“Time Is out of Joint” in Pluripotent Stem Cells: How and Why
Abstract
1. Introduction
2. Hierarchical Organization of Circadian Clock Network
3. Pluripotent Stem Cells: What Is Known and What Is Still Missing
4. Circadian Rhythm in Pluripotent Stem Cells: The Reasons for Silence
5. Differentiation-Coupled Circadian Clock Development from Pluripotent Stem Cells: Moving on the Road
6. Emergence of Ultradian Circadian Oscillations during Ontogenic Differentiation
7. Non-Canonical Role of Circadian Factors in Pluripotency and Metabolic-Driven Reprogramming
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Allada, R.; Bass, J. Circadian Mechanisms in Medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef]
- Kinouchi, K.; Sassone-Corsi, P. Metabolic Rivalry: Circadian Homeostasis and Tumorigenesis. Nat. Rev. Cancer 2020, 20, 645–661. [Google Scholar] [CrossRef]
- Masri, S.; Sassone-Corsi, P. The Emerging Link between Cancer, Metabolism, and Circadian Rhythms. Nat. Med. 2018, 24, 1795–1803. [Google Scholar] [CrossRef]
- Andersen, B.; Duan, J.; Karri, S.S. How and Why the Circadian Clock Regulates Proliferation of Adult Epithelial Stem Cells. Stem Cells 2023, 41, 319–327. [Google Scholar] [CrossRef]
- Bedont, J.L.; Iascone, D.M.; Sehgal, A. The Lineage Before Time: Circadian and Nonclassical Clock Influences on Development. Annu. Rev. Cell Dev. Biol. 2020, 36, 469–509. [Google Scholar] [CrossRef]
- Dierickx, P.; Van Laake, L.W.; Geijsen, N. Circadian Clocks: From Stem Cells to Tissue Homeostasis and Regeneration. EMBO Rep. 2018, 19, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Dierickx, P.; Du Pré, B.; Feyen, D.A.M.; Geijsen, N.; van Veen, T.; Doevendans, P.A.; Van Laake, L.W. Circadian Rhythms in Stem Cell Biology and Function. In Stem Cells and Cardiac Regeneration; Madonna, R., Ed.; Stem Cell Biology and Regenerative Medicine; Springer International Publishing: Cham, Switzerland, 2016; pp. 57–78. ISBN 978-3-319-25427-2. [Google Scholar]
- Gao, W.; Li, R.; Ye, M.; Zhang, L.; Zheng, J.; Yang, Y.; Wei, X.; Zhao, Q. The Circadian Clock Has Roles in Mesenchymal Stem Cell Fate Decision. Stem Cell Res. Ther. 2022, 13, 200. [Google Scholar] [CrossRef] [PubMed]
- Umemura, Y.; Yagita, K. Development of the Circadian Core Machinery in Mammals. J. Mol. Biol. 2020, 432, 3611–3617. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Moriggi, E.; Bauer, C.; Dibner, C.; Brown, S.A. The Circadian Clock Starts Ticking at a Developmentally Early Stage. J. Biol. Rhythm. 2010, 25, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Paulose, J.K.; Rucker, E.B.; Cassone, V.M. Toward the Beginning of Time: Circadian Rhythms in Metabolism Precede Rhythms in Clock Gene Expression in Mouse Embryonic Stem Cells. PLoS ONE 2012, 7, e49555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yagita, K.; Horie, K.; Koinuma, S.; Nakamura, W.; Yamanaka, I.; Urasaki, A.; Shigeyoshi, Y.; Kawakami, K.; Shimada, S.; Takeda, J.; et al. Development of the Circadian Oscillator during Differentiation of Mouse Embryonic Stem Cells in Vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 3846–3851. [Google Scholar] [CrossRef]
- Dierickx, P.; Vermunt, M.W.; Muraro, M.J.; Creyghton, M.P.; Doevendans, P.A.; van Oudenaarden, A.; Geijsen, N.; Van Laake, L.W. Circadian Networks in Human Embryonic Stem Cell-Derived Cardiomyocytes. EMBO Rep. 2017, 18, 1199–1212. [Google Scholar] [CrossRef]
- Umemura, Y.; Maki, I.; Tsuchiya, Y.; Koike, N.; Yagita, K. Human Circadian Molecular Oscillation Development Using Induced Pluripotent Stem Cells. J. Biol. Rhythm. 2019, 34, 525–532. [Google Scholar] [CrossRef]
