Structure, Physicochemical Properties and Biological Activity of Lipopolysaccharide from the Rhizospheric Bacterium Ochrobactrum quorumnocens T1Kr02, Containing d-Fucose Residues
Abstract
1. Introduction
2. Results
2.1. Preparation and Composition of LPS
2.2. Structure of Repeating Units of O-Polysaccharide
A B
Suc(⌋6(~15%)
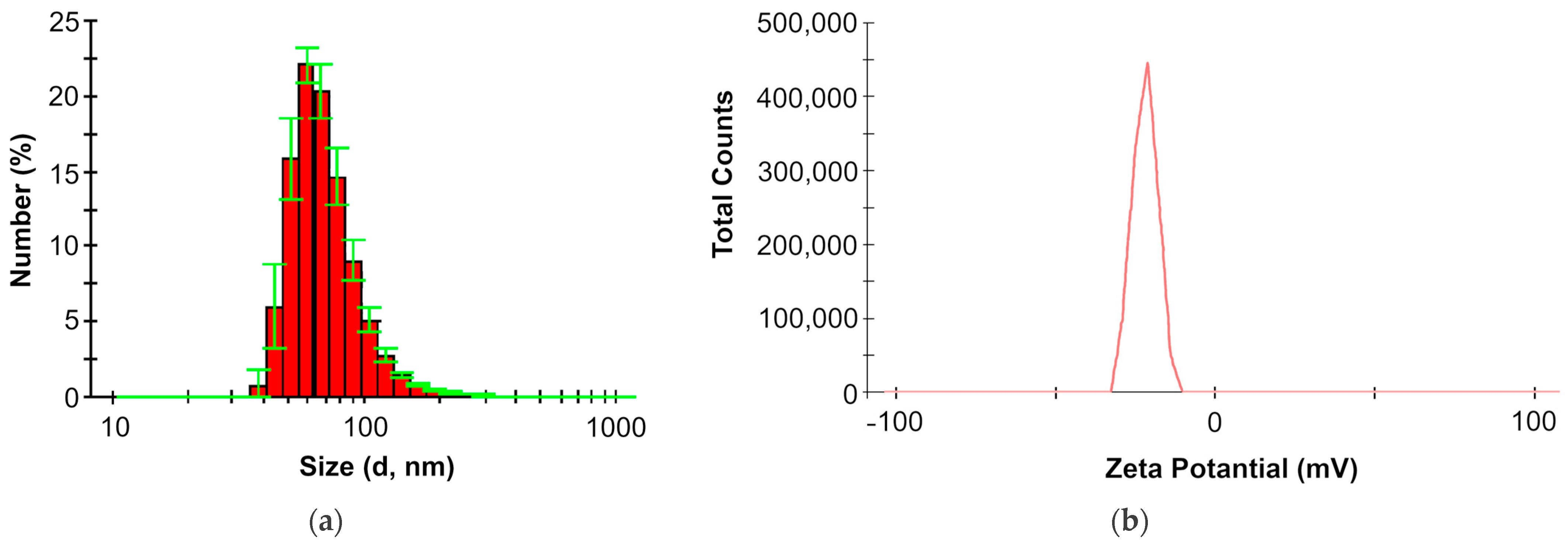
2.3. Physicochemical Properties of LPS in the Aquatic Medium
2.4. Biological Activity of the LPS towards Potato Microplants
3. Discussion
4. Materials and Methods
4.1. Preparation and Characterization of the Qualitative Composition of Lipopolysaccharide and O-Polysaccharide
4.2. Sugar Analysis
4.3. NMR Spectroscopy
4.4. Modeling of Oligosaccharide Structures
4.5. SDS-PAGE Electrophoresis
4.6. Dynamic Light Scatting
4.7. Determination of the Biological Activity of LPS towards Potato Microplants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, Y.; Shen, M.; Song, Q.; Xie, J. Biological activities and pharmaceutical applications of polysaccharide from natural resources: A review. Carbohydr. Polym. 2018, 183, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.S.; Naveed, M.; Jost, N. Polysaccharides; classification, chemical properties, and future perspective applications in fields of pharmacology and biological medicine (a review of current applications and upcoming potentialities). J. Polym. Environ. 2021, 29, 2359–2371. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, V.F.; Silva, G.D.; Taborda, M.; Levin, A.S.; Magri, M.M. Systematic review and meta-analysis of galactomannan antigen testing in serum and bronchoalveolar lavage for the diagnosis of chronic pulmonary aspergillosis: Defining a cutoff. Eur. J. Clin. Microbiol. Infect. Dis. 2023, 42, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tan, J.; Nima, L.; Sang, Y.; Cai, X.; Xue, H. Polysaccharides from fungi: A review on their extraction, purification, structural features, and biological activities. Food Chem. X 2022, 15, 100414. [Google Scholar] [CrossRef] [PubMed]
- Valasques Junior, G.L.; Cedro, P.É.; Mendes, T.P.; dos Anjos Miranda, A.C.; Côrtes Filho, A.B.; Lima, D.M.; Barreto, M.M.; Pinheiro, A.A.; Marques, L.M. Characterization and biological activities of polysaccharides extracted from the filamentous fungal cell wall: An updated literature review. Res. Soc. Dev. 2020, 9, e62191110217. [Google Scholar] [CrossRef]
- Liu, J.; Willför, S.; Xu, C. A review of bioactive plant polysaccharides: Biological activities, functionalization, and biomedical applications. Bioact. Carbohydr. Diet. Fibre. 2015, 5, 31–61. [Google Scholar] [CrossRef]
