Mesophiles vs. Thermophiles: Untangling the Hot Mess of Intrinsically Disordered Proteins and Growth Temperature of Bacteria
Abstract
1. Introduction
1.1. Intrinsically Disordered Proteins and Their Abundance
1.2. Identification of IDP Groups
1.3. Quasi-Independent Contrasts
2. Results and Discussion
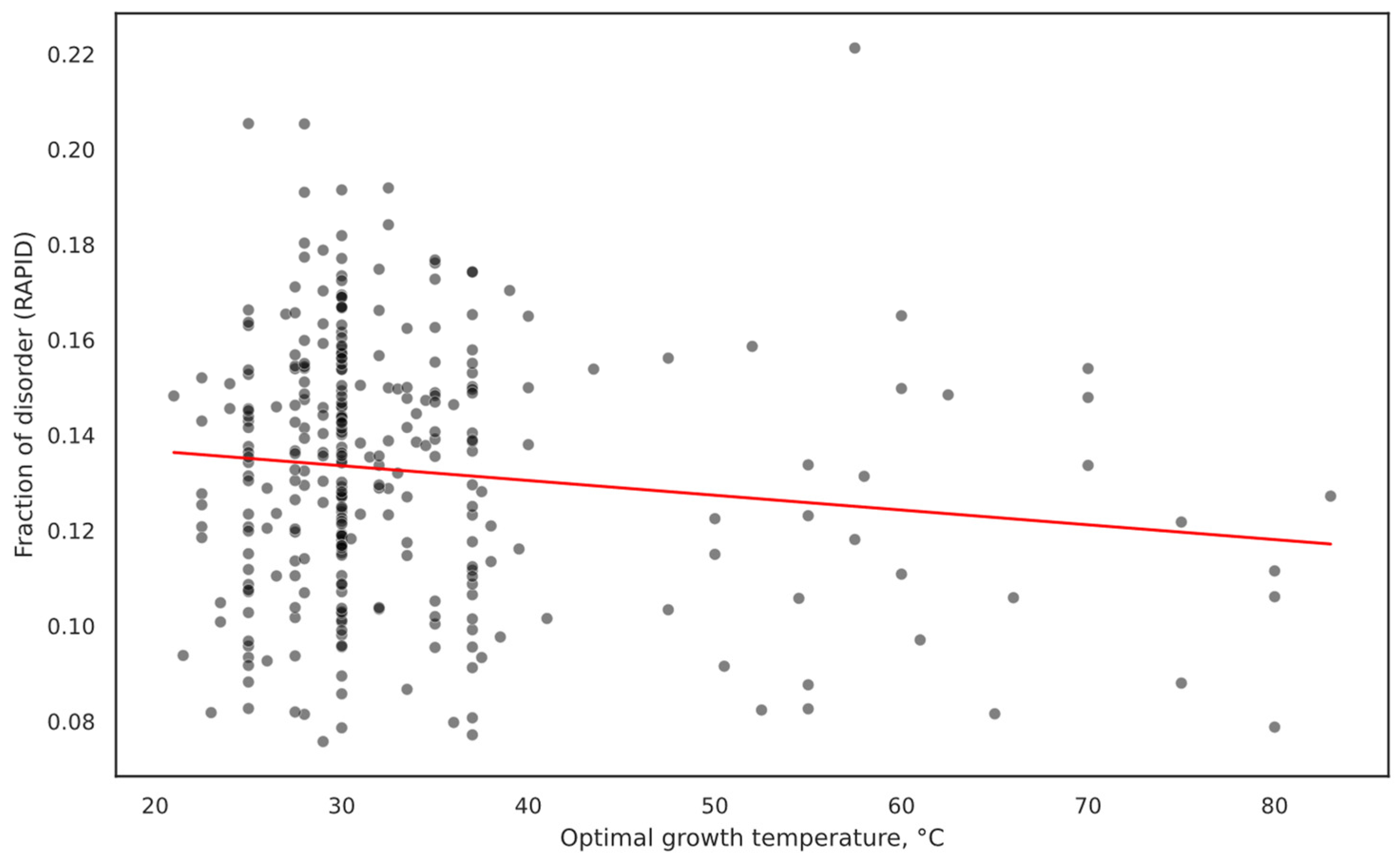
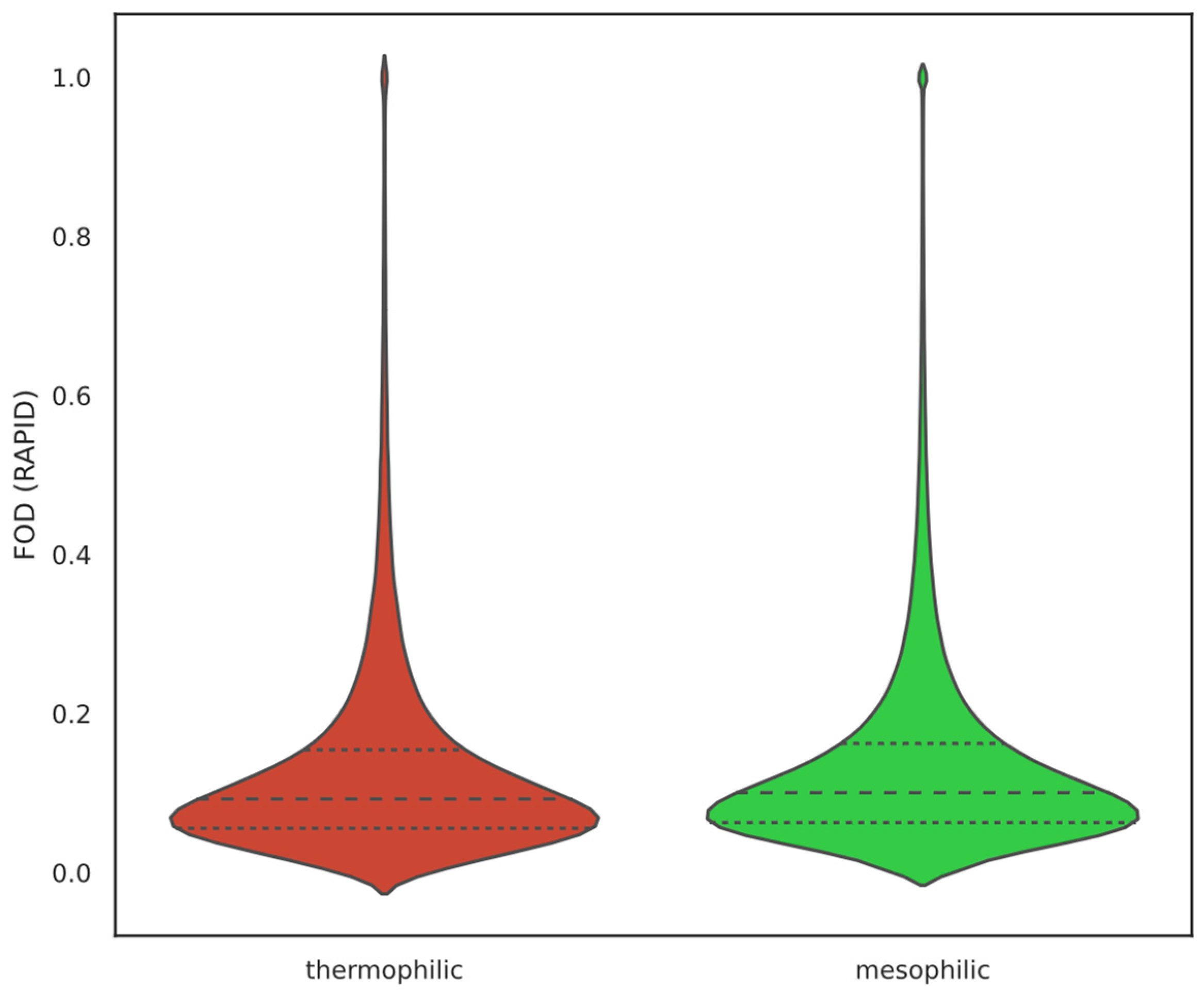
2.1. Overall IDP Abundance in Different Proteomes
2.2. Overall IDP Abundance in Orthologs
2.3. Abundance in Different IDP Classes and Proteins with Different Molecular Functions
2.4. Analysis of Aligned Ortholog Clusters
2.5. Phylogeny Impact on FOD/OGT Relationship
3. Materials and Methods
3.1. Data Sources and Availability
3.2. Protein Clustering
- Proteins from at least 10 different species per cluster;
- At least one candidate IDP (identification described in the disorder calculation subsection);
3.3. Disorder Calculations
- 3.
- RAPID disorder score ≥ 0.5;
- 4.