- Cajochen, C.; Kräuchi, K.; Wirz-Justice, A. Role of Melatonin in the Regulation of Human Circadian Rhythms and Sleep. J. Neuroendocr. 2003, 15, 432–437. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted Feeding Uncouples Circadian Oscillators in Peripheral Tissues from the Central Pacemaker in the Suprachiasmatic Nucleus. Genes Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef]
- Hara, R.; Wan, K.; Wakamatsu, H.; Aida, R.; Moriya, T.; Akiyama, M.; Shibata, S. Restricted Feeding Entrains Liver Clock without Participation of the Suprachiasmatic Nucleus. Genes Cells 2001, 6, 269–278. [Google Scholar] [CrossRef]
- Pezuk, P.; Mohawk, J.A.; Yoshikawa, T.; Sellix, M.T.; Menaker, M. Circadian Organization Is Governed by Extra-SCN Pacemakers. J. Biol. Rhythm. 2010, 25, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Damiola, F.; Schibler, U. A Serum Shock Induces Circadian Gene Expression in Mammalian Tissue Culture Cells. Cell 1998, 93, 929–937. [Google Scholar] [CrossRef]
- Yagita, K.; Tamanini, F.; van Der Horst, G.T.; Okamura, H. Molecular Mechanisms of the Biological Clock in Cultured Fibroblasts. Science 2001, 292, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Numano, R.; Abe, M.; Hida, A.; Takahashi, R.; Ueda, M.; Block, G.D.; Sakaki, Y.; Menaker, M.; Tei, H. Resetting Central and Peripheral Circadian Oscillators in Transgenic Rats. Science 2000, 288, 682–685. [Google Scholar] [CrossRef]
- Yoo, S.-H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.-K.; Oh, W.J.; Yoo, O.J.; et al. PERIOD2::LUCIFERASE Real-Time Reporting of Circadian Dynamics Reveals Persistent Circadian Oscillations in Mouse Peripheral Tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [PubMed]
- Chiou, Y.-Y.; Yang, Y.; Rashid, N.; Ye, R.; Selby, C.P.; Sancar, A. Mammalian Period Represses and De-Represses Transcription by Displacing CLOCK-BMAL1 from Promoters in a Cryptochrome-Dependent Manner. Proc. Natl. Acad. Sci. USA 2016, 113, E6072–E6079. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional Architecture of the Mammalian Circadian Clock. Nat. Rev. Genet. 2017, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Skinner, M.K. E-Box and Cyclic Adenosine Monophosphate Response Elements Are Both Required for Follicle-Stimulating Hormone-Induced Transferrin Promoter Activation in Sertoli Cells*. Endocrinology 1999, 140, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Preitner, N.; Damiola, F.; Lopez-Molina, L.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The Orphan Nuclear Receptor REV-ERBalpha Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A Functional Genomics Strategy Reveals Rora as a Component of the Mammalian Circadian Clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef]
- Ueda, H.R.; Chen, W.; Adachi, A.; Wakamatsu, H.; Hayashi, S.; Takasugi, T.; Nagano, M.; Nakahama, K.; Suzuki, Y.; Sugano, S.; et al. A Transcription Factor Response Element for Gene Expression during Circadian Night. Nature 2002, 418, 534–539. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Mitsui, S.; Yan, L.; Yagita, K.; Miyake, S.; Okamura, H. Role of DBP in the Circadian Oscillatory Mechanism. Mol. Cell Biol. 2000, 20, 4773–4781. [Google Scholar] [CrossRef]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A Circadian Gene Expression Atlas in Mammals: Implications for Biology and Medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Mure, L.S.; Le, H.D.; Benegiamo, G.; Chang, M.W.; Rios, L.; Jillani, N.; Ngotho, M.; Kariuki, T.; Dkhissi-Benyahya, O.; Cooper, H.M.; et al. Diurnal Transcriptome Atlas of a Primate across Major Neural and Peripheral Tissues. Science 2018, 359, eaao0318. [Google Scholar] [CrossRef] [PubMed]
- Fuhr, L.; El-Athman, R.; Scrima, R.; Cela, O.; Carbone, A.; Knoop, H.; Li, Y.; Hoffmann, K.; Laukkanen, M.O.; Corcione, F.; et al. The Circadian Clock Regulates Metabolic Phenotype Rewiring Via HKDC1 and Modulates Tumor Progression and Drug Response in Colorectal Cancer. EBioMedicine 2018, 33, 105–121. [Google Scholar] [CrossRef]