- Minzanova, S.T.; Mironov, V.F.; Arkhipova, D.M.; Khabibullina, A.V.; Mironova, L.G.; Zakirova, Y.M.; Milyukov, V.A. Biological activity and pharmacological application of pectic polysaccharides: A review. Polymers 2018, 10, 1407. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, X.; Chen, T.; Chen, X. A review of the antibacterial activity and mechanisms of plant polysaccharides. Trends Food Sci. Technol. 2022, 123, 264–280. [Google Scholar] [CrossRef]
- Pradhan, B.; Ki, J.S. Biological activity of algal derived carrageenan: A comprehensive review in light of human health and disease. Int. J. Biol. Macromol. 2023, 238, 124085. [Google Scholar] [CrossRef]
- Wang, Y.; Xing, M.; Cao, Q.; Ji, A.; Liang, H.; Song, S. Biological activities of fucoidan and the factors mediating its therapeutic effects: A review of recent studies. Mar. Drugs. 2019, 17, 183. [Google Scholar] [CrossRef]
- Barlow, G.H.; Sanderson, N.D.; McNeill, P.D. Macromolecular properties and biological activity of heparin. Arch. Biochem. Biophys. 1961, 94, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Song, Z.; Hu, W.; Liang, J.; Jing, Y.; He, L.; Huang, S.; Wang, X.; Hou, S.; Xu, T.; et al. Methods of extraction, separation, purification, structural characterization for polysaccharides from aquatic animals and their major pharmacological activities. Crit. Rev. Food Sci. Nutr. 2020, 60, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Dicker, K.T.; Gurski, L.A.; Pradhan-Bhatt, S.; Witt, R.L.; Farach-Carson, M.C.; Jia, X. Hyaluronan: A simple polysaccharide with diverse biological functions. Acta Biomat. 2014, 10, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Sperandeo, P.; Martorana, A.M.; Polissi, A. Lipopolysaccharide biogenesis and transport at the outer membrane of gram-negative bacteria. Biochim. Biophys. Acta Mol. Cell Bio. Lipids. 2017, 1862, 1451–1460. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Sherman, D.J.; Silhavy, T.J.; Ruiz, N.; Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 2016, 14, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2431–2447. [Google Scholar] [CrossRef] [PubMed]
- Mazgaeen, L.; Gurung, P. Recent advances in lipopolysaccharide recognition systems. Int. J. Mol. Sci. 2020, 21, 379. [Google Scholar] [CrossRef] [PubMed]
- Samuel, G.; Reeves, P. Biosynthesis of O-antigens: Genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 2003, 338, 2503–2519. [Google Scholar] [CrossRef]
- Whitfield, C.; Williams, D.M.; Kelly, S.D. Lipopolysaccharide O-antigens—Bacterial glycans made to measure. J. Biol. Chem. 2020, 295, 10593–10609. [Google Scholar] [CrossRef]
