Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma
Abstract
1. Introduction
2. Results
2.1. Patient Demographic and Clinical Characteristics
2.2. Allele and Genotype Distributions
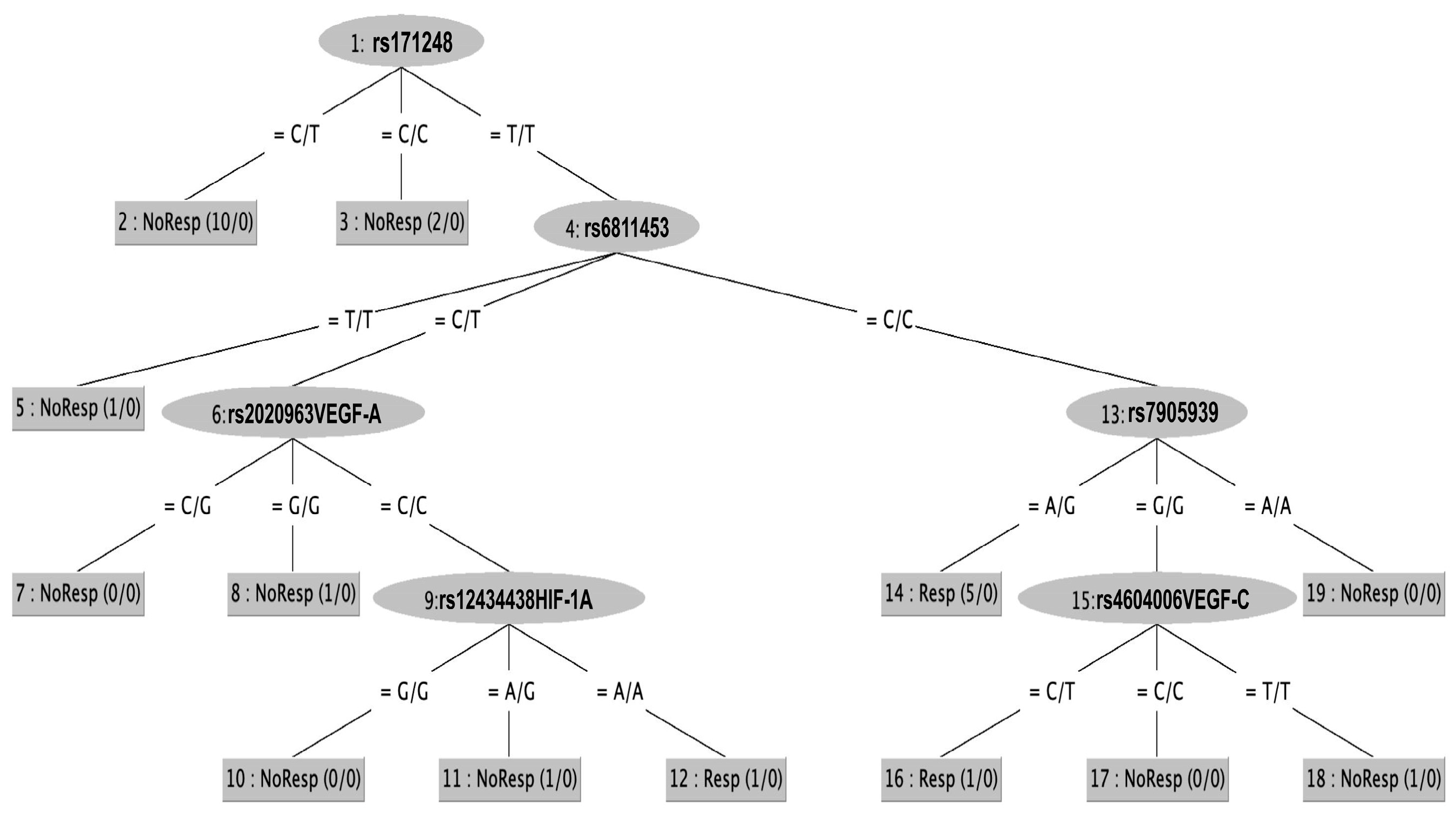
2.3. SNPs and Classification Rules Related to Sorafenib Response
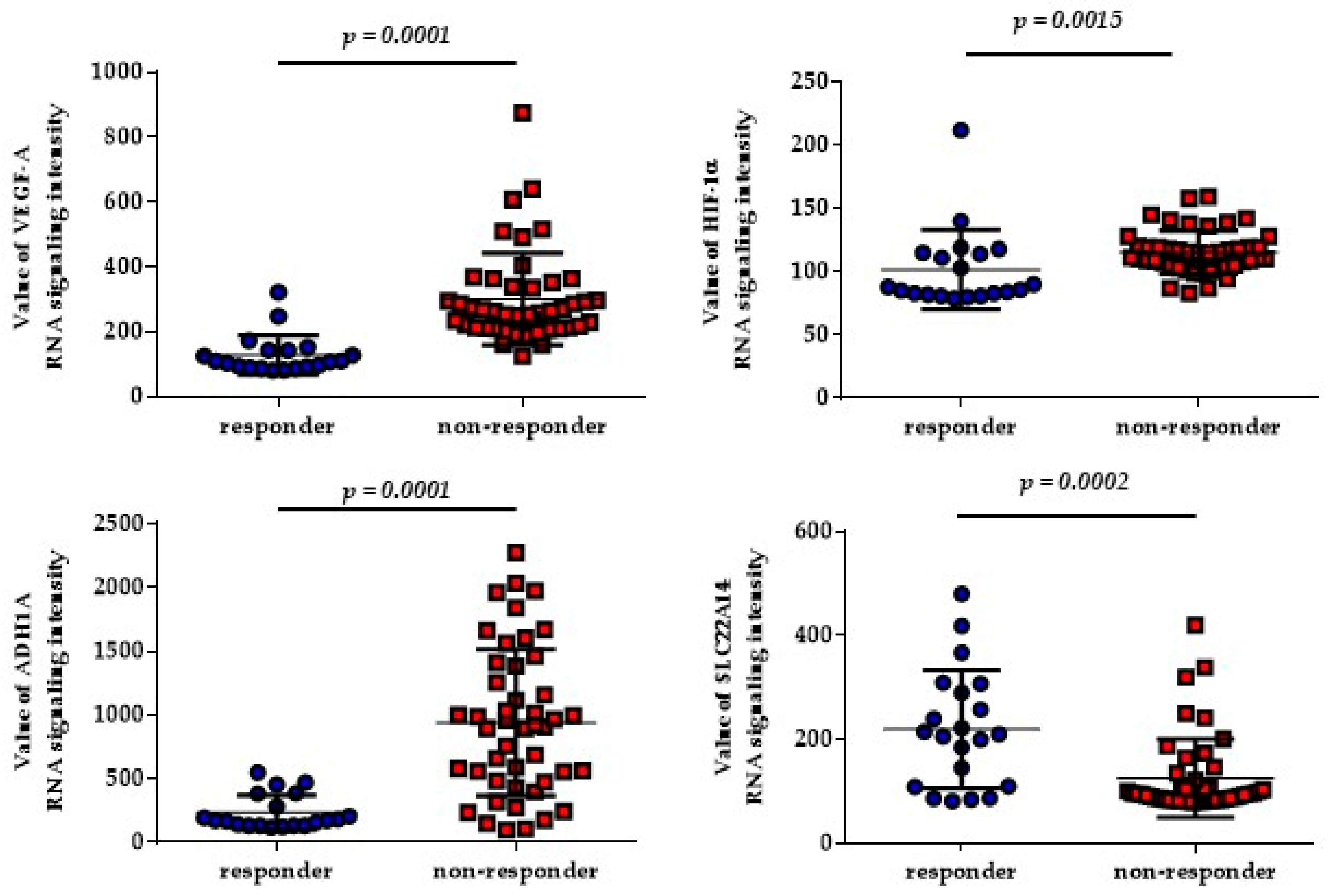
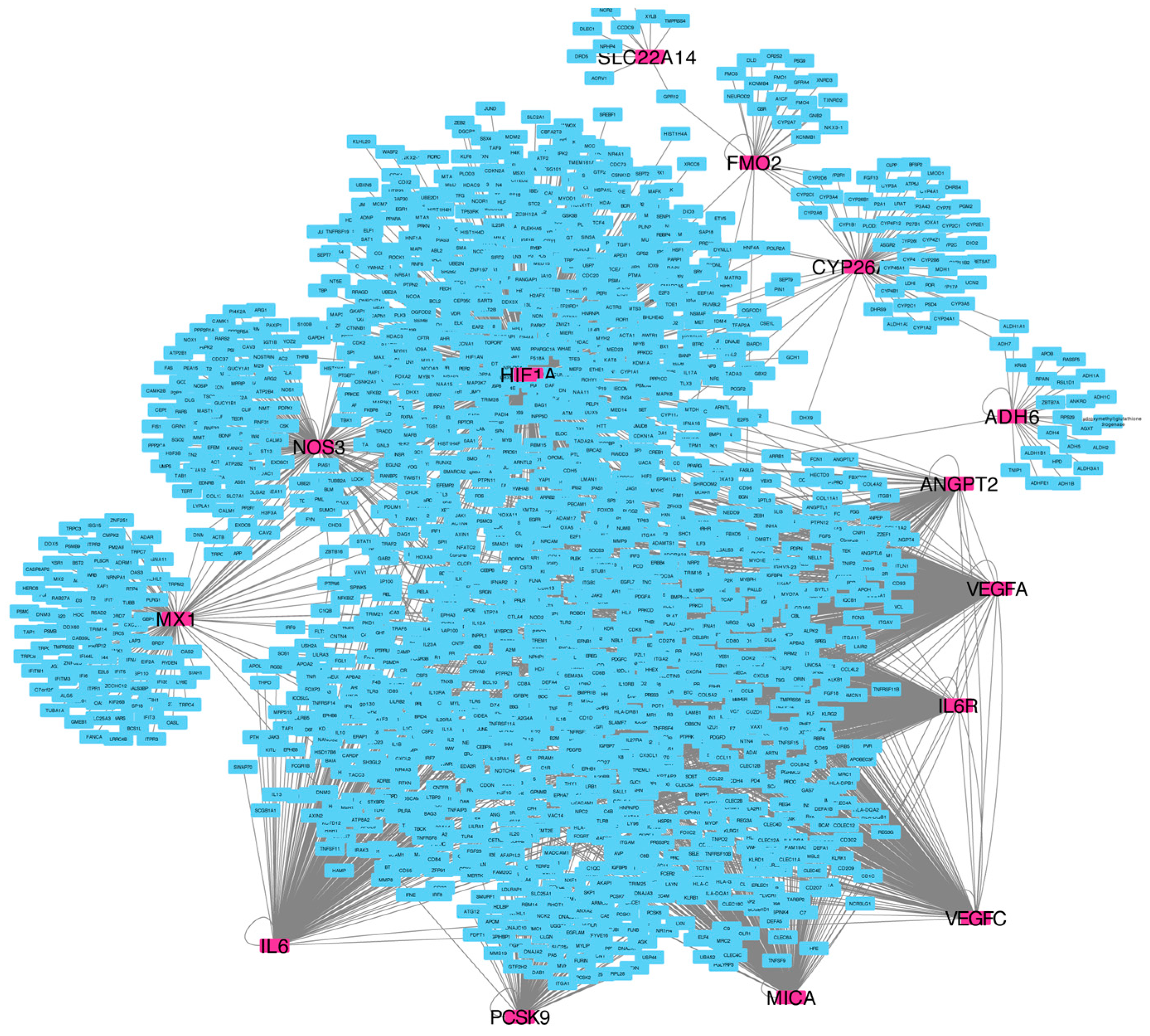
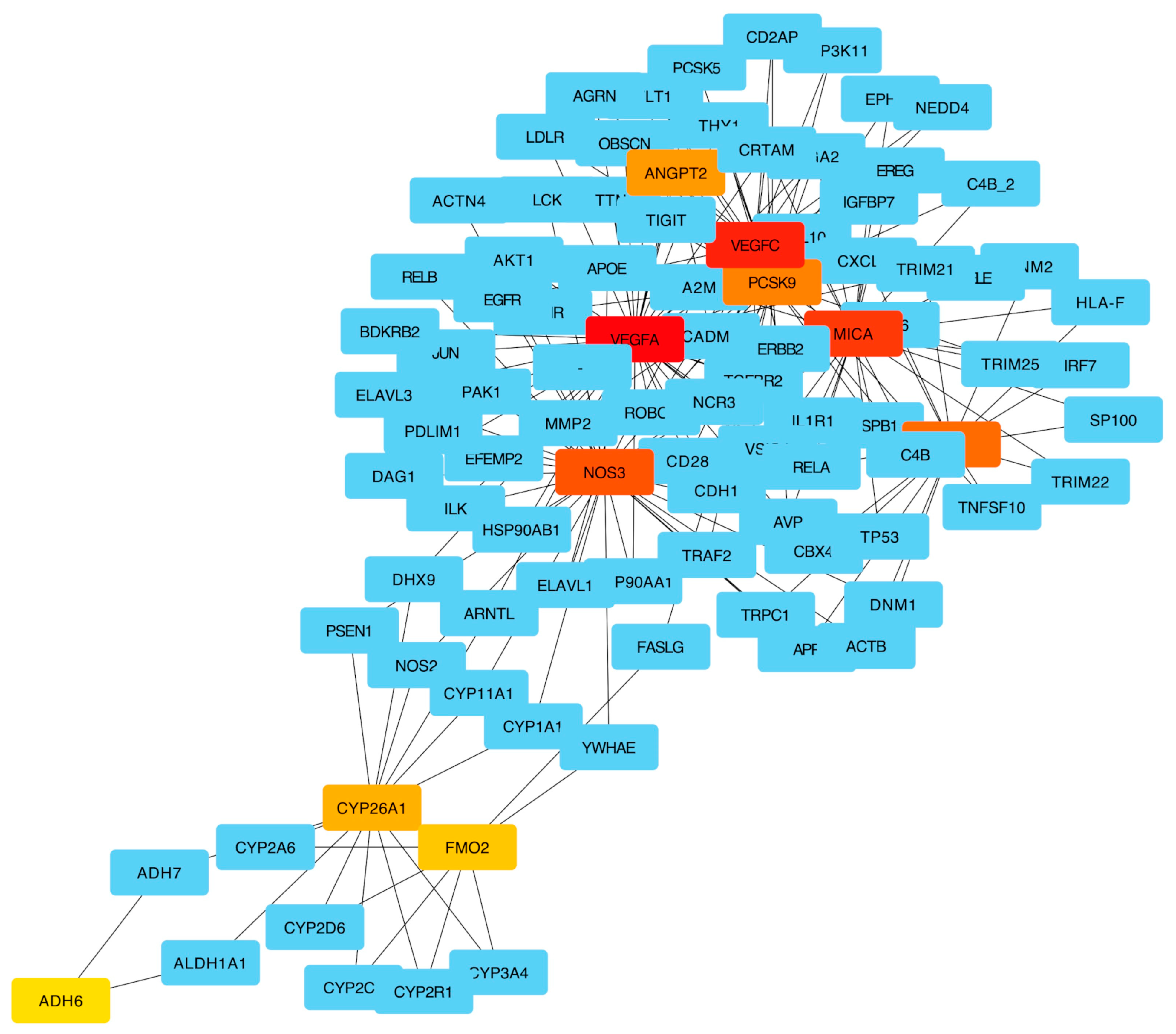
2.4. Pathway Enrichment Results
3. Discussion
4. Materials and Methods
4.1. Study Participants
4.2. Sample Collection and Genotyping
4.3. Genetic Risk Score
4.4. Statistical Analysis
4.5. Classification Rules
4.6. Network and Pathway Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cervello, M.; Emma, M.R.; Augello, G.; Cusimano, A.; Giannitrapani, L.; Soresi, M.; Akula, S.M.; Abrams, S.L.; Steelman, L.S. New landscapes and horizons in hepatocellular carcinoma therapy. Aging 2020, 12, 3053–3094. [Google Scholar] [CrossRef]
- Luo, J.; Gao, B.; Lin, Z.; Fan, H.; Ma, W.; Yu, D.; Yang, Q.; Tian, J.; Yang, X.; Li, B. Efficacy and safety of lenvatinib versus sorafenib in first-line treatment of advanced hepatocellular carcinoma: A meta-analysis. Front. Oncol. 2022, 12, 1010726. [Google Scholar] [CrossRef]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. IMbrave150 Investigators. Atezolizumab plus Bevacizumab in Unresectable Hepatocellular Carcinom. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef]
- Wilhelm, S.; Carter, C.; Lynch, M.; Lowinger, T.; Dumas, J.; Smith, R.A.; Schwartz, B.; Simantov, R.; Kelley, S. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat. Rev. Drug Disc. 2006, 5, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Giacomini, M.M.; Giacomini, C.; Maitland, M.L.; Altman, R.B.; Klein, T.E. PharmGKB summary: Sorafenib pathways. Pharmacogenet. Genom. 2017, 27, 240–246. [Google Scholar] [CrossRef]
- Terada, T.; Noda, S.; Inui, K. Management of dose variability and side effects for individualized cancer pharmacotherapy with tyrosine kinase inhibitors. Pharmacol. Ther. 2015, 152, 125–134. [Google Scholar] [CrossRef]
- Marisi, G.; Cucchetti, A.; Ulivi, P.; Canale, M.; Cabibbo, G.; Solaini, L.; Foschi, F.G.; De Matteis, S.; Ercolani, G.; Valgiusti, M.; et al. Ten Years of Sorafenib in Hepatocellular Carcinoma: Are there any Predictive and/or Prognostic Markers? World J. Gastroenterol. 2018, 24, 4152–4163. [Google Scholar] [CrossRef]
