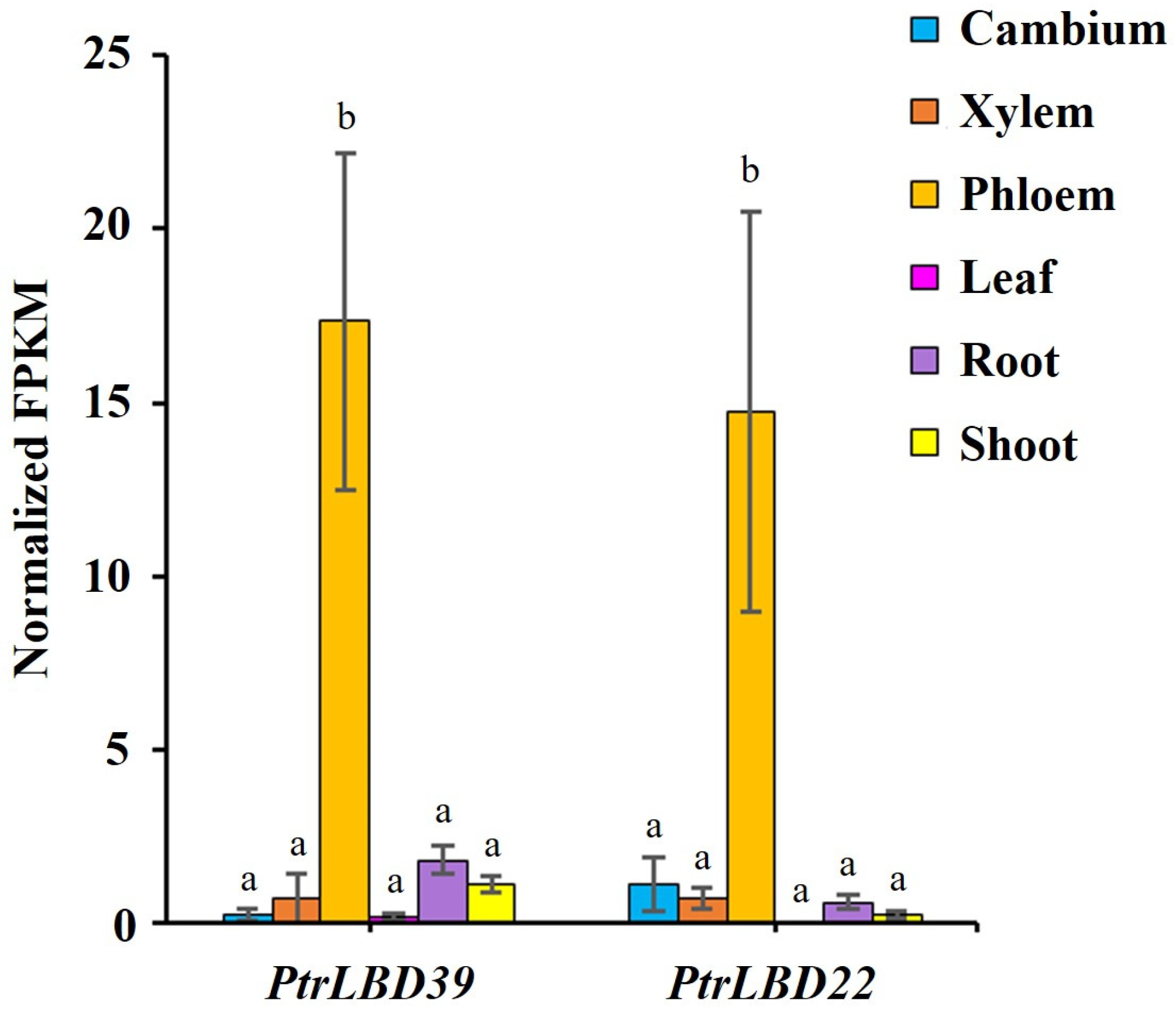
2.1. Tissue-Specific Expression of PtrLBD39 in P. trichocarpa
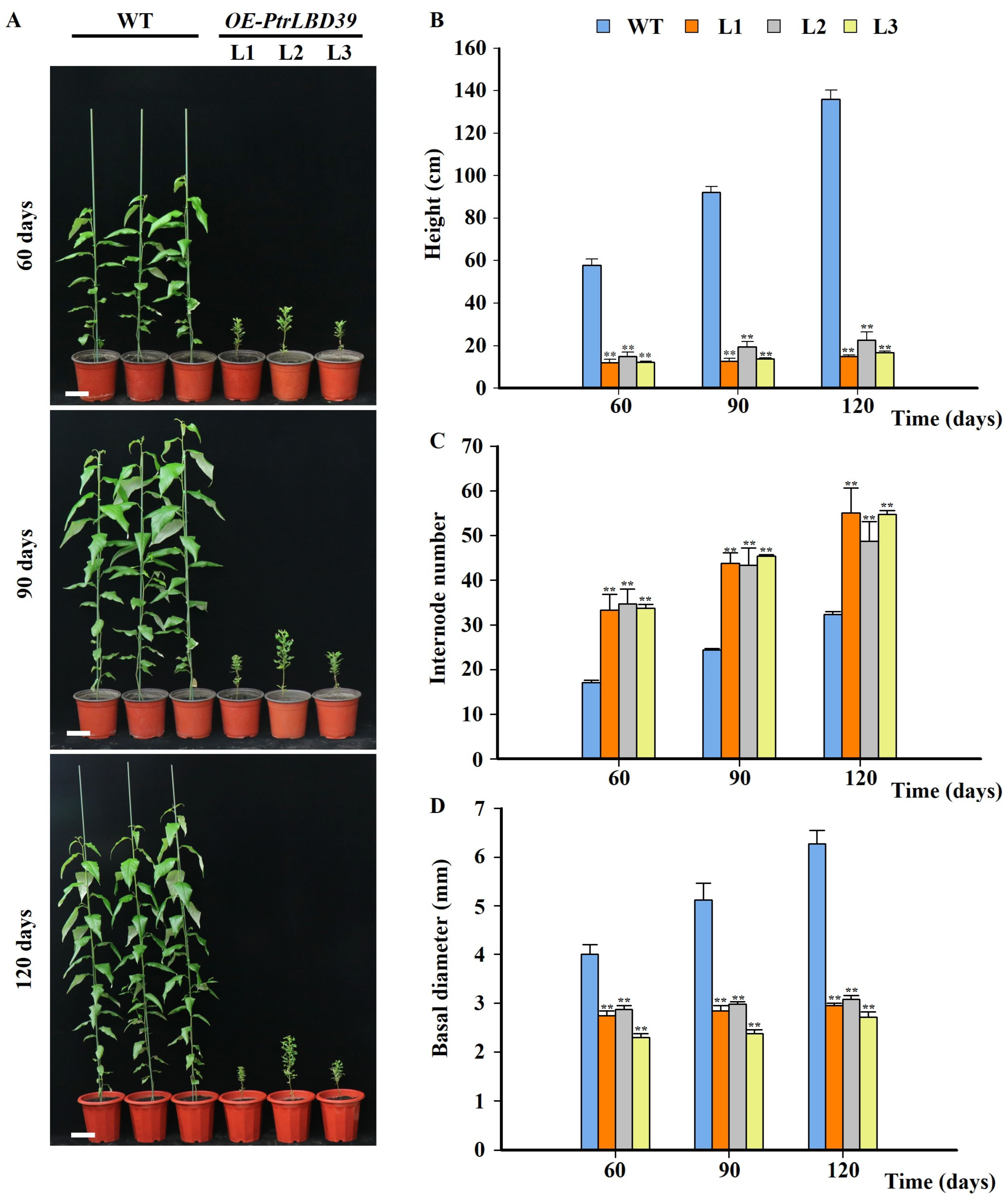
2.2. Ectopic Expression of PtrLBD39 in Transgenic P. trichocarpa Resulted in Stunted Phenotypes
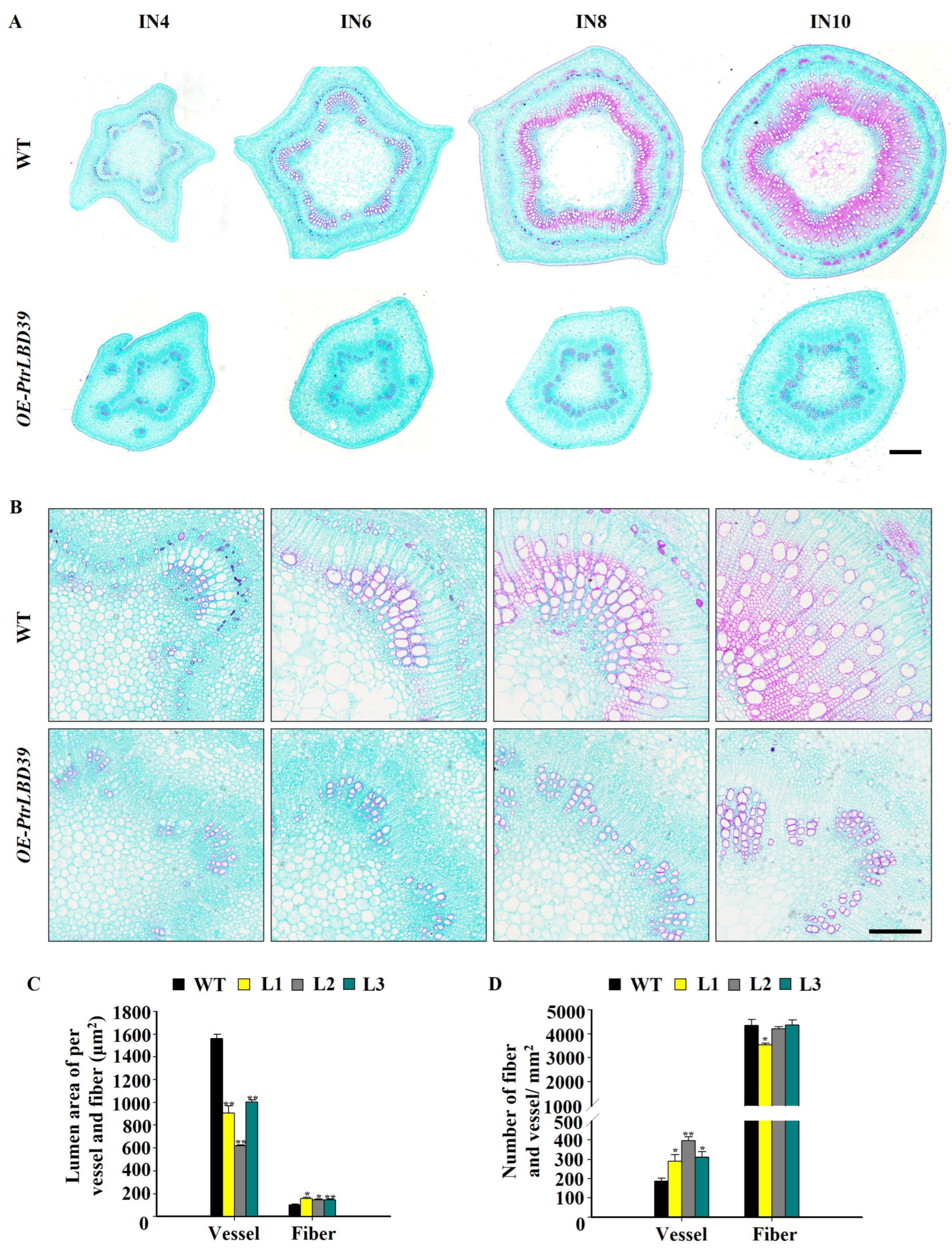
2.3. The Ectopic Expression of PtrLBD39 Significantly Affects the Lignin Structure and Polysaccharide Contents
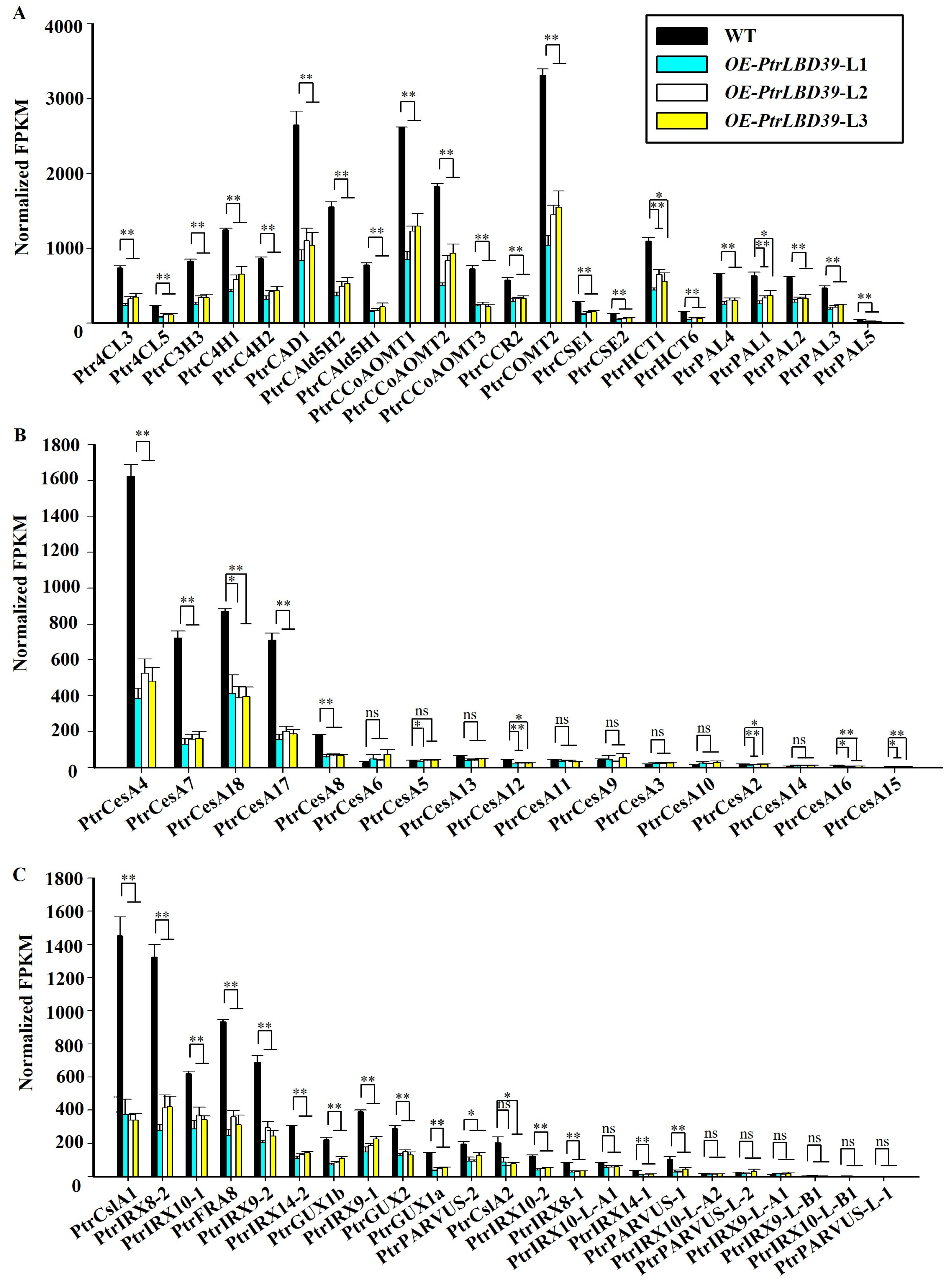
2.4. Identification of Downstream Genes of PtrLBD39
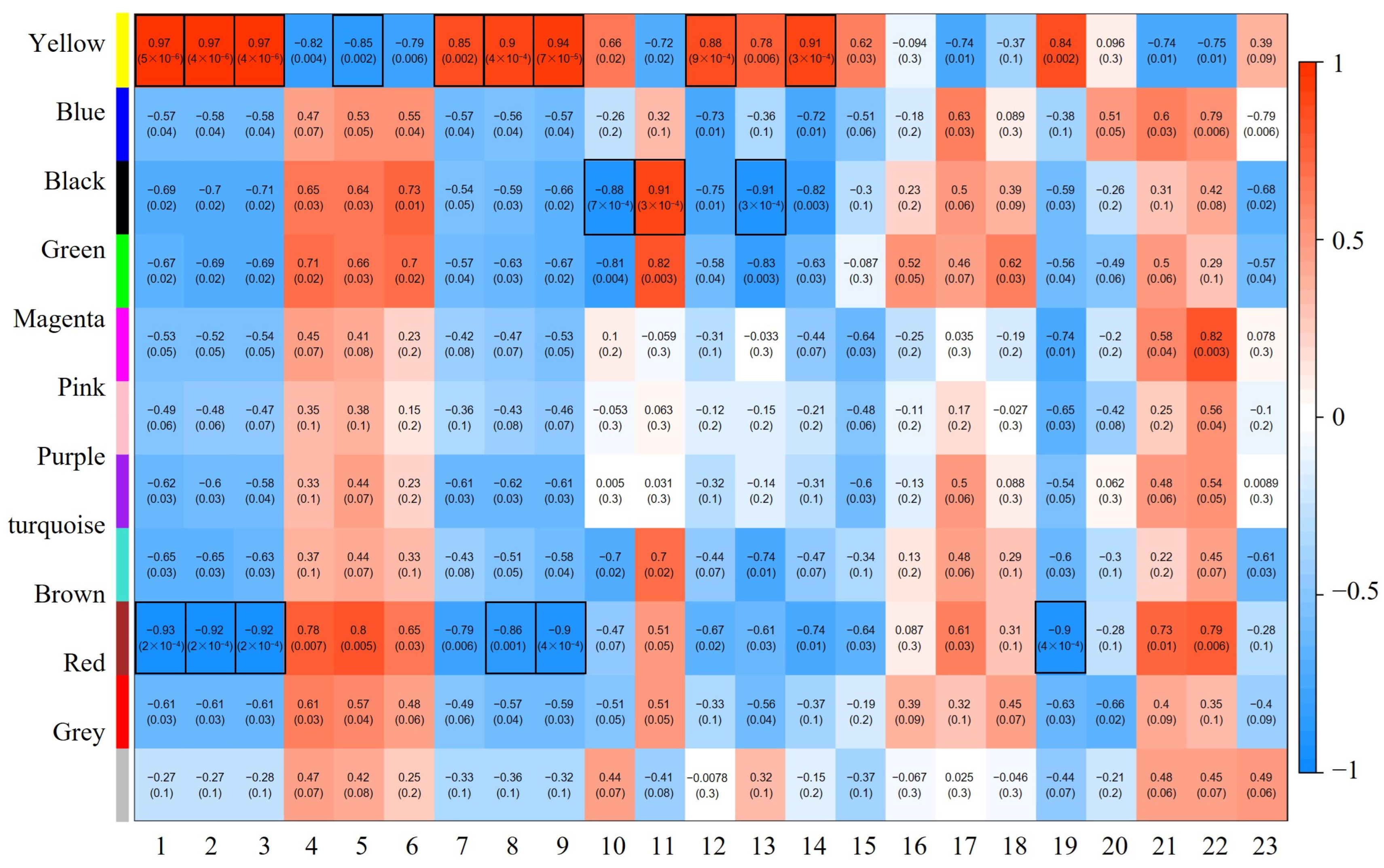
2.5. WGCNA Reveals Gene Modules Associated with Phenotypes
2.6. Functional and Pathway Enrichment Analysis of the Eigengenes in Modules
2.7. WGCNA Reveals the Genes Expression Directly Related to Phenotype in P. trihcocarpa
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Generation of Gene Overexpression Transgenic P. trichocarpa
4.3. RT-qPCR
4.4. Histochemical and Histological Analysis
4.5. Scanning Electron Micrograph Analysis
4.6. Transmission Electron Microscopy
4.7. Wood Chemistry
4.8. RNA-Seq and Data Analysis
4.9. Motif Analysis
4.10. Weighted Gene Co-Expression Network Analysis
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, Z.H.; Zhong, R.Q. Molecular control of wood formation in trees. J. Exp. Bot. 2015, 66, 4119–4131. [Google Scholar] [CrossRef]
- Schuetz, M.; Smith, R.; Ellis, B. Xylem tissue specification, patterning, and differentiation mechanisms. J Exp. Bot. 2013, 64, 11–31. [Google Scholar] [CrossRef]
- Zhang, J.; Nieminen, K.; Serra, J.A.A.; Helariutta, Y. The formation of wood and its control. Curr. Opin. Plant Biol. 2014, 17, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Evert, R.F. Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006; p. 601. [Google Scholar]
- Bowyer, J.L.; Shmulsky, R.; Haygreen, J.G. Forest Products and Wood Science: An Introduction, 4th ed.; Iowa State Press: Ames, IW, USA, 2003. [Google Scholar]
- Zhong, R.Q.; Lee, C.H.; Ye, Z.H. Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 2010, 152, 1044–1055. [Google Scholar] [CrossRef]
- Zhong, R.Q.; McCarthy, R.L.; Lee, C.; Ye, Z.H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011, 157, 1452–1468. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Nishikubo, N.; Xu, B.; Yamaguchi, M.; Mitsuda, N.; Goué, N.; Shi, F.; Ohme-Takagi, M.; Demura, T. A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 2011, 67, 499–512. [Google Scholar] [CrossRef]
