Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene
Abstract
1. Introduction
2. Results
2.1. Genetic Analysis
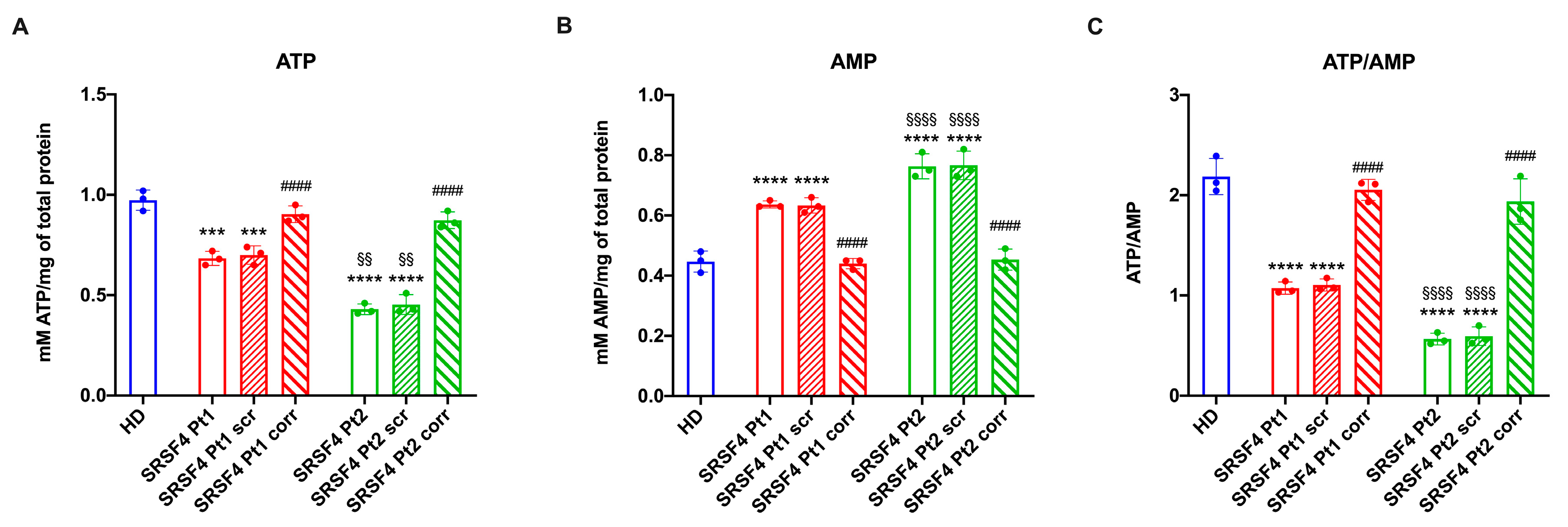
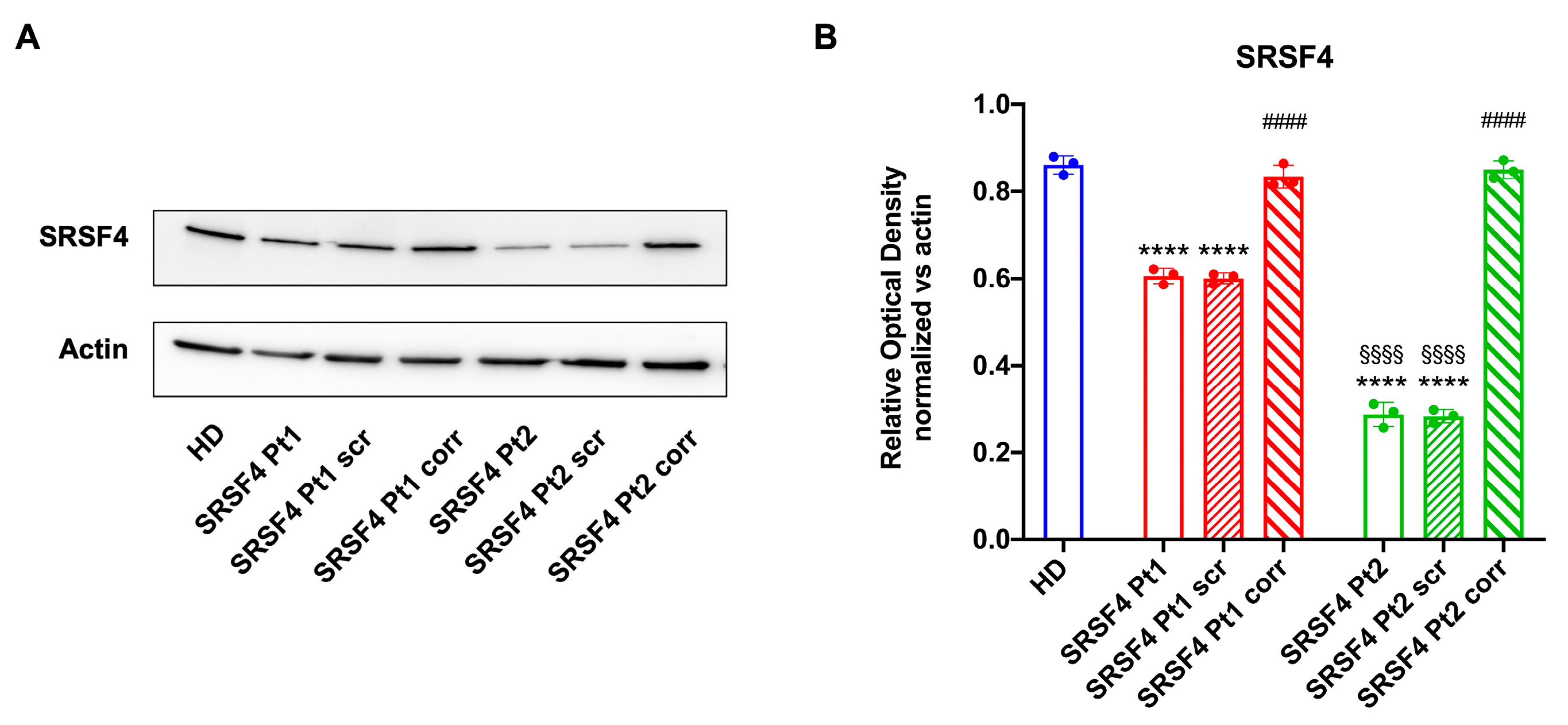
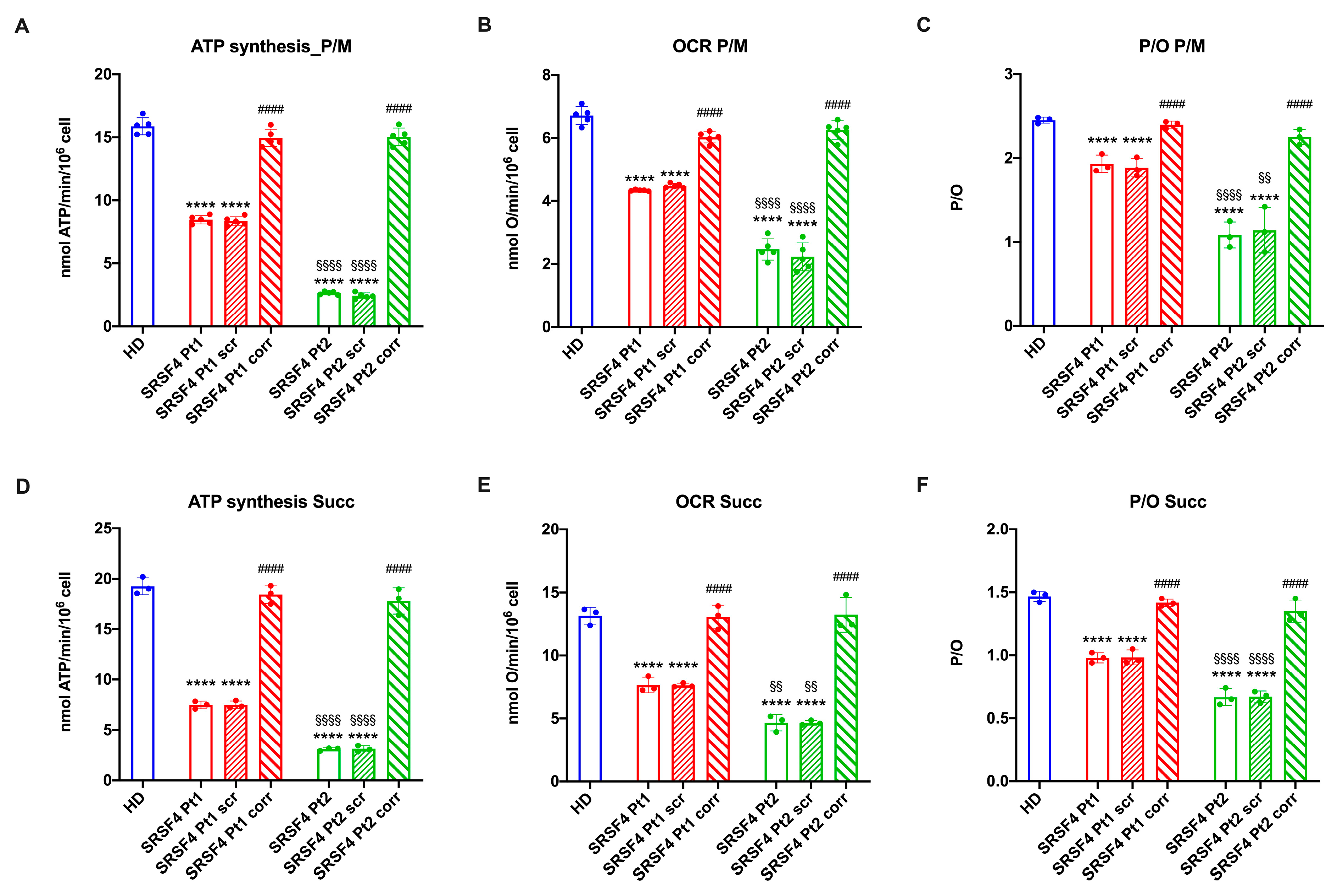
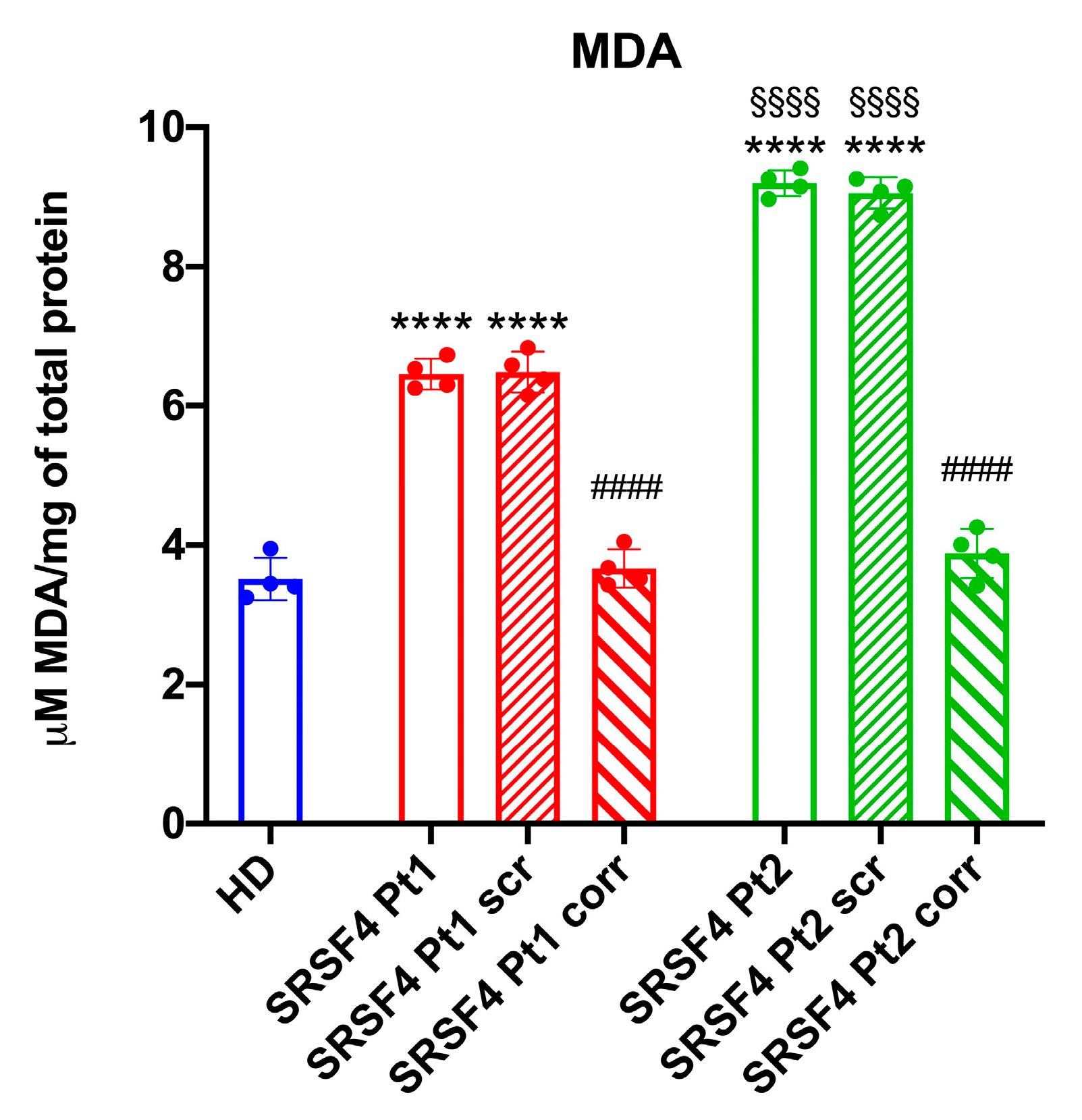
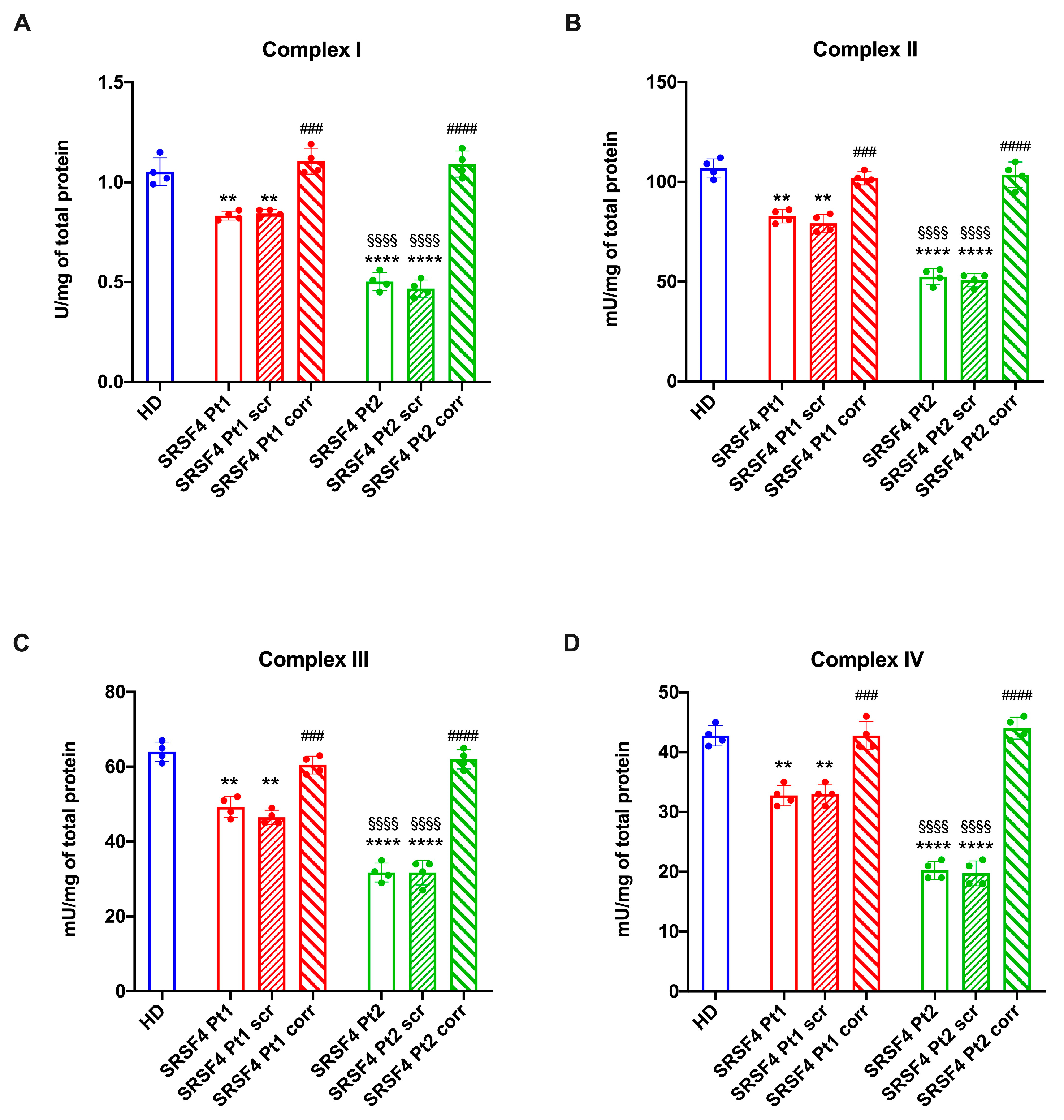
2.2. The SRSF4 Mutation Reduces the Cellular Energy Status and the Aerobic Metabolism by Affecting Oxidative Phosphorylation
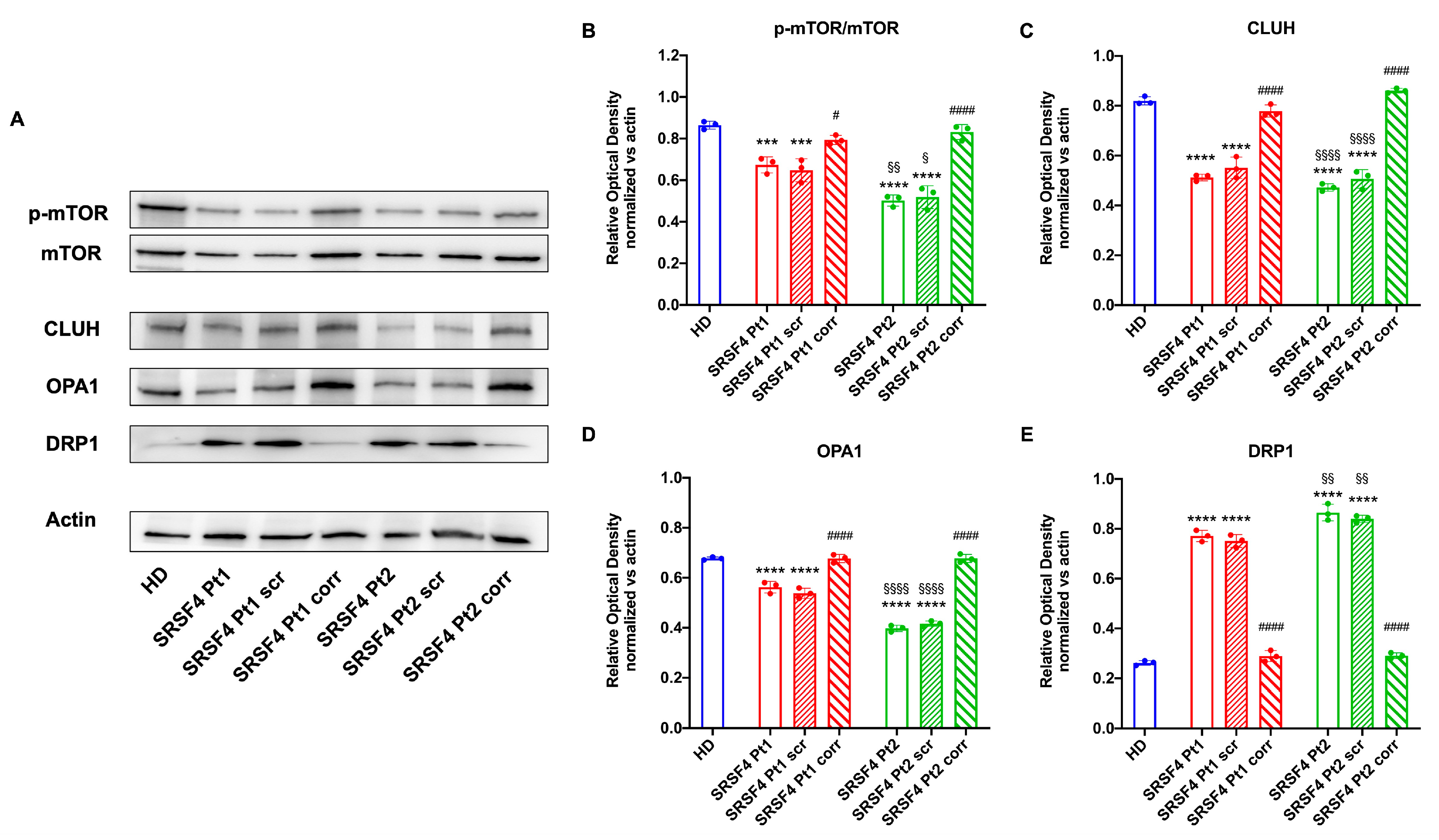
2.3. The SRSF4 Mutation Causes a Reduction in mTOR Phosphorylation and Expression and an Alteration in Mitochondrial Dynamics
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Informed Consent
4.3. Genetic Analysis
4.4. Cell Culture and Transfection
4.5. RNA Extraction, Retrotranscription, and Expression of the SRSF4 Gene
4.6. Evaluation of Aerobic Metabolism Function
4.7. Assessment of the Cellular Energy Status
4.8. Evaluation of Lipid Peroxidation
4.9. Assay of the Electron Transfer Chain Complexes
4.10. Western Blot Analysis
4.11. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fioredda, F.; Rotulo, G.A.; Farruggia, P.; Dagliano, F.; Pillon, M.; Trizzino, A.; Notarangelo, L.; Luti, L.; Lanza, T.; Terranova, P.; et al. Late-onset and long-lasting autoimmune neutropenia: An analysis from the Italian Neutropenia Registry. Blood Adv. 2020, 4, 5644–5649. [Google Scholar] [CrossRef] [PubMed]