- Total number of residues × RAPID disorder score ≥ 100;
- 5.
- IDP type 3, 4, or 5 (strong polyampholytes or positive/negative strong polyelectrolytes);
3.4. Cluster Disorder Alignment
3.5. Quasi-Independent Contrast Calculation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wright, P.E.; Dyson, H.J. Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N.; Obradovic, Z. Functional Anthology of Intrinsic Disorder. 1. Biological Processes and Functions of Proteins with Long Disordered Regions. J. Proteome Res. 2007, 6, 1882–1898. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why Are “Natively Unfolded” Proteins Unstructured under Physiologic Conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A Meta-Predictor of Intrinsically Disordered Amino Acids. Biochim. Biophys. Acta 2010, 1804, 996–1010. [Google Scholar] [CrossRef] [PubMed]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of Protein Disorder Enhanced with Unambiguous Experimental Annotation and Visualization of Evolutionary Conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Katuwawala, A.; Wang, K.; Wu, Z.; Ghadermarzi, S.; Gao, J.; Kurgan, L. flDPnn: Accurate Intrinsic Disorder Prediction with Putative Propensities of Disorder Functions. Nat. Commun. 2021, 12, 4438. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Yan, J.; Fan, X.; Mizianty, M.J.; Xue, B.; Wang, K.; Hu, G.; Uversky, V.N.; Kurgan, L. Exceptionally Abundant Exceptions: Comprehensive Characterization of Intrinsic Disorder in All Domains of Life. Cell. Mol. Life Sci. 2015, 72, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Anjum, F.; Mohammad, T.; Asrani, P.; Shafie, A.; Singh, S.; Yadav, D.K.; Uversky, V.N.; Hassan, M.I. Identification of Intrinsically Disorder Regions in Non-Structural Proteins of SARS-CoV-2: New Insights into Drug and Vaccine Resistance. Mol. Cell Biochem. 2022, 477, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- DeForte, S.; Uversky, V.N. Not an Exception to the Rule: The Functional Significance of Intrinsically Disordered Protein Regions in Enzymes. Mol. BioSystems 2017, 13, 463–469. [Google Scholar] [CrossRef]
- Apic, G.; Gough, J.; Teichmann, S.A. An Insight into Domain Combinations. Bioinformatics 2001, 17 (Suppl. S1), S83–S89. [Google Scholar] [CrossRef]
- Ekman, D.; Björklund, A.K.; Elofsson, A. Quantification of the Elevated Rate of Domain Rearrangements in Metazoa. J. Mol. Biol. 2007, 372, 1337–1348. [Google Scholar] [CrossRef] [PubMed]
- Apic, G.; Gough, J.; Teichmann, S.A. Domain Combinations in Archaeal, Eubacterial and Eukaryotic Proteomes. J. Mol. Biol. 2001, 310, 311–325. [Google Scholar] [CrossRef] [PubMed]