- Li, Q.Z.; Lin, Y.C.; Sun, Y.H.; Song, J.; Chen, H.; Zhang, X.H.; Sederoff, R.R.; Chiang, V.L. Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. USA 2012, 109, 14699–14704. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.P.; Liu, H.Z.; Li, H.Y.; Lin, Y.C.J.; Shi, R.; Yang, C.M.; Gao, J.H.; Zhou, C.G.; Li, Q.Z. Hierarchical transcription factor and chromatin binding network for wood formation in Populus trichocarpa. Plant Cell 2019, 31, 602–626. [Google Scholar] [CrossRef]
- Yeh, C.S.; Wang, Z.F.; Miao, F.; Ma, H.Y.; Kao, C.T.; Hsu, T.S.; Yu, J.H.; Hung, E.T.; Lin, C.C.; Kuan, C.Y.; et al. A novel synthetic-genetic-array-based yeast one-hybrid system for high discovery rate and short processing time. Genome Res. 2019, 29, 1343–1351. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Mao, Y.L.; Guo, Y.J.; Gao, J.H.; Liu, X.Y.; Li, S.; Lin, Y.C.J.; Chen, H.; Wang, J.P.; Chiang, V.L.; et al. MYB transcription factor161 mediates feedback regulation of secondary wall-associated NAC-Domain1 family genes for wood formation. Plant Physiol. 2020, 184, 1389–1406. [Google Scholar] [CrossRef]
- Kubo, M.; Udagawa, M.; Nishikubo, N.; Horiguchi, G.; Yamaguchi, M.; Ito, J.; Mimura, T.; Fukuda, H.; Demura, T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005, 19, 1855–1860. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.Q.; Demura, T.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Yu, J.; Wang, Z.; Sun, Y.; Li, S.; Lin, Y.C.J.; Chiang, V.L.; Li, W.; Wang, J.P. Transcriptional reprogramming of xylem cell wall biosynthesis in tension wood. Plant Physiol. 2021, 186, 250–269. [Google Scholar] [CrossRef]
- Yu, J.; Zhou, C.G.; Li, D.N.; Li, S.; Lin, Y.C.J.; Wang, J.P.; Chiang, V.L.; Li, W. A PtrLBD39-mediated transcriptional network regulates tension wood formation in Populus trichocarpa. Plant Commun. 2022, 3, 100250. [Google Scholar] [CrossRef]
- Björklund, S.; Antti, H.; Uddestrand, I.; Moritz, T.; Sundberg, B. Cross-talk between gibberellin and auxin in development of Populus wood: Gibberellin stimulates polar auxin transport and has a common transcriptome with auxin. Plant J. 2007, 52, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Bin, S.; Cristina, G.R.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2010, 16, 47–52. [Google Scholar] [CrossRef]
- Borghi, L.; Bureau, M.; Simon, R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 2007, 19, 1795–1808. [Google Scholar] [CrossRef]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef]
- Liu, H.J.; Wang, S.F.; Yu, X.B.; Yu, J.; He, X.W.; Zhang, S.L.; Shou, H.X.; Wu, P. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005, 43, 47–56. [Google Scholar] [CrossRef]
- Xu, L.; Xu, Y.; Dong, A.W.; Sun, Y.; Pi, L.M.; Xu, Y.Q.; Huang, H. Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 2003, 130, 4097–4107. [Google Scholar] [CrossRef]