- Kallen, M.E.; Dulau-Florea, A.; Wang, W.; Calvo, K.R. Acquired and germline predisposition to bone marrow failure: Diagnostic features and clinical implications. Semin. Hematol. 2019, 56, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Cifaldi, C.; Brigida, I.; Barzaghi, F.; Zoccolillo, M.; Ferradini, V.; Petricone, D.; Cicalese, M.P.; Lazarevic, D.; Cittaro, D.; Omrani, M.; et al. Targeted NGS Platforms for Genetic Screening and Gene Discovery in Primary Immunodeficiencies. Front. Immunol. 2019, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.-B.; Murphy, M.E.; et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.N.; Liu, Y.; Choi, N.; Oh, J.; Ha, J.; Zheng, X.; Shen, H. Binding of SRSF4 to a novel enhancer modulates splicing of exon 6 of Fas pre-mRNA. Biochem. Biophys. Res. Commun. 2018, 506, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Lucena-Araujo, A.R.; Santana-Lemos, B.A.; Thome, C.H.; Ferreira, G.; Ruggero, D.; Faça, V.M.; Rego, E.M. Early Hematopoietic Progenitors of Dkc1 Hypomorphic Mutant Mice Display Decreased Proliferation Rate and an Impaired Control of Serine/Arginine-Rich Splicing Factor 4(Srsf4) Translation. Blood 2014, 124, 2937. [Google Scholar] [CrossRef]