- Klein, G.; Lindner, B.; Brade, H.; Raina, S. Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: Envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-d-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 2011, 286(50), 42787–42807. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated control of the assembly and diversity of LPS by noncoding sRNAs. BioMed Res. Int. 2015, 153561. [Google Scholar] [CrossRef]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Kutschera, A.; Ranf, S. The multifaceted functions of lipopolysaccharide in plant-bacteria interactions. Biochimie 2019, 59, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Basauri, A.; González-Fernández, C.; Fallanza, M.; Bringas, E.; Fernandez-Lopez, R.; Giner, L.; Moncalián, G.; de la Cruz, F.; Ortiz, I. Biochemical interactions between LPS and LPS-binding molecules. Crit. Rev. Biotechnol. 2020, 40, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Kagan, J.C. A cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol. Cell 2014, 54, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Sigida, E.N.; Zdorovenko, E.L.; Shashkov, A.S.; Dmitrenok, A.S.; Fedonenko, Y.P. Structure of the O-specific polysaccharide of Ochrobactrum endophyticum KCTC 42485T containing 3-(3-hydroxy-2, 3-dimethyl-5-oxoprolyl) amino-3, 6-dideoxy-d-galactose. Carbohydr. Res. 2023, 527, 108810. [Google Scholar] [CrossRef] [PubMed]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1,000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Krzyżanowska, D.M.; Maciąg, T.; Ossowicki, A.; Rajewska, M.; Kaczyński, Z.; Czerwicka, M.; Rąbalski, Ł.; Czaplewska, P.; Jafra, S. Ochrobactrum quorumnocens sp. nov., a quorum quenching bacterium from the potato rhizosphere, and comparative genome analysis with related type strains. PLoS ONE 2019, 14, 0210874. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, Z.; Yang, L.; Hu, M.Y.; Jiang, G.Y.; Chen, J.; Yang, Y.C.; Tian, Y. Ochrobactrum chromiisoli sp. nov., isolated from chromium-contaminated soil. Curr. Microbiol. 2024, 81, 50. [Google Scholar] [CrossRef] [PubMed]
- Riedel, R.; Commichau, F.M.; Benndorf, D.; Hertel, R.; Holzer, K.; Hoelzle, L.E.; Mardoukhi, M.S.; Noack, L.E.; Martienssen, M. Biodegradation of selected aminophosphonates by the bacterial isolate Ochrobactrum sp. BTU1. Microbiol. Res. 2024, 8, 127600. [Google Scholar] [CrossRef] [PubMed]
- Sumayo, M.; Hahm, M.S.; Ghim, S.Y. Determinants of plant growth-promoting Ochrobactrum lupini KUDC1013 involved in induction of systemic resistance against Pectobacterium carotovorum subsp. carotovorum in tobacco leaves. Plant. Pathol. J. 2013, 29, 174. [Google Scholar] [CrossRef]