- Xu, B.; Li, Z.Y.; Zhu, Y.; Wang, H.; Ma, H.; Dong, A.; Huang, H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 2008, 146, 566–575. [Google Scholar] [CrossRef]
- Fan, M.Z.; Xu, C.Y.; Xu, K.; Hu, Y.X. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012, 22, 1169–1180. [Google Scholar] [CrossRef]
- Yordanov, Y.S.; Regan, S.; Busov, V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in Populus. Plant Cell 2010, 22, 3662–3677. [Google Scholar] [CrossRef]
- Lu, Q.; Shao, F.J.; Macmillan, C.; Wilson, I.W.; van der Merwe, K.; Hussey, S.G.; Myburg, A.A.; Dong, X.M.; Qiu, D.Y. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandis reveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef]
- Cao, H.; Liu, C.Y.; Liu, C.X.; Zhao, Y.L.; Xu, R.R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef]
- Han, Z.; Yang, T.; Guo, Y.; Cui, W.H.; Yao, L.J.; Li, G.; Wu, A.M.; Li, J.H.; Liu, L.J. The transcription factor PagLBD3 contributes to the regulation of secondary growth in Populus. J. Exp. Bot. 2021, 72, 7092–7106. [Google Scholar] [CrossRef]
- Smit, M.E.; McGregor, S.R.; Sun, H.; Gough, C.; Bågman, A.M.; Soyars, C.L.; Kroon, J.T.; Gaudinier, A.; Williams, C.J.; Yang, X.Y.; et al. A PXY-mediated transcriptional network integrates signaling mechanisms to control vascular development in Arabidopsis. Plant Cell 2020, 32, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Thitamadee, S.; Machida, Y.; Chua, N.H. Asymmetric Leaves2-Like19/Lateral Organ Boundaries Domain30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell 2008, 20, 3359–3373. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.F.; Zhai, R.; Lin, J.J.; Wang, Z.F.; Meng, D.K.; Li, M.; Mao, Y.L.; Gao, B.Y.; Ma, H.Y.; Zhang, B.F.; et al. Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa. Nat. Plants 2023, 9, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Wang, J.P.; Lin, Y.C.; Li, Q.Z.; Sun, Y.H.; Chen, H.; Sederoff, R.R.; Chiang, V.L. Tissue and cell-type co-expression networks of transcription factors and wood component genes in Populus trichocarpa. Planta 2017, 245, 927–938. [Google Scholar] [CrossRef]
- Suzuki, S.; Li, L.G.; Sun, Y.H.; Chiang, V.L. The cellulose synthase gene superfamily and biochemical functions of xylem specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef]