- Lugano, R.; Ramachandran, M.; Dimberg, A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell. Mol. Life Sci. 2020, 77, 1745–1770. [Google Scholar] [CrossRef]
- Casadei Gardini, A.; Marisi, G.; Faloppi, L.; Scarpi, E.; Foschi, F.G.; Iavarone, M.; Lauletta, G.; Corbelli, J.; Valgiusti, M.; Facchetti, F.; et al. ENOS Polymorphisms and Clinical Outcome in Advanced HCC Patients Receiving Sorafenib: Final Results of the ePHAS Study. Oncotarget 2016, 7, 27988–27999. [Google Scholar] [CrossRef] [PubMed]
- Faloppi, L.; Puzzoni, M.; Casadei Gardini, A.; Silvestris, N.; Masi, G.; Marisi, G.; Vivaldi, C.; Gadaleta, C.D.; Ziranu, P.; Bianconi; et al. Angiogenesis Genotyping and Clinical Outcomes in Patients with Advanced Hepatocellular Carcinoma Receiving Sorafenib: The ALICE-2 Study. Target Oncol. 2020, 15, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Casadei-Gardini, A.; Marisi, G.; Dadduzio, V.; Gramantieri, L.; Faloppi, L.; Ulivi, P.; Foschi, F.G.; Tamburini, E.; Vivaldi, C.; Rizzato, M.D.; et al. Association of NOS3 and ANGPT2 Gene Polymorphisms with Survival in Patients with Hepatocellular Carcinoma Receiving Sorafenib: Results of the Multicenter Prospective INNOVATE Study. Clin. Cancer Res. 2020, 26, 4485–4493. [Google Scholar] [CrossRef] [PubMed]
- Arbitrio, M.; Di Martino, M.T.; Scionti, F.; Agapito, G.; Guzzi, P.H.; Cannataro, M.; Tassone, P.; Tagliaferri, P. DMET (Drug Metabolism Enzymes and Transporters): A pharmacogenomic platform for precision medicine. Oncotarget 2016, 8, 54028–54050. [Google Scholar] [CrossRef] [PubMed]
- Arbitrio, M.; Scionti, F.; Di Martino, M.T.; Caracciolo, D.; Pensabene, L.; Tassone, P.; Tagliaferri, P. Pharmacogenomics Biomarker Discovery and Validation for Translation in Clinical. Practic. Clin. Transl. Sci. 2021, 14, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Cargnin, S.; Viana, M.; Sances, G.; Cantello, R.; Tassorelli, C.; Terrazzino, S. Using a Genetic Risk Score Approach to Predict Headache Response to Triptans in Migraine Without Aura. J. Clin. Pharmacol. 2019, 59, 288–294. [Google Scholar] [CrossRef]
- Du, B.; Zhang, C.; Yue, L.; Ren, B.; Zhao, Q.; Li, D.; He, Y.; Zhang, W. Prediction model for the efficacy of folic acid therapy on hyperhomocysteinaemia based on genetic risk score methods. Br. J. Nutr. 2019, 122, 39–46. [Google Scholar] [CrossRef]