- Ekman, D.; Björklund, A.K.; Frey-Skött, J.; Elofsson, A. Multi-Domain Proteins in the Three Kingdoms of Life: Orphan Domains and Other Unassigned Regions. J. Mol. Biol. 2005, 348, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rost, B. CHOP Proteins into Structural Domain-like Fragments. Proteins 2004, 55, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Burra, P.V.; Kalmar, L.; Tompa, P. Reduction in Structural Disorder and Functional Complexity in the Thermal Adaptation of Prokaryotes. PLoS ONE 2010, 5, e12069. [Google Scholar] [CrossRef] [PubMed]
- Pancsa, R.; Kovacs, D.; Tompa, P. Misprediction of Structural Disorder in Halophiles. Molecules 2019, 24, 479. [Google Scholar] [CrossRef]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. Intrinsically Disordered Proteins Play Diverse Roles in Cell Signaling. Cell Commun. Signal 2022, 20, 20. [Google Scholar] [CrossRef] [PubMed]
- van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef]
- Das, R.K.; Pappu, R.V. Conformations of Intrinsically Disordered Proteins Are Influenced by Linear Sequence Distributions of Oppositely Charged Residues. Proc. Natl. Acad. Sci. USA 2013, 110, 13392–13397. [Google Scholar] [CrossRef]
- Mao, A.H.; Crick, S.L.; Vitalis, A.; Chicoine, C.L.; Pappu, R.V. Net Charge per Residue Modulates Conformational Ensembles of Intrinsically Disordered Proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 8183–8188. [Google Scholar] [CrossRef]
- Xia, X. Phylogeny-Based Comparative Methods. In A Mathematical Primer of Molecular Phylogenetics; Apple Academic Press: Palm Bay, FL, USA, 2020; ISBN 978-0-429-42587-5. [Google Scholar]
- Thole, J.F.; Waudby, C.A.; Pielak, G.J. Disordered Proteins Mitigate the Temperature Dependence of Site-Specific Binding Free Energies. J. Biol. Chem. 2023, 299, 102984. [Google Scholar] [CrossRef]
- Shao, H.; Huang, W.; Avilan, L.; Receveur-Bréchot, V.; Puppo, C.; Puppo, R.; Lebrun, R.; Gontero, B.; Launay, H. A New Type of Flexible CP12 Protein in the Marine Diatom Thalassiosira pseudonana. Cell Commun. Signal. 2021, 19, 38. [Google Scholar] [CrossRef]
- Kolonko, M.; Bystranowska, D.; Taube, M.; Kozak, M.; Bostock, M.; Popowicz, G.; Ożyhar, A.; Greb-Markiewicz, B. The Intrinsically Disordered Region of GCEprotein Adopts a More Fixed Structure by Interacting with the LBD of the Nuclearreceptor FTZ-F1. Cell Commun. Signal. 2020, 18, 180. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Pavlović-Lažetić, G.M.; Mitić, N.S.; Kovačević, J.J.; Obradović, Z.; Malkov, S.N.; Beljanski, M.V. Bioinformatics Analysis of Disordered Proteins in Prokaryotes. BMC Bioinform. 