- Kumar, M.; Thammannagowda, S.; Bulone, V.; Chiang, V.; Han, K.H.; Joshi, C.P.; Mansfield, S.D.; Mellerowicz, E.; Sundberg, B.; Teeri, T.; et al. An update on the nomenclature for the cellulose synthase genes in Populus. Trends Plant Sci. 2009, 14, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, V.B.; Bielach, A.; Hrtyan, M. Redox regulation at the site of primary growth: Auxin, cytokinin and ROS crosstalk. Plant Cell Environ. 2017, 40, 2586–2605. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.D.; Feng, Z.H.; Meng, L.S.; Zhu, J.; Geitmann, A. Arabidopsis ASL11/LBD15 is involved in shoot apical meristem development and regulates WUS expression. Planta 2013, 237, 1367–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Guo, J.S.; Zhou, C.; Zhu, J. Ectopic expression of LBD15 affects lateral branch development and secondary cell wall synthesis in Arabidopsis thaliana. Plant Growth Regul. 2014, 73, 111–120. [Google Scholar] [CrossRef]
- Qin, H.; He, L.N.; Huang, R.F. The coordination of ethylene and other hormones in primary root development. Front. Plant Sci. 2019, 10, 874. [Google Scholar] [CrossRef] [PubMed]
- Baucher, M.; El Jaziri, M.; Vandeputte, O. From primary to secondary growth: Origin and development of the vascular system. J. Exp. Bot. 2007, 58, 3485–3501. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- Wei, L.M.; Wen, S.Y.; Ma, J.K.; Tu, Z.H.; Zhu, S.H.; Zhai, X.Y.; Li, H.G. Overexpression of LtuHB6 from Liriodendron tulipifera causes lobed-leaf formation in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 2022, 28, 1875–1887. [Google Scholar] [CrossRef]
- Konishi, M.; Donner, T.J.; Scarpella, E.; Yanagisawa, S. MONOPTEROS directly activates the auxin-inducible promoter of the Dof5.8 transcription factor gene in Arabidopsis thaliana leaf provascular cells. J. Exp. Bot. 2015, 66, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Eklund, D.M.; Cierlik, I.; Ståldal, V.; Claes, A.R.; Vestman, D.; Chandler, J.; Sundberg, E. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 2011, 157, 2069–2080. [Google Scholar] [CrossRef] [PubMed]
- Sohlberg, J.J.; Myrenås, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J. 2006, 47, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Sohlberg, J.J.; Myrenås, M.; Kuusk, S.; Lagercrantz, U.; Kowalczyk, M.; Sandberg, G.; Sundberg, E. A broadly conserved NERD genetically interacts with the exocyst to affect root growth and cell expansion. J. Exp. Bot. 2018, 69, 3625–3637. [Google Scholar]
- Ohman, D.; Demedts, B.; Kumar, M.; Gerber, L.; Gorzsás, A.; Goeminne, G.; Hedenström, M.; Ellis, B.; Boerjan, W.; Sundberg, B. MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 2013, 73, 63–76. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Song, X.Q.; Zhou, H.J.; Wei, K.L.; Jiang, C.; Wang, J.N.; Cao, Y.; Tang, F.; Zhao, S.T.; Lu, M.Z. KNAT2/6b, a class I KNOX gene, impedes xylem differentiation by regulating NAC domain transcription factors in poplar. New Phytol. 2020, 225, 1531–1544. [Google Scholar] [CrossRef]