- Wagner, A.R.; Weindel, C.G.; West, K.O.; Scott, H.M.; Watson, R.O.; Patrick, K.L. SRSF6 balances mitochondrial-driven innate immune outcomes through alternative splicing of BAX. eLife 2022, 11, e82244. [Google Scholar] [CrossRef]
- Filippi, M.-D.; Ghaffari, S. Mitochondria in the maintenance of hematopoietic stem cells: New perspectives and opportunities. Blood 2019, 133, 1943–1952. [Google Scholar] [CrossRef]
- Kumari, U.; Jun, W.Y.; Bay, B.H.; Lyakhovich, A. Evidence of mitochondrial dysfunction and impaired ROS detoxifyingmachinery in Fanconi anemia cells. Oncogene 2014, 33, 165–172. [Google Scholar] [CrossRef]
- Pagano, G.; Talamanca, A.A.; Castello, G.; d’Ischia, M.; Pallardó, F.V.; Petrović, S.; Porto, B.; Tiano, L.; Zatterale, A. From clinical description, to in vitro and animal studies, and backward to patients: Oxidative stress and mitochondrial dysfunction in Fanconi anemia. Free Radic Biol. Med. 2013, 58, 118–125. [Google Scholar] [CrossRef]
- Ravera, S.; Dufour, C.; Cesaro, S.; Bottega, R.; Faleschini, M.; Cuccarolo, P.; Corsolini, F.; Usai, C.; Columbaro, M.; Cipolli, M.; et al. Evaluation of energy metabolism and calcium homeostasis in cells affected by Shwachman-Diamond syndrome. Sci. Rep. 