- Pacelli, C.; Rotundo, G.; Lecce, L.; Menga, M.; Bidollari, E.; Scrima, R.; Cela, O.; Piccoli, C.; Cocco, T.; Vescovi, A.L.; et al. Parkin Mutation Affects Clock Gene-Dependent Energy Metabolism. Int. J. Mol. Sci. 2019, 20, 2772. [Google Scholar] [CrossRef] [PubMed]
- Cela, O.; Scrima, R.; Pazienza, V.; Merla, G.; Benegiamo, G.; Augello, B.; Fugetto, S.; Menga, M.; Rubino, R.; Fuhr, L.; et al. Clock Genes-Dependent Acetylation of Complex I Sets Rhythmic Activity of Mitochondrial OxPhos. Biochim. Biophys. Acta 2016, 1863, 596–606. [Google Scholar] [CrossRef]
- Laje, R.; Agostino, P.V.; Golombek, D.A. The Times of Our Lives: Interaction Among Different Biological Periodicities. Front. Integr. Neurosci. 2018, 12, 10. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.O.; Herzel, H. Mechanism for 12 Hr Rhythm Generation by the Circadian Clock. Cell Rep. 2013, 3, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Gonzales, N.M.; Jung, S.Y.; Lu, Y.; Putluri, N.; Zhu, B.; Dacso, C.C.; Lonard, D.M.; O’Malley, B.W. Defining the Mammalian Coactivation of Hepatic 12-h Clock and Lipid Metabolism. Cell Rep. 2022, 38, 110491. [Google Scholar] [CrossRef]
- Meng, H.; Gonzales, N.M.; Lonard, D.M.; Putluri, N.; Zhu, B.; Dacso, C.C.; York, B.; O’Malley, B.W. XBP1 Links the 12-Hour Clock to NAFLD and Regulation of Membrane Fluidity and Lipid Homeostasis. Nat. Commun. 2020, 11, 6215. [Google Scholar] [CrossRef]
- Zhu, B.; Zhang, Q.; Pan, Y.; Mace, E.M.; York, B.; Antoulas, A.C.; Dacso, C.C.; O’Malley, B.W. A Cell-Autonomous Mammalian 12 Hr Clock Coordinates Metabolic and Stress Rhythms. Cell Metab. 2017, 25, 1305–1319.e9. [Google Scholar] [CrossRef]
- Evans, M.J.; Kaufman, M.H. Establishment in Culture of Pluripotential Cells from Mouse Embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a Pluripotent Cell Line from Early Mouse Embryos Cultured in Medium Conditioned by Teratocarcinoma Stem Cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Murry, C.E.; Keller, G. Differentiation of Embryonic Stem Cells to Clinically Relevant Populations: Lessons from Embryonic Development. Cell 2008, 132, 661–680. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nagata, N.; Kurokawa, H.; Yamanaka, S. IPS Cells: A Game Changer for Future Medicine. EMBO J. 2014, 33, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced Pluripotent Stem Cells and Their Use in Human Models of Disease and Development. Physiol. Rev. 2019, 99, 79–114. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Regan, S.N.; Xia, Y.; Oostrom, L.A.; Cowan, C.A.; Musunuru, K. Enhanced Efficiency of Human Pluripotent Stem Cell Genome Editing through Replacing TALENs with CRISPRs. Cell Stem Cell 2013, 12, 393–394. [Google Scholar] [CrossRef]
- González, F.; Zhu, Z.; Shi, Z.-D.; Lelli, K.; Verma, N.; Li, Q.V.; Huangfu, D. An ICRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell Stem Cell 2014, 15, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; González, F.; Huangfu, D. The ICRISPR Platform for Rapid Genome Editing in Human Pluripotent Stem Cells. Methods Enzym. 2014, 546, 215–250. [Google Scholar] [CrossRef]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient Induction of Transgene-Free Human Pluripotent Stem Cells Using a Vector Based on Sendai Virus, an RNA Virus That Does Not Integrate into the Host Genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced Pluripotent Stem Cells Generated without Viral Integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac Transposition Reprograms Fibroblasts to Induced Pluripotent Stem Cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef]