- Jin, Y.L.; Yu, C.Y.; Jiang, C.M.; Guo, X.T.; Li, B.; Wang, C.T.; Kong, F.J.; Zhang, H.X.; Wang, H.H. PtiCYP85A3, a BR C-6 oxidase gene, plays a critical role in brassinosteroid-mediated tension wood formation in poplar. Front. Plant Sci. 2020, 11, 468. [Google Scholar] [CrossRef]
- Choi, S.J.; Lee, Z.; Kim, S.; Jeong, E.; Shim, J.S. Modulation of lignin biosynthesis for drought tolerance in plants. Front. Plant Sci. 2023, 14, 1116426. [Google Scholar] [CrossRef]
- Youn, B.; Camacho, R.; Moinuddin, S.G.A.; Lee, C.; Davin, L.B.; Lewis, N.G.; Kang, C.H. Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Org. Biomol. Chem. 2006, 4, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.M.; Tunlaya-Anukit, S.; Chen, H.C.; Li, Q.Z.; Liu, J.; Lin, C.Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef] [PubMed]
- Christians, M.J.; Gingerich, D.J.; Hansen, M.; Binder, B.M.; Kieber, J.J.; Vierstra, R.D. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 2009, 57, 332–345. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.X.; Zhu, L.; Liu, T.Q.; Leng, X.Y.; Zhu, Z.X.; Xie, W.; Lv, H.T.; Jin, Z.X.; Wu, P.; Zhang, Z.C. Identification of a novel OsCYP2 allele that was involved in rice response to low temperature stress. Phyton Int. J. Exp. Bot. 2023, 92, 1743–1763. [Google Scholar] [CrossRef]
- Islam, N.S. Maize ZmWRKY28: A target to regulate shade avoidance response under high planting density. J. Exp. Bot. 2023, 74, 2937–2939. [Google Scholar] [CrossRef]
- Yuan, T.T.; Xu, H.H.; Li, J.; Lu, Y.T. Auxin abolishes SHI-RELATED SEQUENCE5-mediated inhibition of lateral root development in Arabidopsis. New Phytol. 2020, 225, 297–309. [Google Scholar] [CrossRef]
- Skirycz, A.; Radziejwoski, A.; Busch, W.; Hannah, M.A.; Czeszejko, J.; Kwasniewski, M.; Zanor, M.I.; Lohmann, J.U.; De Veylder, L.; Witt, I.; et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. Plant J. 2008, 56, 779–792. [Google Scholar] [CrossRef]
- Li, L.; Zhou, Y.H.; Cheng, X.F.; Sun, J.Y.; Marita, J.M.; Ralph, J.; Chiang, V.L. Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc. Natl. Acad. Sci. USA 2003, 100, 4939–4944. [Google Scholar] [CrossRef]
- Saito, K.; Watanabe, Y.; Shirakawa, M.; Matsushita, Y.; Imai, T.; Koike, T.; Sano, Y.; Funada, R.; Fukazawa, K.; Fukushima, K. Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant J. 2012, 69, 542–552. [Google Scholar] [CrossRef]