- Gagno, S.; D’Andrea, M.R.; Mansutti, M.; Zanusso, C.; Puglisi, F.; Dreussi, E.; Montico, M.; Biason, P.; Cecchin, E.; Iacono, D.; et al. A New Genetic Risk Score to Predict the Outcome of Locally Advanced or Metastatic Breast Cancer Patients Treated With First-Line Exemestane: Results From a Prospective Study. Clin. Breast Cancer 2019, 19, 137–145. [Google Scholar] [CrossRef]
- Farzaneh, Z.; Vosough, M.; Agarwal, T.; Farzaneh, M. Critical signaling pathways governing hepatocellular carcinoma behavior; small molecule-based approaches. Cancer Cell. Int. 2021, 21, 208. [Google Scholar] [CrossRef]
- Kobayashi, K.; Higai, K.; Matsuo, K.; Bannai, Y.; Horie, H.; Otakara, Y.; Li, W.; Koike, K.; Yanagino, S.; Watanabe, K.; et al. Quantitative Measurements of Sorafenib in Patients with Advanced Hepatocellular Carcinoma. Int. J. Res. Stud. Med. Health Sci. 2018, 3, 14–19. [Google Scholar]
- Ruggiero, D.; Dalmasso, C.; Nutile, T.; Sorice, R.; Dionisi, L.; Aversano, M.; Bröet, P.; Leutenegger, A.L.; Bourgain, C.; Ciullo, M. Genetics of VEGF serum variation in human isolated populations of cilento: Importance of VEGF polymorphisms. PLoS ONE 2011, 6, e16982. [Google Scholar] [CrossRef]
- Al-Habboubi, H.H.; Mahdi, N.; Abu-Hijleh, T.M.; Abu-Hijleh, F.M.; Sater, M.S.; Almawi, W.Y. The relation of vascular endothelial growth factor (VEGF) gene polymorphisms on VEGF levels and the risk of vasoocclusive crisis in sickle cell disease. Eur. J. Haematol. 2012, 89, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; de Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. SHARP Investigators Study Group. SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 24, 378–390. [Google Scholar] [CrossRef]
- Gardini, A.C.; Faloppi, L.; Aprile, G.; Brunetti, O.; Caparello, C.; Corbelli, J.; Chessa, L.; Bruno, D.; Ercolani, G.; Leonetti, A.; et al. Multicenter prospective study of angiogenesis polymorphism validation in HCC patients treated with sorafenib. An INNOVATE study protocol. Tumori 2018, 104, 476–479. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Liu, Y.; Xiao, W.; Zhao, N.; Zhu, C.; Yu, D.; Zhao, Y. Prognostic SLC family genes promote cell proliferation, migration, and invasion in hepatocellular carcinoma. Acta Biochim. Biophys. Sin. 2021, 53, 1065–1075. [Google Scholar] [CrossRef]
- Arimany-Nardi, C.; Minuesa, G.; Keller, T.; Erkizia, I.; Koepsell, H.; Martinez-Picado, J.; Pastor-Anglada, M. Role of Human Organic Cation Transporter 1 (hOCT1) Polymorphisms in Lamivudine (3TC) Uptake and Drug-Drug Interactions. Front. Pharmacol. 2016, 7, 175. [Google Scholar] [CrossRef]