- Jia, F.; Wilson, K.D.; Sun, N.; Gupta, D.M.; Huang, M.; Li, Z.; Panetta, N.J.; Chen, Z.Y.; Robbins, R.C.; Kay, M.A.; et al. A Nonviral Minicircle Vector for Deriving Human IPS Cells. Nat. Methods 2010, 7, 197–199. [Google Scholar] [CrossRef]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified MRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. A Decade of Transcription Factor-Mediated Reprogramming to Pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic Stem Cell States: Naive to Primed Pluripotency in Rodents and Humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef] [PubMed]
- Kunitomi, A.; Hirohata, R.; Arreola, V.; Osawa, M.; Kato, T.M.; Nomura, M.; Kawaguchi, J.; Hara, H.; Kusano, K.; Takashima, Y.; et al. Improved Sendai Viral System for Reprogramming to Naive Pluripotency. Cell Rep. Methods 2022, 2, 100317. [Google Scholar] [CrossRef] [PubMed]
- Io, S.; Kabata, M.; Iemura, Y.; Semi, K.; Morone, N.; Minagawa, A.; Wang, B.; Okamoto, I.; Nakamura, T.; Kojima, Y.; et al. Capturing Human Trophoblast Development with Naive Pluripotent Stem Cells in Vitro. Cell Stem Cell 2021, 28, 1023–1039.e13. [Google Scholar] [CrossRef]
- Duggal, G.; Warrier, S.; Ghimire, S.; Broekaert, D.; Van der Jeught, M.; Lierman, S.; Deroo, T.; Peelman, L.; Van Soom, A.; Cornelissen, R.; et al. Alternative Routes to Induce Naïve Pluripotency in Human Embryonic Stem Cells. Stem Cells 2015, 33, 2686–2698. [Google Scholar] [CrossRef]
- Liu, Y.; Goldberg, A.J.; Dennis, J.E.; Gronowicz, G.A.; Kuhn, L.T. One-Step Derivation of Mesenchymal Stem Cell (MSC)-like Cells from Human Pluripotent Stem Cells on a Fibrillar Collagen Coating. PLoS ONE 2012, 7, e33225. [Google Scholar] [CrossRef]
- Agriesti, F.; Tataranni, T.; Pacelli, C.; Scrima, R.; Laurenzana, I.; Ruggieri, V.; Cela, O.; Mazzoccoli, C.; Salerno, M.; Sessa, F.; et al. Nandrolone Induces a Stem Cell-like Phenotype in Human Hepatocarcinoma-Derived Cell Line Inhibiting Mitochondrial Respiratory Activity. Sci. Rep. 2020, 10, 2287. [Google Scholar] [CrossRef]
- Hanna, H.; Mir, L.M.; Andre, F.M. In Vitro Osteoblastic Differentiation of Mesenchymal Stem Cells Generates Cell Layers with Distinct Properties. Stem Cell Res. Ther. 2018, 9, 203. [Google Scholar] [CrossRef]
- Zou, L.; Luo, Y.; Chen, M.; Wang, G.; Ding, M.; Petersen, C.C.; Kang, R.; Dagnaes-Hansen, F.; Zeng, Y.; Lv, N.; et al. A Simple Method for Deriving Functional MSCs and Applied for Osteogenesis in 3D Scaffolds. Sci. Rep. 2013, 3, 2243. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, H.; Kaitsuka, T.; Tomizawa, K. Response to Stimulations Inducing Circadian Rhythm in Human Induced Pluripotent Stem Cells. Cells 2020, 9, 620. [Google Scholar] [CrossRef]
- O’Connell, E.J.; Martinez, C.-A.; Liang, Y.G.; Cistulli, P.A.; Cook, K.M. Out of Breath, out of Time: Interactions between HIF and Circadian Rhythms. Am. J. Physiol. -Cell Physiol. 2020, 319, C533–C540. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, C.; Agriesti, F.; Scrima, R.; Falzetti, F.; Di Ianni, M.; Capitanio, N. To Breathe or Not to Breathe: The Haematopoietic Stem/Progenitor Cells Dilemma. Br. J. Pharmacol. 2013, 169, 1652–1671. [Google Scholar] [CrossRef]
- Lu, C.; Yang, Y.; Zhao, R.; Hua, B.; Xu, C.; Yan, Z.; Sun, N.; Qian, R. Role of Circadian Gene Clock during Differentiation of Mouse Pluripotent Stem Cells. Protein Cell 2016, 7, 820–832. [Google Scholar] [CrossRef]
- Chetty, S.; Pagliuca, F.W.; Honore, C.; Kweudjeu, A.; Rezania, A.; Melton, D.A. A Simple Tool to Improve Pluripotent Stem Cell Differentiation. Nat. Methods 2013, 10, 553–556. [Google Scholar] [CrossRef]
- Pauklin, S.; Vallier, L. The Cell-Cycle State of Stem Cells Determines Cell Fate Propensity. Cell 2013, 155, 135–147. [Google Scholar] [CrossRef]
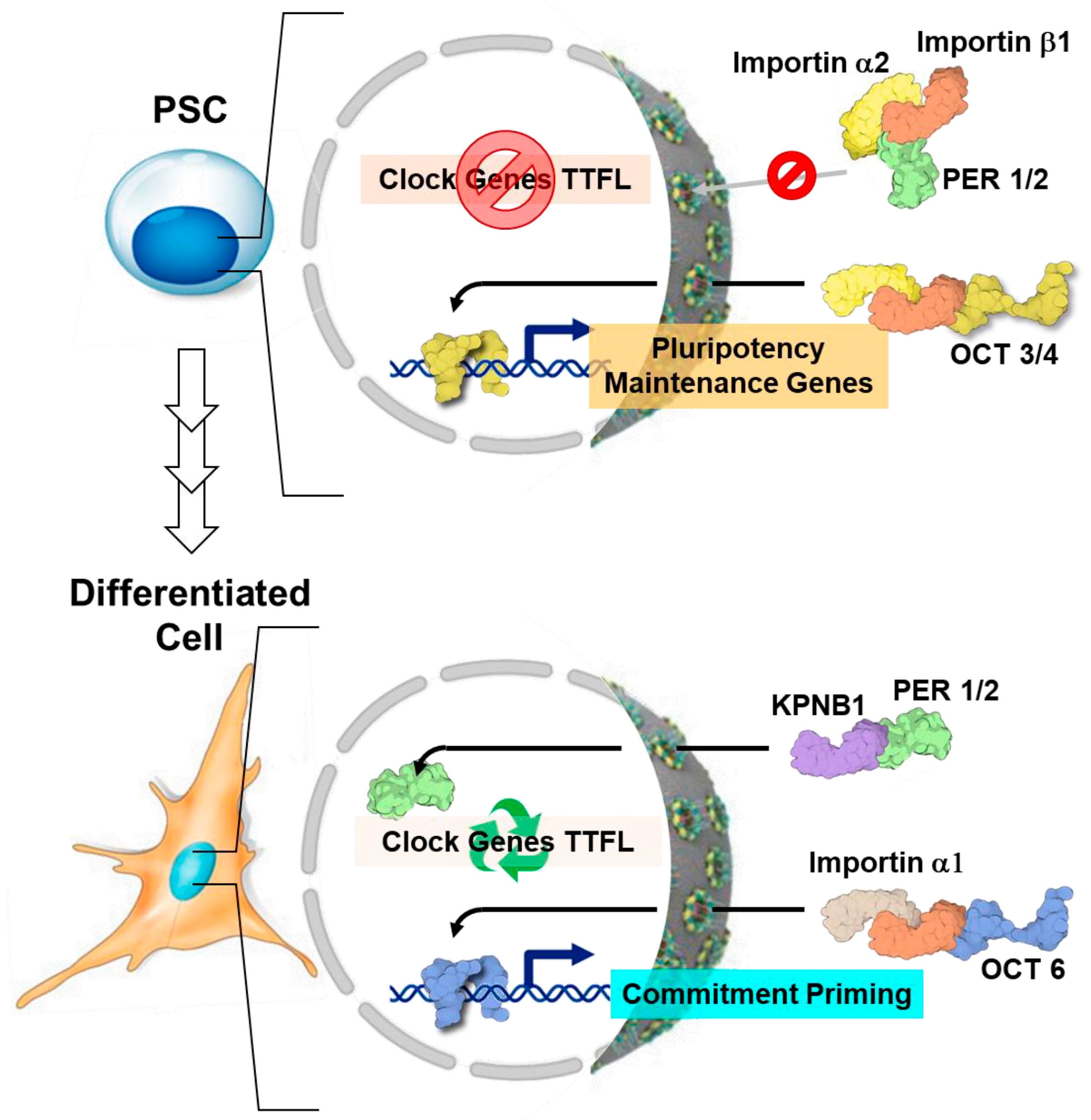
- Umemura, Y.; Koike, N.; Matsumoto, T.; Yoo, S.-H.; Chen, Z.; Yasuhara, N.; Takahashi, J.S.; Yagita, K. Transcriptional Program of Kpna2/Importin-A2 Regulates Cellular Differentiation-Coupled Circadian Clock Development in Mammalian Cells. Proc. Natl. Acad. Sci. USA 2014, 111, E5039–E5048. [Google Scholar] [CrossRef]