2016, 6, 25441. [Google Scholar] [CrossRef]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich proteins plicing factors (SRproteins). Genes Dev. 2010, 24, 1073–1074. [Google Scholar] [CrossRef] [PubMed]
- Correia-Melo, C.; Marques, F.D.; Anderson, R.; Hewitt, G.; Hewitt, R.; Cole, J.; Carroll, B.M.; Miwa, S.; Birch, J.; Merz, A.; et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016, 35, 724–742. [Google Scholar] [CrossRef] [PubMed]
- Morita, M.; Gravel, S.-P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Schatton, D.; Martinelli, P.; Hansen, H.; Pla-Martin, D.; Barth, E.; Becker, C.; Altmueller, J.; Frommolt, P.; Sardiello, M.; et al. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J. Cell Biol. 2014, 207, 213–223. [Google Scholar] [CrossRef]
- Signa, S.; Bertoni, A.; Penco, F.; Caorsi, R.; Cafaro, A.; Cangemi, G.; Volpi, S.; Gattorno, M.; Schena, F. Adenosine Deaminase 2 Deficiency (DADA2): A Crosstalk Between Innate and Adaptive Immunity. Front. Immunol. 2022, 13, 3511. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.S.; Kozyra, E.J.; Wlodarski, M.W. Germline predisposition in myeloid neoplasms: Unique genetic and clinical features of GATA2 deficiency and SAMD9/SAMD9L syndromes. Best. Pract. Res. Clin. Haematol. 2020, 33, 101197. [Google Scholar] [CrossRef] [PubMed]