- Zhao, Q.; Zheng, B.; Meng, S.; Xu, Y.; Guo, J.; Chen, L.J.; Xiao, J.; Zhang, W.; Tan, Z.R.; Tang, J.; et al. Increased expression of SLC46A3 to oppose the progression of hepatocellular carcinoma and its effect on sorafenib therapy. Biomed. Pharmacother. 2019, 114, 108864. [Google Scholar] [CrossRef] [PubMed]
- Herraez, E.; Lozano, E.; Macias, R.I.; Vaquero, J.; Bujanda, L.; Banales, J.M.; Marin, J.J.; Briz, O. Expression of SLC22A1 variants may affect the response of hepatocellular carcinoma and cholangiocarcinoma to sorafenib. Hepatology 2013, 58, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Geier, A.; Macias, R.I.; Bettinger, D.; Weiss, J.; Bantel, H.; Jahn, D.; Al-Abdulla, R.; Marin, J.J. The lack of the organic cation transporter OCT1 at the plasma membrane of tumor cells precludes a positive response to sorafenib in patients with hepatocellular carcinoma. Oncotarget 2017, 8, 15846–15857. [Google Scholar] [CrossRef]
- Liu, X.; Huang, R.; Liu, X.; You, H.; Kong, F.; Tang, R. Prognostic implications of alcohol dehydrogenases in hepatocellular carcinoma. BMC Cancer 2020, 20, 1204. [Google Scholar] [CrossRef]
- Osanai, M.; Lee, G.H. Increased expression of the retinoic acid-metabolizing enzyme CYP26A1 during the progression of cervical squamous neoplasia and head and neck cancer. BMC Res. Notes 2014, 7, 697. [Google Scholar] [CrossRef]
- Osanai, M.; Lee, G.H. The retinoic acid-metabolizing enzyme CYP26A1 upregulates fascin and promotes the malignant behavior of breast carcinoma cells. Oncol. Rep. 2015, 34, 850–858. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, Y.; Zou, Y.; Yu, Y. CYP26A1 Is a Novel Cancer Biomarker of Pancreatic Carcinoma: Evidence from Integration Analysis and In Vitro Experiments. Dis. Markers 2022, 2022, 5286820. [Google Scholar] [CrossRef]
- Kanki, K.; Akechi, Y.; Ueda, C.; Tsuchiya, H.; Shimizu, H.; Ishijima, N.; Toriguchi, K.; Hatano, E.; Endo, K.; Hirooka, Y.; et al. Biological and clinical implications of retinoic acid-responsive genes in human hepatocellular carcinoma cells. J. Hepatol. 2013, 59, 1037–1044. [Google Scholar] [CrossRef] [PubMed]
- Lotan, R. Retinoids as modulators of tumor cells invasion and metastasis. Semin. Cancer Biol. 1991, 2, 197–208. [Google Scholar]