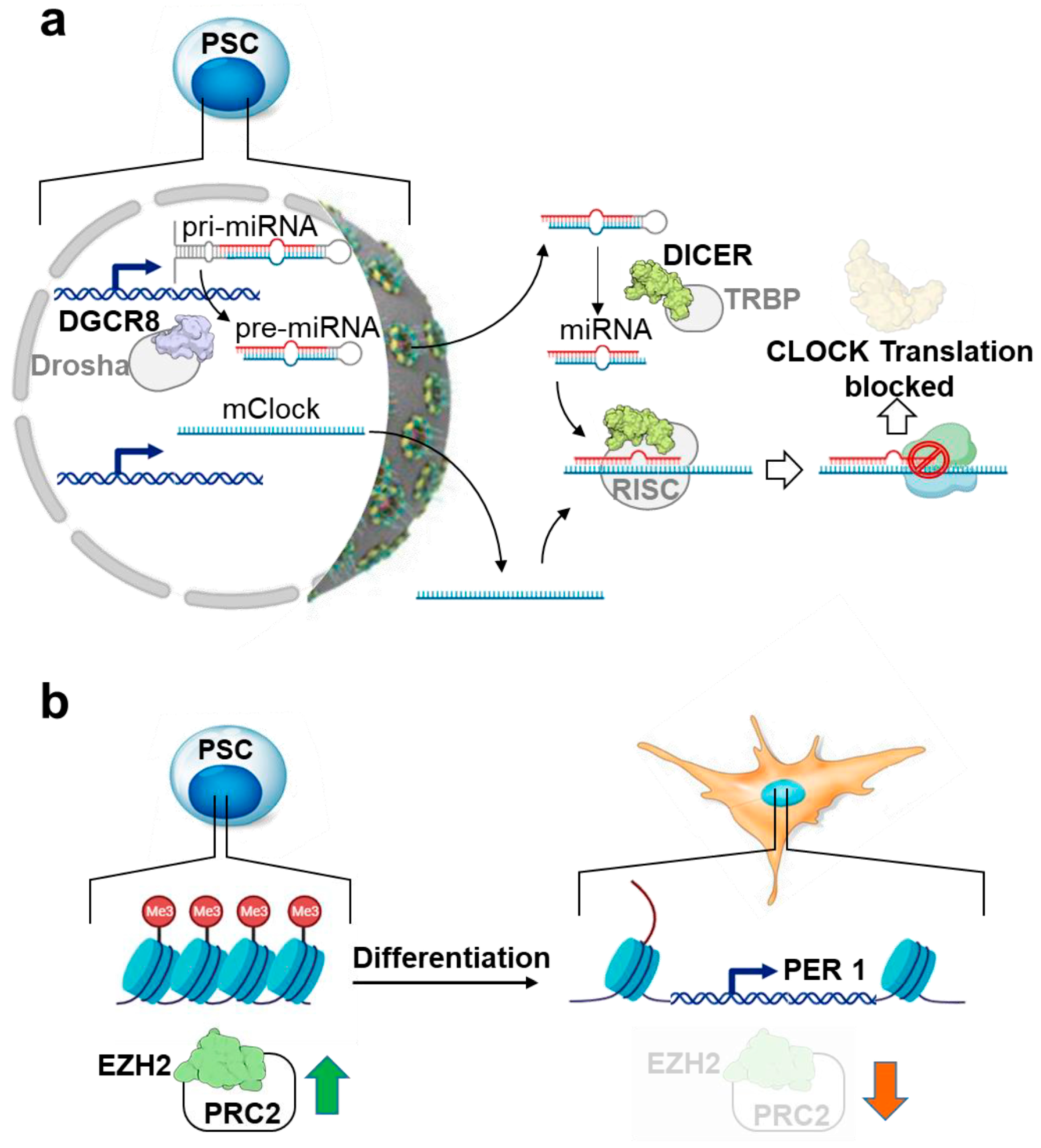
- Umemura, Y.; Koike, N.; Ohashi, M.; Tsuchiya, Y.; Meng, Q.J.; Minami, Y.; Hara, M.; Hisatomi, M.; Yagita, K. Involvement of Posttranscriptional Regulation of Clock in the Emergence of Circadian Clock Oscillation during Mouse Development. Proc. Natl. Acad. Sci. USA 2017, 114, E7479–E7488. [Google Scholar] [CrossRef]
- Hirano, A.; Fu, Y.-H.; Ptáček, L.J. The Intricate Dance of Post-Translational Modifications in the Rhythm of Life. Nat. Struct. Mol. Biol. 2016, 23, 1053–1060. [Google Scholar] [CrossRef]
- Eide, E.J.; Vielhaber, E.L.; Hinz, W.A.; Virshup, D.M. The Circadian Regulatory Proteins BMAL1 and Cryptochromes Are Substrates of Casein Kinase Iε*. J. Biol. Chem. 2002, 277, 17248–17254. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Etchegaray, J.P.; Cagampang, F.R.; Loudon, A.S.; Reppert, S.M. Posttranslational Mechanisms Regulate the Mammalian Circadian Clock. Cell 2001, 107, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Okamoto-Uchida, Y.; Izawa, J.; Nishimura, A.; Hattori, A.; Suzuki, N.; Hirayama, J. Post-Translational Modifications Are Required for Circadian Clock Regulation in Vertebrates. Curr. Genom. 2019, 20, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Iitaka, C.; Miyazaki, K.; Akaike, T.; Ishida, N. A Role for Glycogen Synthase Kinase-3β in the Mammalian Circadian Clock*. J. Biol. Chem. 2005, 280, 29397–29402. [Google Scholar] [CrossRef]
- Maier, B.; Wendt, S.; Vanselow, J.T.; Wallach, T.; Reischl, S.; Oehmke, S.; Schlosser, A.; Kramer, A. A Large-Scale Functional RNAi Screen Reveals a Role for CK2 in the Mammalian Circadian Clock. Genes Dev. 2009, 23, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Narasimamurthy, R.; Virshup, D.M. The Phosphorylation Switch That Regulates Ticking of the Circadian Clock. Mol. Cell 2021, 81, 1133–1146. [Google Scholar] [CrossRef] [PubMed]
- Yasuhara, N.; Yamagishi, R.; Arai, Y.; Mehmood, R.; Kimoto, C.; Fujita, T.; Touma, K.; Kaneko, A.; Kamikawa, Y.; Moriyama, T.; et al. Importin Alpha Subtypes Determine Differential Transcription Factor Localization in Embryonic Stem Cells Maintenance. Dev. Cell 2013, 26, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jang, A.R.; Francey, L.J.; Sehgal, A.; Hogenesch, J.B. KPNB1 Mediates PER/CRY Nuclear Translocation and Circadian Clock Function. Elife 2015, 4, e08647. [Google Scholar] [CrossRef] [PubMed]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. CLOCK and NPAS2 Have Overlapping Roles in the Suprachiasmatic Circadian Clock. Nat. Neurosci. 2007, 10, 543–545. [Google Scholar] [CrossRef] [PubMed]
- DeBruyne, J.P.; Weaver, D.R.; Reppert, S.M. Peripheral Circadian Oscillators Require CLOCK. Curr. Biol. 2007, 17, R538–R539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murgo, E.; Colangelo, T.; Bellet, M.M.; Malatesta, F.; Mazzoccoli, G. Role of the Circadian Gas-Responsive Hemeprotein NPAS2 in Physiology and Pathology. Biology 2023, 12, 1354. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A Comparison of Genetically Matched Cell Lines Reveals the Equivalence of Human IPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef]
- Li, M.; Liu, G.-H.; Izpisua Belmonte, J.C. Navigating the Epigenetic Landscape of Pluripotent Stem Cells. Nat. Rev. Mol. Cell Biol. 2012, 13, 524–535. [Google Scholar] [CrossRef]
- Etchegaray, J.-P.; Yang, X.; DeBruyne, J.P.; Peters, A.H.F.M.; Weaver, D.R.; Jenuwein, T.; Reppert, S.M. The Polycomb Group Protein EZH2 Is Required for Mammalian Circadian Clock Function. J. Biol. Chem. 2006, 281, 21209–21215. [Google Scholar] [CrossRef]