- Meyts, I.; Aksentijevich, I. Deficiency of Adenosine Deaminase 2 (DADA2): Up dates on the Phenotype, Genetics, Pathogenesis, and Treatment. J. Clin. Immunol. 2018, 38, 569–578. [Google Scholar] [CrossRef]
- Ganapathi, K.A.; Townsley, D.M.; Hsu, A.P.; Arthur, D.C.; Zerbe, C.S.; Cuellar-Rodriguez, J.; Hickstein, D.D.; Rosenzweig, S.D.; Braylan, R.C.; Young, N.S.; et al. GATA2 deficiency-associated bone marrow disorder differs from idiopathic aplastic anemia. Blood 2015, 125, 56–70. [Google Scholar] [CrossRef]
- Yoshida, M.; Tanase-Nakao, K.; Shima, H.; Shirai, R.; Yoshida, K.; Osumi, T.; Deguchi, T.; Mori, M.; Arakawa, Y.; Takagi, M.; et al. Prevalence of germline GATA2 and SAMD9/9L variants in paediatric haematological disorders with monosomy 7. Br. J. Haematol. 2020, 191, 835–843. [Google Scholar] [CrossRef]
- Miano, M.; Cappelli, E.; Pezzulla, A.; Venè, R.; Grossi, A.; Terranova, P.; Palmisani, E.; Maggiore, R.; Guardo, D.; Lanza, T.; et al. FAS-mediated apoptosis impairment in patients with ALPS/ALPS-like phenotype carrying variants on CASP10 gene. Br. J. Haematol. 2019, 187, 502–508. [Google Scholar] [CrossRef]