- Lencioni, R.; Llovet, J.M. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin. Liver Dis. 2010, 30, 52–60. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef]
- European Association for the Study of The Liver; European Organisation for Research And Treatment of Cancer. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2012, 56, 908–943. [Google Scholar] [CrossRef] [PubMed]
- Scionti, F.P.L.; Di Martino, M.T.; Arbitrio, M.; Tagliaferri, P. Ethical Perspectives on Pharmacogenomic Profiling; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Scionti, F.; Di Martino, M.T.; Sestito, S.; Nicoletti, A.; Falvo, F.; Roppa, K.; Arbitrio, M.; Guzzi, P.H.; Agapito, G.; Pisani, A.; et al. Genetic variants associated with Fabry disease progression despite enzyme replacement therapy. Oncotarget 2017, 8, 107558–107564. [Google Scholar] [CrossRef]
- Scionti, F.; Agapito, G.; Caracciolo, D.; Riillo, C.; Grillone, K.; Cannataro, M.; Di Martino, M.T.; Tagliaferri, P.; Tassone, P.; Arbitrio, M. Risk Alleles for Multiple Myeloma Susceptibility in ADME Genes. Cells 2022, 11, 189. [Google Scholar] [CrossRef] [PubMed]
- Arbitrio, M.; Scionti, F.; Altomare, E.; Di Martino, M.T.; Agapito, G.; Galeano, T.; Staropoli, N.; Iuliano, E.; Grillone, F.; Fabiani, F.; et al. Polymorphic Variants in NR1I3 and UGT2B7 Predict Taxane Neurotoxicity and Have Prognostic Relevance in Patients With Breast Cancer: A Case-Control Study. Clin. Pharmacol. Ther. 2019, 106, 422–431. [Google Scholar] [CrossRef]
- Faloppi, L.; Casadei Gardini, A.; Masi, G.; Silvestris, N.; Loretelli, C.; Ulivi, P.; Vivaldi, C.; Bianconi, M.; Giampieri, R. Angiogenesis polymorphisms profile in the prediction of clinical outcome of advanced HCC patients receiving sorafenib: Combined analysis of VEGF and HIF-1α Final results of the ALICE-2 study. J. Clin. Oncol. 2016, 34, 280. [Google Scholar] [CrossRef]
- Agapito, G.; Milano, M.; Cannataro, M. A statistical network pre-processing method to improve relevance and significance of gene lists in microarray gene expression studies. BMC Bioinform. 2022, 23, 393. [Google Scholar] [CrossRef]
- Kotlyar, M.; Pastrello, C.; Malik, Z.; Jurisica, I. IID 2018 update: Context-specific physical protein-protein interactions in human, model organisms and domesticated species. Nucleic Acids Res. 2019, 47, 581–589. [Google Scholar] [CrossRef]