- Sigida, E.N.; Kargapolova, K.Y.; Shashkov, A.S.; Zdorovenko, E.L.; Ponomaryova, T.S.; Meshcheryakova, A.A.; Tkachenko, O.V.; Burygin, G.L.; Knirel, Y.A. Structure, gene cluster of the O antigen and biological activity of the lipopolysaccharide from the rhizospheric bacterium Ochrobactrum cytisi IPA7.2. Int. J. Biol. Macromol. 2020, 154, 1375–1381. [Google Scholar] [CrossRef]
- Pac, M.; Komaniecka, I.; Zamlynska, K.; Turska-Szewczuk, A.; Choma, A. Structure of the O-specific polysaccharide from the legume endosymbiotic bacterium Ochrobactrum cytisi strain ESC1T. Carbohydr. Res. 2015, 413, 37–40. [Google Scholar] [CrossRef]
- Szpakowska, N.; Kowalczyk, A.; Jafra, S.; Kaczyński, Z. The chemical structure of polysaccharides isolated from the Ochrobactrum rhizosphaerae PR17T. Carbohydr. Res. 2020, 497, 108136. [Google Scholar] [CrossRef]
- Lipkind, G.M.; Shashkov, A.S.; Knirel, Y.A.; Vinogradov, E.V.; Kochetkov, N.K. A computer-assisted structural analysis of regular polysaccharides on the basis of 13C-n.m.r. data. Carbohydr. Res. 1988, 75, 59–75. [Google Scholar] [CrossRef]
- Bock, K.; Pedersen, C. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 1983, 41, 27–66. [Google Scholar] [CrossRef]
- Altona, C.; Haasnoot, C.A.G. Prediction of anti and gauche vicinal proton-proton coupling constants in carbohydrates: A simple additivity rule for pyranose rings. Org. Magn. Reson. 1980, 13, 417–429. [Google Scholar] [CrossRef]
- Cyr, N.; Perlin, A.S. The conformations of furanosides. A 13C nuclear magnetic resonance study. Can. J. Chem. 1979, 57, 2504–2511. [Google Scholar] [CrossRef]
- Angyal, S.J. Hudson’s rules of isorotation as applied to furanosides, and the conformations of methyl aldofuranosides. Carbohydr. Res. 1979, 77, 37–50. [Google Scholar] [CrossRef]
- Kochetkov, N.K.; Chizhov, O.S.; Shashkov, A.S. Dependence of 13C chemical shifts on the spatial interaction of protons, and its application in structural and conformational studies of oligo- and poly-saccharides. Carbohydr. Res. 1984, 133, 173–185. [Google Scholar] [CrossRef]
- Shashkov, A.S.; Lipkind, G.; Knirel, Y.A.; Kochetkov, N.K. Stereochemical factors determining the effects of glycosylation the 13C chemical shifts in carbohydrates. Magn. Reson. Chem. 1988, 26, 735–747. [Google Scholar] [CrossRef]
- Kawaharada, Y.; Kiyota, H.; Eda, S.; Minamisawa, K.; Mitsui, H. Structural characterization of neutral and anionic glucans from Mesorhizobium loti. Carbohydr. Res. 2008, 343, 2422–2427. [Google Scholar] [CrossRef]
- Velasco, J.; Bengoechea, J.A.; Brandenburg, K.; Lindner, B.; Seydel, U.; González, D.; Zahringer, U.; Moreno, E.; Moriyón, I. Brucella abortus and its closest phylogenetic relative, Ochrobactrum spp., differ in outer membrane permeability and cationic peptide resistance. Infect. Immun. 2000, 68, 3210–3218. [Google Scholar] [CrossRef]