- Gimeno-Valiente, F.; López-Rodas, G.; Castillo, J.; Franco, L. Alternative Splicing, Epigenetic Modifications and Cancer: A Dangerous Triangle, or a Hopeful One? Cancers 2022, 14, 560. [Google Scholar] [CrossRef]
- Morita, M.; Prudent, J.; Basu, K.; Goyon, V.; Katsumura, S.; Hulea, L.; Pearl, D.; Siddiqui, N.; Strack, S.; McGuirk, S.; et al. mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 2017, 67, 922–935.e5. [Google Scholar] [CrossRef]
- Morita, M.; Gravel, S.-P.; Chénard, V.; Sikström, K.; Zheng, L.; Alain, T.; Gandin, V.; Avizonis, D.; Arguello, M.; Zakaria, C. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013, 18, 698–711. [Google Scholar] [CrossRef]
- Ravera, S.; Podestà, M.; Sabatini, F.; Fresia, C.; Columbaro, M.; Bruno, S.; Fulcheri, E.; Ramenghi, L.A.; Frassoni, F. Mesenchymal stem cells from preterm to term newborns undergo a significant switch from anaerobic glycolysis to the oxidative phosphorylation. Cell. Mol. Life Sci. 2018, 75, 889–903. [Google Scholar] [CrossRef]
- Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1817, 1833–1838. [Google Scholar] [CrossRef]
- Marchi, S.; Patergnani, S.; Pinton, P. The endoplasmic reticulum–Mitochondria connection: Onetouch, multiple functions. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Nasr, W.; Filippi, M.-D. Acquired and hereditary bone marrow failure: A mitochondrial perspective. Front. Oncol. 2022, 12, 1048746. [Google Scholar] [CrossRef] [PubMed]