2011, 12, 66. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Sato, Y.; Okano, K.; Kimura, H.; Honda, K. TEMPURA: Database of Growth TEMPeratures of Usual and RAre Prokaryotes. Microbes Environ. 2020, 35, ME20074. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Mizianty, M.J.; Filipow, P.L.; Uversky, V.N.; Kurgan, L. RAPID: Fast and Accurate Sequence-Based Prediction of Intrinsic Disorder Content on Proteomic Scale. Biochim. Biophys. Acta 2013, 1834, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, L.; Hu, G.; Wang, K.; Ghadermarzi, S.; Zhao, B.; Malhis, N.; Erdős, G.; Gsponer, J.; Uversky, V.N.; Dosztányi, Z. Tutorial: A Guide for the Selection of Fast and Accurate Computational Tools for the Prediction of Intrinsic Disorder in Proteins. Nat. Protoc. 2023, 18, 3157–3172. [Google Scholar] [CrossRef]
- Necci, M.; Piovesan, D.; Tosatto, S.C.E. Critical Assessment of Protein Intrinsic Disorder Prediction. Nat. Methods 2021, 18, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]

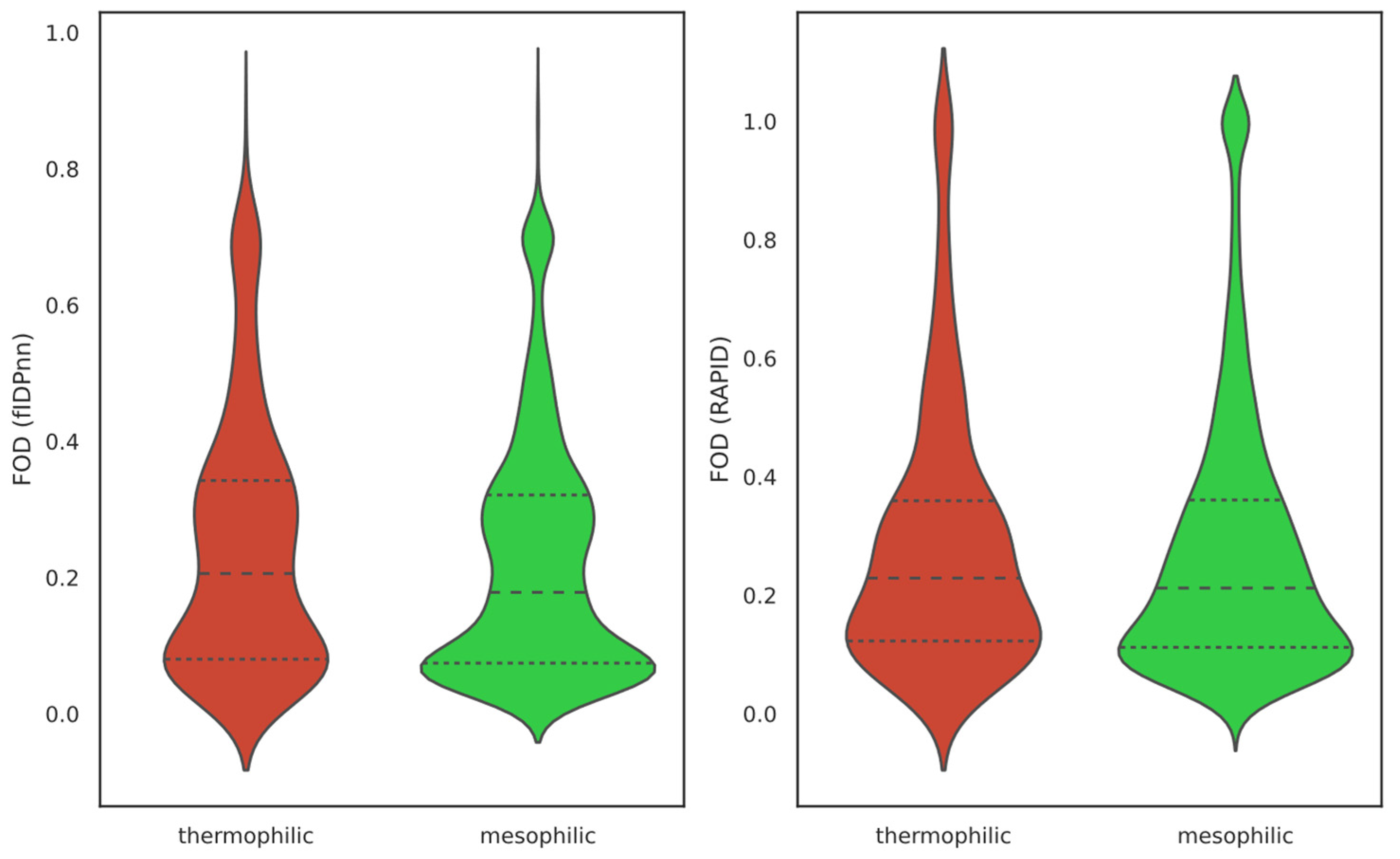

| IDP Class | Thermophilic FOD | Mesophilic FOD |
|---|---|---|
| Weak polyampholytes/polyelectrolytes | 0.303 ± 0.054; n = 21 | 0.192 ± 0.007; n = 609 |
| Boundary proteins | 0.181 ± 0.007; n = 422 | 0.176 ± 0.002; n = 7830 |
| Strong polyampholytes | 0.358 ± 0.014; n = 180 | 0.357 ± 0.004; n = 2145 |
| Negative strong polyelectrolytes | - | - |
| Positive strong polyelectrolytes | 0.682 ± 0.016; n = 13 | 0.706 ± 0.005; n = 256 |
| IDP Function Tag | Thermophilic FOD | Mesophilic FOD |
|---|---|---|
| Activator | - | 0.339 ± 0.010; n = 33 |
| Nuclease | - | 0.110 ± 0.003; n = 18 |
| Chaperone | 0.090 ± 0.010; n = 18 | 0.114 ± 0.003; n = 516 |
| DNA-binding | 0.276 ± 0.032; n = 35 | 0.264 ± 0.008; n = 477 |
| Elongation factor | 0.125 ± 0.034; n = 33 | 0.070 ± 0.005; n = 367 |
| Excision nuclease | 0.053 ± 0.005; n = 7 | 0.054 ± 0.001; n = 192 |
| Hydrolase | 0.064 ± 0.005; n = 14 | 0.108 ± 0.009; n = 209 |
| Initiation factor | 0.217 ± 0.026; n = 3 | 0.209 ± 0.004; n = 56 |
| Isomerase | 0.090 ± 0.024; n = 3 | 0.076 ± 0.001; n = 91 |
| Ligase | 0.060 ± 0.008; n = 12 | 0.058 ± 0.002; n = 336 |
| Lyase | 0.077 ± 0.005; n = 11 | 0.076 ± 0.001; n = 230 |
| Multifunctional enzyme | 0.055 ± 0.000; n = 1 | 0.052 ± 0.001; n = 13 |
| Oxidoreductase | 0.102 ± 0.018; n = 13 | 0.073 ± 0.002; n = 245 |
| Peroxidase | 0.082 ± 0.007; n = 3 | 0.097 ± 0.004; n = 59 |
| Protease | 0.082 ± 0.008; n = 7 | 0.080 ± 0.001; n = 284 |
| RNA-binding | 0.165 ± 0.033; n = 17 | 0.126 ± 0.005; n = 388 |
| Receptor | 0.095 ± 0.000; n = 1 | 0.112 ± 0.016; n = 10 |