- Guenther, M.G.; Frampton, G.M.; Soldner, F.; Hockemeyer, D.; Mitalipova, M.; Jaenisch, R.; Young, R.A. Chromatin Structure and Gene Expression Programs of Human Embryonic and Induced Pluripotent Stem Cells. Cell Stem Cell 2010, 7, 249–257. [Google Scholar] [CrossRef]
- Kaneko, H.; Kaitsuka, T.; Tomizawa, K. Artificial Induction of Circadian Rhythm by Combining Exogenous BMAL1 Expression and Polycomb Repressive Complex 2 Inhibition in Human Induced Pluripotent Stem Cells. Cell Mol. Life Sci. 2023, 80, 200. [Google Scholar] [CrossRef]
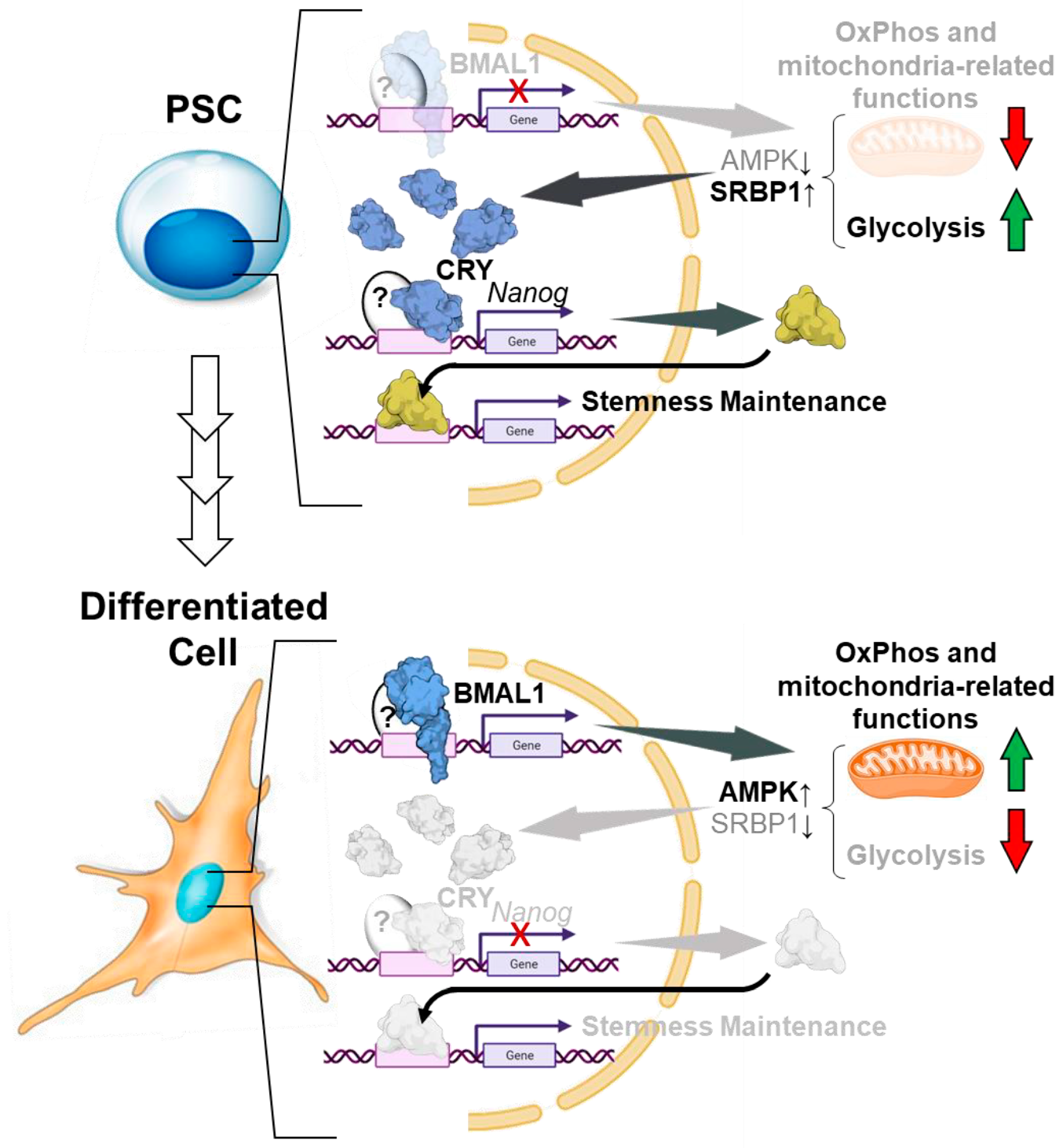
- Ameneiro, C.; Moreira, T.; Fuentes-Iglesias, A.; Coego, A.; Garcia-Outeiral, V.; Escudero, A.; Torrecilla, D.; Mulero-Navarro, S.; Carvajal-Gonzalez, J.M.; Guallar, D.; et al. BMAL1 Coordinates Energy Metabolism and Differentiation of Pluripotent Stem Cells. Life Sci. Alliance 2020, 3, e201900534. [Google Scholar] [CrossRef]
- Gallardo, A.; Molina, A.; Asenjo, H.G.; Martorell-Marugán, J.; Montes, R.; Ramos-Mejia, V.; Sanchez-Pozo, A.; Carmona-Sáez, P.; Lopez-Onieva, L.; Landeira, D. The Molecular Clock Protein Bmal1 Regulates Cell Differentiation in Mouse Embryonic Stem Cells. Life Sci. Alliance 2020, 3, e201900535. [Google Scholar] [CrossRef]
- Thakur, S.; Storewala, P.; Basak, U.; Jalan, N.; Pethe, P. Clocking the Circadian Genes in Human Embryonic Stem Cells. Stem Cell Investig. 2020, 7, 9. [Google Scholar] [CrossRef]
- Zhao, J.; Kilman, V.L.; Keegan, K.P.; Peng, Y.; Emery, P.; Rosbash, M.; Allada, R. Drosophila Clock Can Generate Ectopic Circadian Clocks. Cell 2003, 113, 755–766. [Google Scholar] [CrossRef]
- Lerner, I.; Bartok, O.; Wolfson, V.; Menet, J.S.; Weissbein, U.; Afik, S.; Haimovich, D.; Gafni, C.; Friedman, N.; Rosbash, M.; et al. Clk Post-Transcriptional Control Denoises Circadian Transcription Both Temporally and Spatially. Nat. Commun. 2015, 6, 7056. [Google Scholar] [CrossRef] [PubMed]
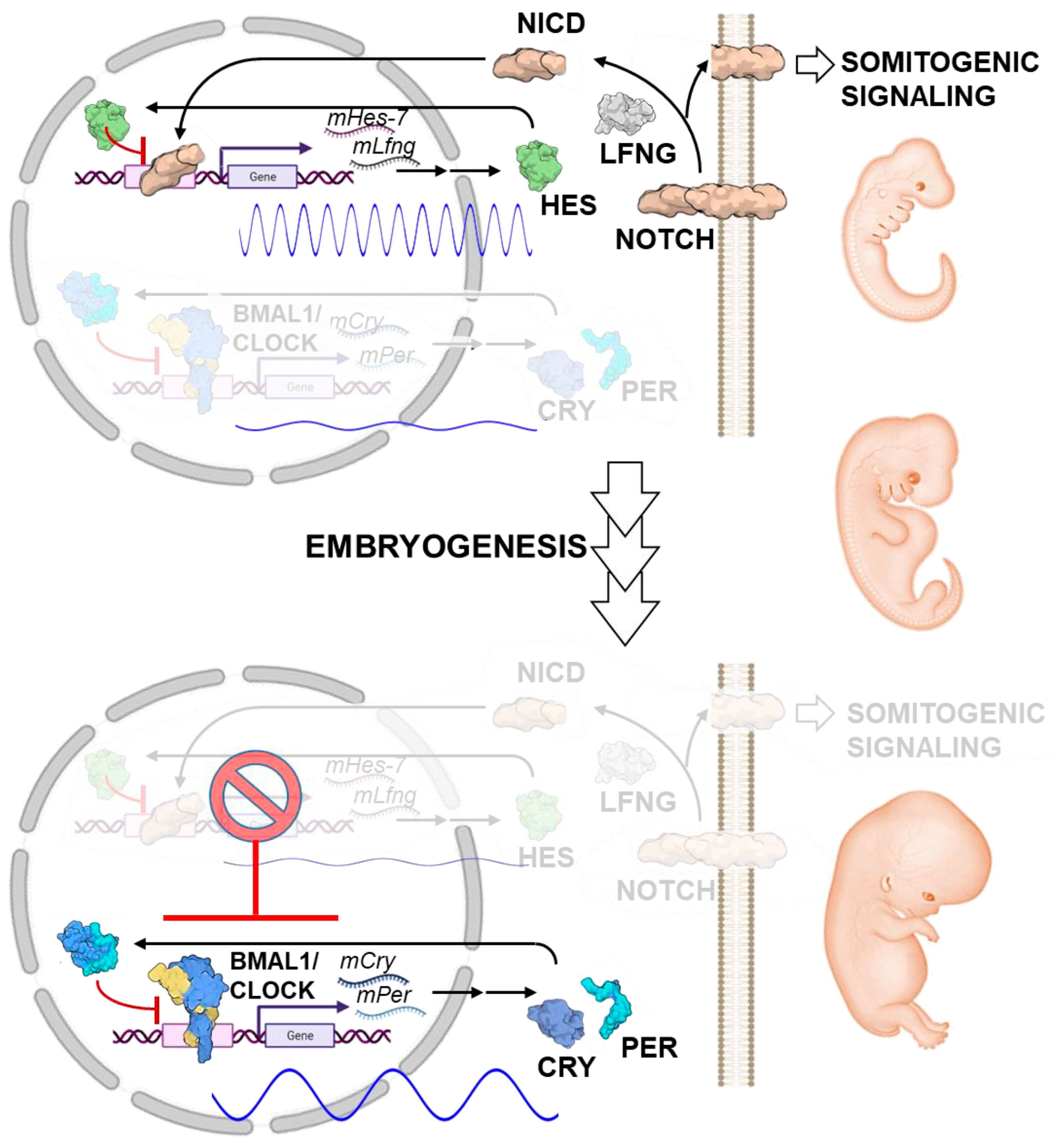
- Harima, Y.; Imayoshi, I.; Shimojo, H.; Kobayashi, T.; Kageyama, R. The Roles and Mechanism of Ultradian Oscillatory Expression of the Mouse Hes Genes. Semin. Cell Dev. Biol. 2014, 34, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Hubaud, A.; Pourquié, O. Signalling Dynamics in Vertebrate Segmentation. Nat. Rev. Mol. Cell Biol. 2014, 15, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Umemura, Y.; Koike, N.; Tsuchiya, Y.; Watanabe, H.; Kondoh, G.; Kageyama, R.; Yagita, K. Circadian Key Component CLOCK/BMAL1 Interferes with Segmentation Clock in Mouse Embryonic Organoids. Proc. Natl. Acad. Sci. USA 2022, 119, e2114083119. [Google Scholar] [CrossRef]
- Bessho, Y.; Hirata, H.; Masamizu, Y.; Kageyama, R. Periodic Repression by the BHLH Factor Hes7 Is an Essential Mechanism for the Somite Segmentation Clock. Genes Dev. 2003, 17, 1451–1456. [Google Scholar] [CrossRef]
- Takashima, Y.; Ohtsuka, T.; González, A.; Miyachi, H.; Kageyama, R. Intronic Delay Is Essential for Oscillatory Expression in the Segmentation Clock. Proc. Natl. Acad. Sci. USA 2011, 108, 3300–3305. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, L.; Grant, G.R.; Paschos, G.; Song, W.-L.; Musiek, E.S.; Lee, V.; McLoughlin, S.C.; Grosser, T.; Cotsarelis, G.; et al. Timing of Expression of the Core Clock Gene Bmal1 Influences Its Effects on Aging and Survival. Sci. Transl. Med. 2016, 8, 324ra16. [Google Scholar] [CrossRef]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early Aging and Age-Related Pathologies in Mice Deficient in BMAL1, the Core Componentof the Circadian Clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef]
- Hanna, J.; Cheng, A.W.; Saha, K.; Kim, J.; Lengner, C.J.; Soldner, F.; Cassady, J.P.; Muffat, J.; Carey, B.W.; Jaenisch, R. Human Embryonic Stem Cells with Biological and Epigenetic Characteristics Similar to Those of Mouse ESCs. Proc. Natl. Acad. Sci. USA 2010, 107, 9222–9227. [Google Scholar] [CrossRef]