- Leclercq, S.O.; Cloeckaert, A.; Zygmunt, M.S. Taxonomic organization of the family Brucellaceae based on a phylogenomic approach. Front. Microbiol. 2020, 10, 3083. [Google Scholar] [CrossRef]
- Velasco, J.; Moll, H.; Vinogradov, E.V.; Moriyón, I.; Zähringer, U. Determination of the O-specific polysaccharide structure in the lipopolysaccharide of Ochrobactrum anthropi LMG 3331. Carbohydr. Res. 1996, 287, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Zdorovenko, E.L.; Kadykova, A.A.; Shashkov, A.S.; Varbanets, L.D.; Bulyhina, T.V. Pantoea agglomerans P1a lipopolysaccharide: Structure of the O-specific polysaccharide and lipid A and biological activity. Carbohydr. Res. 2019, 484, 107767. [Google Scholar] [CrossRef] [PubMed]
- Zdorovenko, E.L.; Ovod, V.V.; Zatonsky, G.V.; Shashkov, A.S.; Kocharova, N.A.; Knirel, Y.A. Location of the O-methyl groups in the O polysaccharide of Pseudomonas syringae pv. phaseolicola. Carbohydr. Res. 2001, 330, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Nakazawa, F.; Ito, T.; Hoshino, T.; Hoshino, E. The structure of the antigenic polysaccharide produced by Eubactrium saburreum T15. Carbohydr. Res. 2003, 338, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ding, P.; Perepelov, A.V.; Xu, Y.; Wang, Y.; Knirel, Y.A.; Wang, L.; Feng, L. Characterization of the dTDP-d-fucofuranose biosynthetic pathway in Escherichia coli O52. Mol. Microbiol. 2008, 70, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Helander, I.M.; Haikara, A.; Sadovskaya, I.; Vinogradov, E.; Salkinoja-Salonen, M.S. Lipopolysaccharides of anaerobic beer spoilage bacteria of the genus Pectinatus—Lipopolysaccharides of a Gram-positive genus. FEMS Microbiol. Rev. 2004, 28, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Cullimore, J.; Gough, C. Lipochitooligosaccharide perception and the basis of partner recognition in root endosymbioses. In Molecular Microbial Ecology of the Rhizosphere; de Bruijn, F.J., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; pp. 483–494. [Google Scholar] [CrossRef]
- Ovtsyna, A.O.; Lugtenberg, B.J.; Tikhonovich, I.A.; Spaink, H.P. The role of Nod factor modifications for nodulation on Afghan peas. In Biological Nitrogen Fixation for the 21st Century; Elmerich, C., Kondorosi, A., Newton, W.E., Eds.; Kluwer: Dordrecht, The Netherlands, 1998; p. 251. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.T.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Tkachenko, O.V.; Burygin, G.L.; Evseeva, N.V.; Fedonenko, Y.P.; Matora, L.Y.; Lobachev, Y.V.; Shchyogolev, S.Y. Morphogenesis of wheat calluses by the treatment with lipopolysaccharides from Azospirillum spp. Plant Cell Tissue Organ. Cult. 2021, 147, 147–155. [Google Scholar] [CrossRef]