| Repressor | 0.331 ± 0.086; n = 4 | 0.263 ± 0.017; n = 51 |
| Ribosomal protein | 0.467 ± 0.018; n = 110 | 0.437 ± 0.004; n = 1859 |
| Rotamase | 0.191 ± 0.030; n = 4 | 0.209 ± 0.010; n = 36 |
| Serine protease | 0.062 ± 0.000; n = 1 | 0.063 ± 0.001; n = 49 |
| Sigma factor | 0.142 ± 0.011; n = 18 | 0.149 ± 0.003; n = 154 |
| Topoisomerase | 0.075 ± 0.007; n = 6 | 0.087 ± 0.001; n = 151 |
| Transferase | 0.087 ± 0.012; n = 27 | 0.073 ± 0.003; n = 713 |
| Translocase | 0.049 ± 0.004; n = 7 | 0.076 ± 0.004; n = 127 |
| rRNA-binding | 0.294 ± 0.010; n = 96 | 0.284 ± 0.002; n = 1700 |
| tRNA-binding | 0.338 ± 0.015; n = 55 | 0.297 ± 0.005; n = 686 |
| Protein Name | FOD (Thermophilic) | FOD (Mesophilic) | Absolute FOD Difference | Cluster Members |
|---|---|---|---|---|
| Rubredoxin | 0.085 | 0.389 | 0.304 | A0A291P6P4, A0A410H536, A0A1B2LXP3… |
| Acyl carrier | 0.534 | 0.392 | 0.142 | A0A291P5S3, A0A410H1W4, A0A386X534… |
| Spore protein | 0.647 | 0.786 | 0.139 | A0A0D8BNT0, A0A0D8BRQ7, M5R4X2… |
| LRSP * bL19 | 0.357 | 0.239 | 0.118 | A0A1U9K6D3, A0A1B9NF78, A0A1B0ZK26… |
| SRSP bS21 | 0.596 | 0.484 | 0.112 | A0A0D5YVA4, A0A1Z4BT12, A0A1L3J4J5… |
| SRSP uS14 | 0.476 | 0.370 | 0.106 | A0A0P0DDQ1, A0A0D5YRD0, A0A0S2I2L9… |
| LRSP bL28 | 0.512 | 0.610 | 0.098 | A0A0D8BU85, M5QWZ7, A0A1D7QW46… |
| Cupin | 0.385 | 0.287 | 0.097 | A0A0K2SHK7, A0A0D5NPB9, A0A4P6K4Z4… |
| LRSP uL24 | 0.323 | 0.414 | 0.090 | A0A0D8BQ30, M5QVZ2, A0A1D7QZW9… |
| SRSP uS14 | 0.501 | 0.417 | 0.083 | A0A291PBX7, A0A7C9NQP7, A0A3T1DHB4… |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruglikov, A.; Xia, X. Mesophiles vs. Thermophiles: Untangling the Hot Mess of Intrinsically Disordered Proteins and Growth Temperature of Bacteria. Int. J. Mol. Sci. 2024, 25, 2000. https://doi.org/10.3390/ijms25042000
Kruglikov A, Xia X. Mesophiles vs. Thermophiles: Untangling the Hot Mess of Intrinsically Disordered Proteins and Growth Temperature of Bacteria. International Journal of Molecular Sciences. 2024; 25(4):2000. https://doi.org/10.3390/ijms25042000
Chicago/Turabian StyleKruglikov, Alibek, and Xuhua Xia. 2024. "Mesophiles vs. Thermophiles: Untangling the Hot Mess of Intrinsically Disordered Proteins and Growth Temperature of Bacteria" International Journal of Molecular Sciences 25, no. 4: 2000. https://doi.org/10.3390/ijms25042000
APA StyleKruglikov, A., & Xia, X. (2024). Mesophiles vs. Thermophiles: Untangling the Hot Mess of Intrinsically Disordered Proteins and Growth Temperature of Bacteria. International Journal of Molecular Sciences, 25(4), 2000. https://doi.org/10.3390/ijms25042000