- Ware, C.B.; Nelson, A.M.; Mecham, B.; Hesson, J.; Zhou, W.; Jonlin, E.C.; Jimenez-Caliani, A.J.; Deng, X.; Cavanaugh, C.; Cook, S.; et al. Derivation of Naive Human Embryonic Stem Cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4484–4489. [Google Scholar] [CrossRef]
- Cliff, T.S.; Dalton, S. Metabolic Switching and Cell Fate Decisions: Implications for Pluripotency, Reprogramming and Development. Curr. Opin. Genet. Dev. 2017, 46, 44–49. [Google Scholar] [CrossRef]
- Dahan, P.; Lu, V.; Nguyen, R.M.T.; Kennedy, S.A.L.; Teitell, M.A. Metabolism in Pluripotency: Both Driver and Passenger? J. Biol. Chem. 2019, 294, 5420–5429. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, F.; Matsubara, C.; Myung, J.; Yoritaka, T.; Kamimura, N.; Tsutsumi, S.; Kanai, A.; Suzuki, Y.; Sassone-Corsi, P.; Aburatani, H.; et al. Genome-Wide Profiling of the Core Clock Protein BMAL1 Targets Reveals a Strict Relationship with Metabolism. Mol. Cell Biol. 2010, 30, 5636–5648. [Google Scholar] [CrossRef] [PubMed]
- Reinke, H.; Asher, G. Crosstalk between Metabolism and Circadian Clocks. Nat. Rev. Mol. Cell Biol. 2019, 20, 227–241. [Google Scholar] [CrossRef]
- Wu, Y.; Tang, D.; Liu, N.; Xiong, W.; Huang, H.; Li, Y.; Ma, Z.; Zhao, H.; Chen, P.; Qi, X.; et al. Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab. 2017, 25, 73–85. [Google Scholar] [CrossRef]
- Nishimura, K.; Fukuda, A.; Hisatake, K. Mechanisms of the Metabolic Shift during Somatic Cell Reprogramming. Int. J. Mol. Sci. 2019, 20, 2254. [Google Scholar] [CrossRef]
- Scrima, R.; Cela, O.; Agriesti, F.; Piccoli, C.; Tataranni, T.; Pacelli, C.; Mazzoccoli, G.; Capitanio, N. Mitochondrial Calcium Drives Clock Gene-Dependent Activation of Pyruvate Dehydrogenase and of Oxidative Phosphorylation. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118815. [Google Scholar] [CrossRef]
- Scrima, R.; Cela, O.; Merla, G.; Augello, B.; Rubino, R.; Quarato, G.; Fugetto, S.; Menga, M.; Fuhr, L.; Relógio, A.; et al. Clock-Genes and Mitochondrial Respiratory Activity: Evidence of a Reciprocal Interplay. Biochim. Biophys. Acta 2016, 1857, 1344–1351. [Google Scholar] [CrossRef]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. MYC Disrupts the Circadian Clock and Metabolism in Cancer Cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Vander Heiden, M.G.; Jacks, T. Circadian Rhythm Disruption Promotes Lung Tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Tohyama, S.; Fujita, J.; Hishiki, T.; Matsuura, T.; Hattori, F.; Ohno, R.; Kanazawa, H.; Seki, T.; Nakajima, K.; Kishino, Y.; et al. Glutamine Oxidation Is Indispensable for Survival of Human Pluripotent Stem Cells. Cell Metab. 2016, 23, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Hishida, T.; Kinouchi, K.; Hatanaka, F.; Li, Y.; Nguyen, Q.; Chen, Y.; Wang, P.H.; Kessenbrock, K.; Li, W.; et al. The Circadian Clock CRY1 Regulates Pluripotent Stem Cell Identity and Somatic Cell Reprogramming. Cell Rep. 2023, 42, 112590. [Google Scholar] [CrossRef] [PubMed]
- Amano, T.; Matsushita, A.; Hatanaka, Y.; Watanabe, T.; Oishi, K.; Ishida, N.; Anzai, M.; Mitani, T.; Kato, H.; Kishigami, S.; et al. Expression and Functional Analyses of Circadian Genes in Mouse Oocytes and Preimplantation Embryos: Cry1 Is Involved in the Meiotic Process Independently of Circadian Clock Regulation. Biol. Reprod. 2009, 80, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, G.; Qu, M.; Gimple, R.C.; Wu, Q.; Qiu, Z.; Prager, B.C.; Wang, X.; Kim, L.J.Y.; Morton, A.R.; et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov. 2019, 9, 1556–1573. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Kondratov, R.V.; Jamasbi, R.J.; Geusz, M.E. Circadian Clock Genes Are Essential for Normal Adult Neurogenesis, Differentiation, and Fate Determination. PLoS ONE 2015, 10, e0139655. [Google Scholar] [CrossRef]