- Lee, S.; Liu, Y.; Li, J.; Friedman, C.; Lussier, Y.A. Discovery of protein interaction networks shared by diseases. Pac. Symp. Biocomput. 2007, 76–87. [Google Scholar]
- Paul, S.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. Cytohubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. S4), S11. [Google Scholar] [CrossRef] [PubMed]
- Agapito, G.; Cannataro, U. Using BioPAX-Parser (BiP) to enrich lists of genes or proteins with pathway data. BMC Bioinform. 2021, 22, 376. [Google Scholar] [CrossRef]
- Rahmati, S.; Abovsky, M.; Pastrello, C.M.; Kotlyar, M.; Lu, R.; Cumbaa, C.A.; Rahman, P.; Chandran, V.; Jurisica, I. pathDIP 4: An extended pathway annotations and enrichment analysis resource for human, model organisms and domesticated species. Nucleic Acids Res. 2020, 48, 479–488. [Google Scholar] [CrossRef]

| SNP (Gene) | R n (%) | NR n (%) | p |
|---|---|---|---|
| rs2010963 (VEGF-A) | |||
| Allele | |||
| G | 2 (11.1) | 25 (50.0) | - |
| C | 16 (88.9%) | 25 (50.0) | 0.004 |
| Genotype | |||
| GG | 0 (0.0) | 7 (28.0) | - |
| GC | 2 (22.2) | 11 (44.0) | 0.056 |
| CC | 7 (77.8) | 7 (28.0) | 0.046 |
| rs4604006 (VEGF-C) | |||
| Allele | |||
| C | 13 (72.2) | 34 (68.0) | - |
| T | 5 (27.8) | 16 (32.0) | 0.739 |
| Genotype | |||
| CC | 5 (57.0) | 11 (44.0) | - |
| CT | 3 (28.5) | 12 (48.0) | 0.474 |
| TT | 1 (14.5) | 2 (8.0) | 0.943 |
| rs12434438 (HIF-1α) | |||
| Allele | |||
| A | 14 (77.8) | 30 (60.0) | - |
| G | 4 (22.2) | 20 (40.0) | 0.175 |
| Genotype | |||
| AA | 5 (55.6) | 9 (36.0) | - |
| AG | 4 (44.4) | 12 (48.0) | 0.522 |
| GG | 0 (0.0) | 4 (16.0) | 0.159 |
| rs55633437 (ANGPT2) | |||
| Allele | |||
| G | 16 (88.9) | 38 (76.0) | - |
| T | 2 (11.1) | 12 (24.0) | 0.246 |
| Genotype | |||
| GG | 7 (77.8) | 14 (56.0) | - |
| GT | 2 (22.2) | 10 (40.0) | 0.301 |
| TT | 0 (0.0) | 1 (4.0) | 0.484 |
| rs2070744 (NOS3) | |||
| Allele | |||
| C | 6 (33.3) | 19 (38.0) | - |
| T | 12 (66.7) | 31 (62.0) | 0.724 |
| Genotype | |||
| CC | 0 (0) | 4 (16.0) | - |
| CT | 6 (66.7) | 11 (44.0) | 0.159 |
| TT | 3 (33.3) | 10 (40.0) | 0.289 |
| SNP (Gene) | R n (%) | NR n (%) | p |
|---|---|---|---|
| rs6811453(ADH1A) | |||
| Allele | |||
| T | 1 (7.2) | 18 (56.2) | - |
| C | 13 (92.8) | 14 (43.8) | 0.002 |
| Genotype | |||
| TT | 0 (0.0) | 5 (24.0) | - |
| CT | 1 (14.0) | 8 (48.0) | - |
| CC | 6 (86.0) | 3 (28.0) | 0.005 |
| rs10008281 (ADH6) | |||
| Allele | |||
| G | 6 (42.8) | 28 (87.5) | - |
| T | 8 (57.2) | 4 (12.5) | 0.003 |
| Genotype | |||