- Gui, J.; Lam, P.Y.; Tobimatsu, Y.; Sun, J.; Huang, C.; Cao, S.M.; Zhong, Y.; Umezawa, T.; Li, L.G. Fiber-specific regulation of lignin biosynthesis improves biomass quality in Populus. New Phytol. 2020, 226, 1074–1087. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J.; Hallett, J.P.; Leak, D.J.; Liotta, C.L.; et al. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef]
- Groover, A.T.; Mansfield, S.D.; DiFazio, S.P.; Dupper, G.; Fontana, J.R.; Millar, R.; Wang, Y. The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 2006, 61, 917–932. [Google Scholar] [CrossRef]
- Wessels, B.; Seyfferth, C.; Escamez, S.; Vain, T.; Antos, K.; Vahala, J.; Delhomme, N.; Kangasjarvi, J.; Eder, M.; Felten, J.; et al. An AP2/ERF transcription factor ERF139 coordinates xylem cell expansion and secondary cell wall deposition. New Phytol. 2019, 224, 1585–1599. [Google Scholar] [CrossRef]
- Felten, J.; Vahala, J.; Love, J.; Gorzsás, A.; Rüggeberg, M.; Delhomme, N.; Lesniewska, J.; Kangasjärvi, J.; Hvidsten, T.R.; Mellerowicz, E.J. Ethylene signaling induces gelatinous layers with typical features of tension wood in hybrid aspen. New Phytol. 2018, 218, 999–1014. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinins. Arab. Book 2014, 12, e0168. [Google Scholar] [CrossRef]
- Skalák, J.; Vercruyssen, L.; Claeys, H.; Hradilová, J.; Cerny, M.; Novák, O.; Placková, L.; Saiz-Fernández, I.; Skaláková, P.; Coppens, F.; et al. Multifaceted activity of cytokinin in leaf development shapes its size and structure in Arabidopsis. Plant J. 2019, 97, 805–824. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.; Park, G.; Cho, H.; Choi, D.; Umeda, M.; Choi, Y.; Hwang, D.; Hwang, I. CYTOKININ-RESPONSIVE GROWTH REGULATOR regulates cell expansion and cytokinin-mediated cell cycle progression. Plant Physiol. 2021, 186, 1734–1746. [Google Scholar] [CrossRef]
- Castro-Camba, R.; Sánchez, C.; Vidal, N.; Vielba, J.M. Plant development and crop yield: The role of gibberellins. Plants 2022, 11, 2650. [Google Scholar] [CrossRef]
- Lange, M.J.P.; Lange, T. Gibberellin biosynthesis and the regulation of plant development. Plant Biol. 2006, 8, 281–290. [Google Scholar] [CrossRef]
- Wu, H.; Bai, B.B.; Lu, X.D.; Li, H.Y. A gibberellin-deficient maize mutant exhibits altered plant height, stem strength and drought tolerance. Plant Cell Rep. 2003, 42, 1687–1699. [Google Scholar] [CrossRef]
- Hedden, P.; Spomsel, V. A century of gibberellin research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef]