- Ansó, E.; Weinberg, S.E.; Diebold, L.P.; Thompson, B.J.; Malinge, S.; Schumacker, P.T.; Liu, X.; Zhang, Y.; Shao, Z.; Steadman, M.; et al. The mitochondrial respiratory chain is essential for haematopoietic stem cell function. Nat. Cell Biol. 2017, 19, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Grossi, A.; Miano, M.; Lanciotti, M.; Fioredda, F.; Guardo, D.; Palmisani, E.; Terranova, P.; Santamaria, G.; Caroli, F.; Caorsi, R.; et al. Targeted NGS Yields Plentiful Ultra-Rare Variants in Inborn Errors of Immunity Patients. Genes 2021, 12, 1299. [Google Scholar] [CrossRef]
- Hinkle, P.C. P/O ratios of mitochondrial oxidative phosphorylation. Biochim. Biophys. Acta (BBA) Bioenerg. 2005, 1706, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Colla, R.; Izzotti, A.; Ciucis, C.D.; Fenoglio, D.; Ravera, S.; Speciale, A.; Ricciarelli, R.; Furfaro, A.L.; Pulliero, A.; Passalacqua, M.; et al. Glutathione-mediated antioxidant response and aerobic metabolism: Two crucial factors involved in determining the multi-drug resistance of high-risk neuroblastoma. Oncotarget 2016, 7, 70715–70737. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miano, M.; Bertola, N.; Grossi, A.; Dell’Orso, G.; Regis, S.; Rusmini, M.; Uva, P.; Vozzi, D.; Fioredda, F.; Palmisani, E.; et al. Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene. Int. J. Mol. Sci. 2024, 25, 2083. https://doi.org/10.3390/ijms25042083
Miano M, Bertola N, Grossi A, Dell’Orso G, Regis S, Rusmini M, Uva P, Vozzi D, Fioredda F, Palmisani E, et al. Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene. International Journal of Molecular Sciences. 2024; 25(4):2083. https://doi.org/10.3390/ijms25042083
Chicago/Turabian StyleMiano, Maurizio, Nadia Bertola, Alice Grossi, Gianluca Dell’Orso, Stefano Regis, Marta Rusmini, Paolo Uva, Diego Vozzi, Francesca Fioredda, Elena Palmisani, and et al. 2024. "Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene" International Journal of Molecular Sciences 25, no. 4: 2083. https://doi.org/10.3390/ijms25042083
APA StyleMiano, M., Bertola, N., Grossi, A., Dell’Orso, G., Regis, S., Rusmini, M., Uva, P., Vozzi, D., Fioredda, F., Palmisani, E., Lupia, M., Lanciotti, M., Grilli, F., Corsolini, F., Arcuri, L., Giarratana, M. C., Ceccherini, I., Dufour, C., Cappelli, E., & Ravera, S. (2024). Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene. International Journal of Molecular Sciences, 25(4), 2083. https://doi.org/10.3390/ijms25042083