| TT | 2 (28.6) | 0 (0.0) | - |
| GT | 4 (57.1) | 4 (25.0) | - |
| GG | 1 (14.3) | 12 (75.0) | 0.019 |
| rs11401 (SULT1A2) | |||
| Allele | |||
| A | 7 (50.0) | 27 (84.3) | - |
| G | 7 (50.0) | 5 (15.7) | 0.026 |
| Genotype | |||
| AA | 1 (15.5) | 12 (75.0) | 0.019 |
| AG | 5 (71.0) | 3 (18.7) | 0.019 |
| GG | 1 (14.5) | 1 (6.3) | - |
| rs7905939 (CYP26A1) | |||
| Allele | |||
| G | 6 (42.8) | 6 (18.7) | - |
| A | 8 (57.2) | 26 (81.3) | 0.143 |
| Genotype | |||
| AA | 0 (0.0) | 1 (6.3) | - |
| AG | 6 (85.7) | 4 (25.0) | - |
| GG | 1 (14.3) | 11 (68.7) | 0.027 |
| rs2297595 (DPYD) | |||
| Allele | |||
| G | 9 (64.2) | 30 (93.7) | - |
| A | 5 (35.8) | 2 (6.3) | 0.020 |
| Genotype | |||
| AA | 2 (28.6) | 14 (87.5) | 0.011 |
| AG | 5 (71.4) | 2 (12.5) | 0.011 |
| GG | 0 (0.0) | 0 (0.0%) | - |
| rs1801265 (DPYD) | |||
| Allele | |||
| C | 9 (64.2%) | 29 (90.6) | 0.044 |
| T | 5 (35.8) | 3 (9.4) | - |
| Genotype | |||
| TT | 2 (28.6) | 13 (81.3) | 0.026 |
| CT | 5 (71.4) | 3 (18.7) | 0.026 |
| CC | 0 (0.0) | 0 (0.0) | - |
| rs2020863 (FMO2) | |||
| Allele | |||
| G | 4 (28.5) | 0 (0.0) | 0.006 |
| A | 10 (71.5) | 32 (100.0) | - |
| Genotype | |||
| AA | 3 (42.9%) | 16 (100.0) | 0.004 |
| AG | 4 (57.1) | 0 (0.0) | 0.004 |
| GG | 0 (0.0) | 0 (0.0) | - |
| rs171248 (SLC22A14) | |||
| Allele | |||
| T | 14 (100.0) | 18 (56.2) | 0.004 |
| C | 0 (0.0) | 14 (43.8) | - |
| Genotype | |||
| TT | 7 (100.0) | 4 (25.5) | 0.001 |
| CT | 0 (0.0) | 10 (62.5) | 0.007 |
| CC | 0 (0.0) | 2 (12.5) | - |
| rs149738 (SLC22A14) | |||
| Allele | |||
| T | 14 (100.0) | 18 (56.2) | 0.004 |
| C | 0 (0.0) | 14 (43.8) | - |
| Genotype | |||
| TT | 7 (100.0) | 4 (25.5) | 0.001 |
| CT | 0 (0.0) | 10 (62.5) | 0.007 |
| CC | 0 (0.0) | 2 (12.5) | - |
| rs183574 (SLC22A14) | |||
| Allele | |||
| C | 0 (0.0) | 14 (43.7) | - |
| T | 14 (100.0) | 18 (56.3) | 0.004 |
| Genotype | |||
| TT | 7 (100.0) | 4 (25.5) | 0.001 |
| TC | 0 (0.0) | 10 (62.5) | 0.007 |
| CC | 0 (0.0) | 2 (12.5) | - |
| If rs171248 = CT then NR If rs171248 = CC then NR If rs171248 = TT & rs6811453 = TT then NR If ss171248 = TT & rs6811453 = CT & rs2010963 = CG then NR If rs171248 = TT & rs6811453 = CT & rs2010963 = GG then NR If rs171248 = TT & rs6811453 = CT & rs2010963 = CC & rs12434438 = GG then NR If rs171248 = TT & rs6811453 = CT & rs2010963 = CC & rs12434438 = AG then NR If rs171248 = TT & rs6811453 = CC & rs7905939 = GG & rs4604006 = CC then NR If rs171248 = TT & rs6811453 = CC & rs7905939 = GG & rs4604006 = TT then NR If rs171248 = TT & rs6811453 = CC & rs7905939 = AA then NR |