- Sorce, C.; Giovannelli, A.; Sebastiani, L.; Anfodillo, T. Hormonal signals involved in the regulation of cambial activity, xylogenesis and vessel patterning in trees. Plant Cell Rep. 2013, 32, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Serra, J.A.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and auxin display distinct but interconnected distribution and signaling profiles to stimulate cambial activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef]
- Miyashima, S.; Roszak, P.; Sevilem, I.; Toyokura, K.; Blob, B.; Heo, J.O.; Mellor, N.; Help-Rinta-Rahko, H.; Otero, S.; Smet, W.; et al. Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 2019, 565, 490–494. [Google Scholar] [CrossRef]
- Hu, J.; Su, H.L.; Cao, H.; Wei, H.B.; Fu, X.K.; Jiang, X.M.; Song, Q.; He, X.H.; Xu, C.Z.; Luo, K.M. AUXIN RESPONSE FACTOR7 integrates gibberellin and auxin signaling via interactions between DELLA and AUX/IAA proteins to regulate cambial activity in poplar. Plant Cell 2022, 34, 2688–2707. [Google Scholar] [CrossRef]
- Smetana, O.; Mäkilä, R.; Lyu, M.; Amiryousefi, A.; Rodríguez, F.S.; Wu, M.F.; Solé-Gil, A.; Gavarrón, M.L.; Siligato, R.; Miyashima, S.; et al. High levels of auxin signalling define the stem-cell organizer of the vascular cambium. Nature 2019, 565, 485–489. [Google Scholar] [CrossRef]
- Li, W.; Lin, Y.C.J.; Chen, Y.L.; Zhou, C.; Li, S.; De Ridder, N.; Oliveira, D.M.; Zhang, L.; Zhang, B.; Wang, J.P.; et al. Woody plant cell walls: Fundamentals and utilization. Mol. Plant 2024, 17, 112–140. [Google Scholar] [CrossRef]
- Geng, P.; Zhang, S.; Liu, J.Y.; Zhao, C.H.; Wu, J.; Cao, Y.P.; Fu, C.X.; Han, X.; He, H.; Zhao, Q. MYB20, MYB42, MYB43, and MYB85 regulate phenylalanine and lignin biosynthesis during secondary cell wall formation. Plant Physiol. 2020, 182, 1272–1283. [Google Scholar] [CrossRef]
- Zhong, R.Q.; Lee, C.H.; Zhou, J.L.; McCarthy, R.L.; Ye, Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef]
- Liebsch, D.; Sunaryo, W.; Holmlund, M.; Norberg, M.; Zhang, J.; Hall, H.C.; Helizon, H.; Jin, X.; Helariutta, Y.; Nilsson, O.; et al. Class I KNOX transcription factors promote differentiation of cambial derivatives into xylem fibers in the Arabidopsis hypocotyl. Development 2014, 141, 4311–4319. [Google Scholar] [CrossRef]
- Sugimoto, H.; Tanaka, T.; Muramoto, N.; Kitagawa-Yogo, R.; Mitsukawa, N. Transcription factor NTL9 negatively regulates Arabidopsis vascular cambium development during stem secondary growth. Plant Physiol. 2022, 190, 1731–1746. [Google Scholar] [CrossRef]
- Li, S.; Lin, Y.C.J.; Wang, P.; Zhang, B.; Li, M.; Chen, S.; Shi, R.; Tunlaya-Anukit, S.; Liu, X.; Wang, Z.; et al. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 2019, 31, 663–686. [Google Scholar] [CrossRef]