- Dow, M.; Newman, M.A.; Von Roepenack, E. The induction and modulation of plant defense responses by bacterial lipopolysaccharides. Annu. Rev. Phytopathol. 2000, 38, 241–261. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, J.N.; Blanco-Ulate, B.; Muszyński, A.; Figueroa-Balderas, R.; Morales-Cruz, A.; Azadi, P.; Dobruchowska, J.M.; Castro, C.; Cantu, D.; Roper, M.C. Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 2018, 9, 390. [Google Scholar] [CrossRef] [PubMed]
- del Carmen Orozco-Mosqueda, M.; Fadiji, A.E.; Babalola, O.O.; Santoyo, G. Bacterial elicitors of the plant immune system: An overview and the way forward. Plant Stress 2023, 4, 100138. [Google Scholar] [CrossRef]
- Hernaández-Esquivel, A.A.; Castro-Mercado, E.; Valencia-Cantero, E.; Alexandre, G.; García-Pineda, E. Application of Azospirillum brasilense lipopolysaccharides to promote early wheat plant growth and analysis of related biochemical responses. Front. Sustain. Food S 2020, 4, 579976. [Google Scholar] [CrossRef]
- Evseeva, N.V.; Matora, L.Y.; Burygin, G.L.; Dmitrienko, V.V.; Shchyogolev, S.Y. Effect of Azospirillum brasilense Sp245 lipopolysaccharide on the functional activity of wheat root meristematic cells. Plant Soil 2011, 346, 181–188. [Google Scholar] [CrossRef]
- Chávez-Herrera, E.; Hernández-Esquivel, A.A.; Castro-Mercado, E.; García-Pineda, E. Effect of Azospirillum brasilense Sp245 lipopolysaccharides on wheat plant development. J. Plant Growth Regul. 2018, 37, 859–866. [Google Scholar] [CrossRef]
- Ranf, S.; Gisch, N.; Schäffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Sánchez-Carballo, P.M.; Zähringer, U.; Hückelhoven, R.; Lee, J.; et al. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433. [Google Scholar] [CrossRef]
- Desaki, Y.; Kouzai, Y.; Ninomiya, Y.; Iwase, R.; Shimizu, Y.; Seko, K.; Molinaro, A.; Minami, E.; Shibuya, N.; Kaku, H.; et al. OsCERK1 plays a crucial role in the lipopolysaccharide-induced immune response of rice. New Phytol. 2018, 217, 1042–1049. [Google Scholar] [CrossRef]
- Hernández-Esquivel, A.A.; Castro-Mercado, E.; García-Pineda, E. Comparative effects of Azospirillum brasilense Sp245 and Pseudomonas aeruginosa PAO1 lipopolysaccharides on wheat seedling growth and peroxidase activity. J. Plant Growth Regul. 2021, 40, 1903–1911. [Google Scholar] [CrossRef]