| If rs171248 = TT & rs6811453 = CT & rs2010963 = CC & rs12434438 = AA then R If rs171248 = TT & rs6811453 = CC & rs7905939 = AG then R If rs171248 = TT & rs6811453 = CC & rs7905939 = GG & rs4604006VEGF-C = CT then R |
| Rank | Name | Score |
|---|---|---|
| 1 | VEGFA | 730,700.53 |
| 2 | MICA | 379,197.11 |
| 3 | NOS3 | 339,569.49 |
| 4 | VEGFC | 313,274.47 |
| 5 | MX1 | 249,727.84 |
| 6 | PCSK9 | 194,571.69 |
| 7 | CYP26A1 | 79,311.53 |
| 8 | ANGPT2 | 112,225.26 |
| 9 | FMO2 | 69,997.19 |
| 10 | DHX9 | 65,954.87 |
| 11 | ARNTL | 65,954.87 |
| 12 | ADH6 | 51,195.00 |
| 13 | RELA | 47,118.36 |
| 14 | CXCL10 | 43,934.98 |
| 15 | FASLG | 32,946.27 |
| 16 | YWHAE | 29,395.65 |
| 17 | ALDH1A1 | 26,473.00 |
| 18 | ADH7 | 26,473.00 |
| 19 | GPR12 | 23,300.00 |
| 20 | TGFBR2 | 21,738.55 |
| 21 | SLC22A14 | 21,060.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannitrapani, L.; Di Gaudio, F.; Cervello, M.; Scionti, F.; Ciliberto, D.; Staropoli, N.; Agapito, G.; Cannataro, M.; Tassone, P.; Tagliaferri, P.; et al. Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma. Int. J. Mol. Sci. 2024, 25, 2197. https://doi.org/10.3390/ijms25042197
Giannitrapani L, Di Gaudio F, Cervello M, Scionti F, Ciliberto D, Staropoli N, Agapito G, Cannataro M, Tassone P, Tagliaferri P, et al. Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2024; 25(4):2197. https://doi.org/10.3390/ijms25042197
Chicago/Turabian StyleGiannitrapani, Lydia, Francesca Di Gaudio, Melchiorre Cervello, Francesca Scionti, Domenico Ciliberto, Nicoletta Staropoli, Giuseppe Agapito, Mario Cannataro, Pierfrancesco Tassone, Pierosandro Tagliaferri, and et al. 2024. "Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma" International Journal of Molecular Sciences 25, no. 4: 2197. https://doi.org/10.3390/ijms25042197
APA StyleGiannitrapani, L., Di Gaudio, F., Cervello, M., Scionti, F., Ciliberto, D., Staropoli, N., Agapito, G., Cannataro, M., Tassone, P., Tagliaferri, P., Seidita, A., Soresi, M., Affronti, M., Bertino, G., Russello, M., Ciriminna, R., Lino, C., Spinnato, F., Verderame, F., ... Arbitrio, M. (2024). Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma. International Journal of Molecular Sciences, 25(4), 2197. https://doi.org/10.3390/ijms25042197