- Li, S.J.; Zhen, C.; Xu, W.J.; Wang, C.; Cheng, Y.X. Simple, rapid and efficient transformation of genotype Nisqually-1: A basic tool for the first sequenced model tree. Sci. Rep. 2017, 7, 2638. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Abraham, P.; Giannone, R.J.; Adams, R.M.; Kalluri, U.; Tuskan, G.A.; Hettich, R.L. Putting the pieces together: High-performance LC-MS/MS provides network, pathway and protein-level perspectives in Populus. Mol. Cell. Proteom. 2013, 12, 106–119. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]

| Wood Composition | WT | L1 | L2 | L3 |
|---|---|---|---|---|
| Glucose | 54.00 ± 0.34 | 48.16 ± 0.56 ** | 47.66 ± 0.62 ** | 47.5 ± 0.8 ** |
| Xylose | 14.84 ± 0.72 | 11.69 ± 0.37 * | 11.66 ± 0.28 * | 11.72 ± 0.19 * |
| Total carbohydrate | 72.51 ± 1.39 | 63.09 ± 1.30 * | 61.30 ± 0.52 ** | 61.39 ± 0.88 ** |
| Acid insoluble lignin | 18.90 ± 0.66 | 19.33 ± 0.09 | 18.67 ± 0.44 | 19.02 ± 0.31 |
| Acid soluble lignin | 2.21 ± 0.06 | 3.01 ± 0.15 * | 2.52 ± 0.10 | 3.29 ± 0.09 ** |
| Total lignin | 21.12 ± 0.66 | 22.34 ± 0.24 | 21.19 ± 0.54 | 22.30 ± 0.26 |
| Lignin Composition | WT | L1 | L2 | L3 |
|---|---|---|---|---|
| S-Lignin | 63.7 ± 0.5% | 45.5 ± 0.7% ** | 59.7 ± 0.1% ** | 59.46 ± 0.6% ** |
| G-Lignin | 31.2 ± 0.2% | 51.2 ± 0.8% ** | 36.5 ± 0.3% ** | 37.4 ± 0.4% ** |
| H-Lignin | 5.0 ± 0.3% | 3.2 ± 0.3% * | 3.8 ± 0.1% * | 3.1 ± 0.2% ** |
| S/G ratio | 2.0 ± 0.03 | 0.8 ± 0.02 ** | 1.63 ± 0.01 ** | 1.59 ± 0.03 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Gao, B.; Li, D.; Li, S.; Chiang, V.L.; Li, W.; Zhou, C. Ectopic Expression of PtrLBD39 Retarded Primary and Secondary Growth in Populus trichocarpa. Int. J. Mol. Sci. 2024, 25, 2205. https://doi.org/10.3390/ijms25042205
Yu J, Gao B, Li D, Li S, Chiang VL, Li W, Zhou C. Ectopic Expression of PtrLBD39 Retarded Primary and Secondary Growth in Populus trichocarpa. International Journal of Molecular Sciences. 2024; 25(4):2205. https://doi.org/10.3390/ijms25042205
Chicago/Turabian StyleYu, Jing, Boyuan Gao, Danning Li, Shuang Li, Vincent L. Chiang, Wei Li, and Chenguang Zhou. 2024. "Ectopic Expression of PtrLBD39 Retarded Primary and Secondary Growth in Populus trichocarpa" International Journal of Molecular Sciences 25, no. 4: 2205. https://doi.org/10.3390/ijms25042205
APA StyleYu, J., Gao, B., Li, D., Li, S., Chiang, V. L., Li, W., & Zhou, C. (2024). Ectopic Expression of PtrLBD39 Retarded Primary and Secondary Growth in Populus trichocarpa. International Journal of Molecular Sciences, 25(4), 2205. https://doi.org/10.3390/ijms25042205