- Matsuo, T.; Yamaguchi, S.; Mitsui, S.; Emi, A.; Shimoda, F.; Okamura, H. Control Mechanism of the Circadian Clock for Timing of Cell Division in Vivo. Science 2003, 302, 255–259. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Kang, T.-H.; Reardon, J.T.; Lee, J.H.; Ozturk, N. Circadian Clock Control of the Cellular Response to DNA Damage. FEBS Lett. 2010, 584, 2618–2625. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.A.; McNair, C.M.; McCann, J.J.; Alshalalfa, M.; Shostak, A.; Severson, T.M.; Zhu, Y.; Bergman, A.; Gordon, N.; Mandigo, A.C.; et al. The Circadian Cryptochrome, CRY1, Is a pro-Tumorigenic Factor That Rhythmically Modulates DNA Repair. Nat. Commun. 2021, 12, 401. [Google Scholar] [CrossRef]
- Jang, H.; Lee, G.Y.; Selby, C.P.; Lee, G.; Jeon, Y.G.; Lee, J.H.; Cheng, K.K.Y.; Titchenell, P.; Birnbaum, M.J.; Xu, A.; et al. SREBP1c-CRY1 Signalling Represses Hepatic Glucose Production by Promoting FOXO1 Degradation during Refeeding. Nat. Commun. 2016, 7, 12180. [Google Scholar] [CrossRef]
- Lamia, K.A.; Sachdeva, U.M.; DiTacchio, L.; Williams, E.C.; Alvarez, J.G.; Egan, D.F.; Vasquez, D.S.; Juguilon, H.; Panda, S.; Shaw, R.J.; et al. AMPK Regulates the Circadian Clock by Cryptochrome Phosphorylation and Degradation. Science 2009, 326, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Lévi, F.A.; Okyar, A.; Hadadi, E.; Innominato, P.F.; Ballesta, A. Circadian Regulation of Drug Responses: Toward Sex-Specific and Personalized Chronotherapy. Annu. Rev. Pharmacol. Toxicol. 2024, 64, 89–114. [Google Scholar] [CrossRef]
- Rasmussen, E.S.; Takahashi, J.S.; Green, C.B. Time to Target the Circadian Clock for Drug Discovery. Trends Biochem. Sci. 2022, 47, 745–758. [Google Scholar] [CrossRef]
- Weger, M.; Weger, B.D.; Gachon, F. Understanding Circadian Dynamics: Current Progress and Future Directions for Chronobiology in Drug Discovery. Expert Opin. Drug Discov. 2023, 18, 893–901. [Google Scholar] [CrossRef]
- Malhan, D.; Schoenrock, B.; Yalçin, M.; Blottner, D.; Relógio, A. Circadian Regulation in Aging: Implications for Spaceflight and Life on Earth. Aging Cell 2023, 22, e13935. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Goodell, M.A.; Rando, T.A. Ageing and Rejuvenation of Tissue Stem Cells and Their Niches. Nat. Rev. Mol. Cell Biol. 2023, 24, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Hu, J.; Li, N.; Gao, J.; Huo, R.; Peng, X.; Zhang, N.; Liu, Y.; Zhao, H.; Liu, R.; et al. The Mechanism of Stem Cell Aging. Stem Cell Rev. Rep. 2022, 18, 1281–1293. [Google Scholar] [CrossRef]
- Sancar, A.; Van Gelder, R.N. Clocks, Cancer, and Chronochemotherapy. Science 2021, 371, eabb0738. [Google Scholar] [CrossRef]
- Yang, Y.; Lindsey-Boltz, L.A.; Vaughn, C.M.; Selby, C.P.; Cao, X.; Liu, Z.; Hsu, D.S.; Sancar, A. Circadian Clock, Carcinogenesis, Chronochemotherapy Connections. J. Biol. Chem. 2021, 297, 101068. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agriesti, F.; Cela, O.; Capitanio, N. “Time Is out of Joint” in Pluripotent Stem Cells: How and Why. Int. J. Mol. Sci. 2024, 25, 2063. https://doi.org/10.3390/ijms25042063
Agriesti F, Cela O, Capitanio N. “Time Is out of Joint” in Pluripotent Stem Cells: How and Why. International Journal of Molecular Sciences. 2024; 25(4):2063. https://doi.org/10.3390/ijms25042063
Chicago/Turabian StyleAgriesti, Francesca, Olga Cela, and Nazzareno Capitanio. 2024. "“Time Is out of Joint” in Pluripotent Stem Cells: How and Why" International Journal of Molecular Sciences 25, no. 4: 2063. https://doi.org/10.3390/ijms25042063
APA StyleAgriesti, F., Cela, O., & Capitanio, N. (2024). “Time Is out of Joint” in Pluripotent Stem Cells: How and Why. International Journal of Molecular Sciences, 25(4), 2063. https://doi.org/10.3390/ijms25042063