- Duvick, J.P.; Sequeira, L. Interaction of Pseudomonas solanacearum lipopolysaccharide and extracellular polysaccharide with agglutinin from potato tubers. App. Environ. Microbiol. 1984, 48, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Esposito, N.; Ovchinnikova, O.G.; Barone, A.; Zoina, A.; Holst, O.; Evidente, A. Host and non-host plant response to bacterial wilt in potato: Role of the lipopolysaccharide isolated from Ralstonia solanacearum and molecular analysis of plant–pathogen interaction. Chem. Biodivers. 2008, 5, 2662–2675. [Google Scholar] [CrossRef] [PubMed]
- Paul, F.; Morin, A.; Monsan, P. Microbial polysaccharides with actual potential industrial applications. Biotechnol. Adv. 1986, 4, 245–259. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Niemann, E.G.; Fendrik, I. Close association of Azospirillum and diazotrophic rods with different root zones of Kallar grass. Appl. Environ. Microbiol. 1986, 52, 520–526. [Google Scholar] [CrossRef]
- Westphal, O. Bacterial lipopolysaccharides: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 5, 83. [Google Scholar]
- Arbatsky, N.P.; Wang, M.; Shashkov, A.S.; Feng, L.; Knirel, Y.A.; Wang, L. Structure of the O-polysaccharide of Cronobacter sakazakii O1 containing 3-(N-acetyl-l-alanyl)amino-3,6-dideoxy-d-glucose. Carbohydr. Res. 2010, 345, 2095–2098. [Google Scholar] [CrossRef]
- Karkhanis, Y.D.; Zeltner, J.Y.; Jackson, J.J.; Carlo, D.J. A new and improved microassay to determine 2-keto-3-deoxyoctonate in lipopolysaccharide of Gram-negative bacteria. Anal. Biochem. 1978, 85, 595–601. [Google Scholar] [CrossRef]
- Murphy, J.A.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta. 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Zdorovenko, E.L.; Kadykova, A.A.; Shashkov, A.S.; Varbanets, L.D.; Bulyhina, T.V.; Knirel, Y.A. Lipopolysaccharide of Pantoea agglomerans 7969: Chemical identification, function and biological activity. Carbohydr. Polym. 2017, 165, 351–358. [Google Scholar] [CrossRef]
- Sawardeker, J.S.; Sloneker, J.H.; Jeanes, A. Quantitative determination of monosaccharides as their alditol acetates by gas liquid chromatography. Anal. Chem. 1965, 37, 1602–1603. [Google Scholar] [CrossRef]
- Leontein, K.; Lindberg, B.; Lönngren, J. Assignment of absolute configuration of sugars by g.l.c. of their acetylated glycosides formed from chiral alcohols. Carbohydr. Res. 1978, 62, 359–362. [Google Scholar] [CrossRef]
- Damerell, D.; Ceroni, A.; Maass, K.; Ranzinger, R.; Dell, A.; Haslam, S.M. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: Updates and new developments. Biol. Chem. 2012, 393, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, P.J.; Brown, T.M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 1983, 154, 269–277. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]


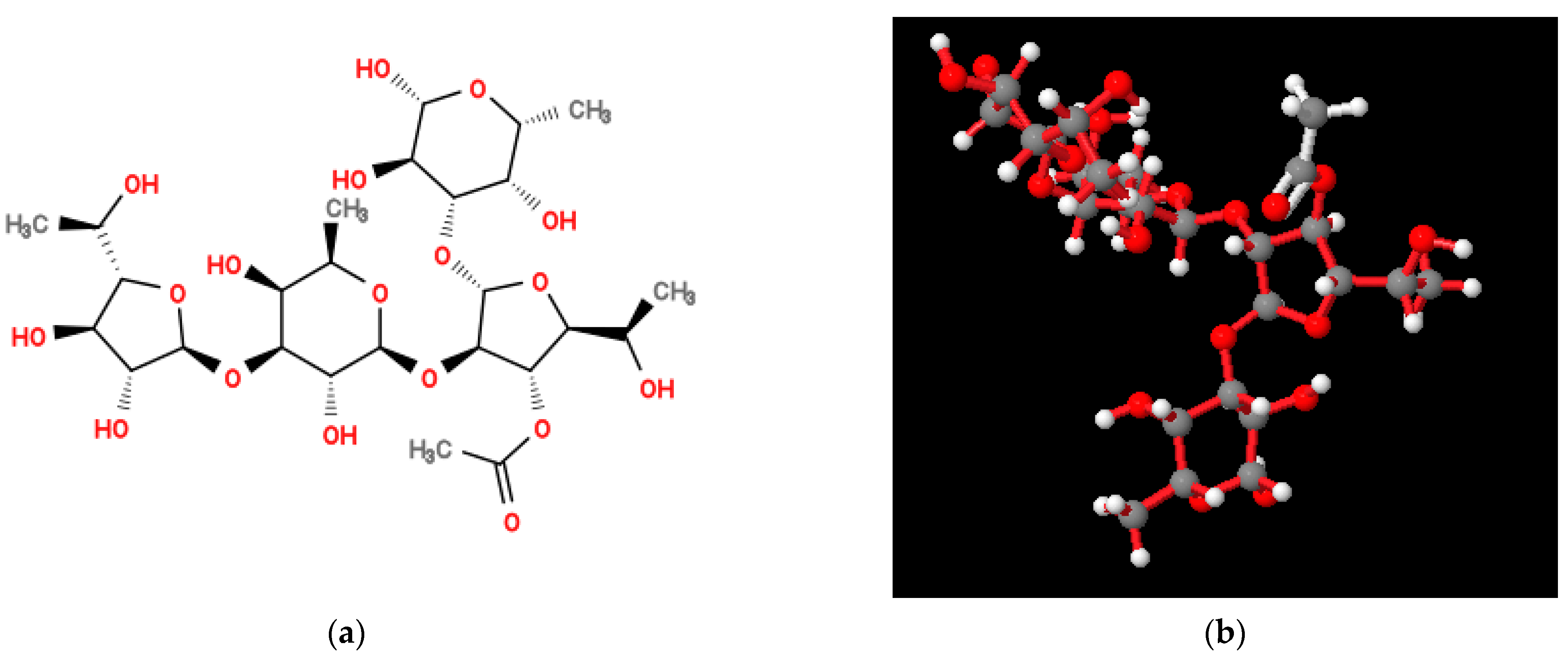
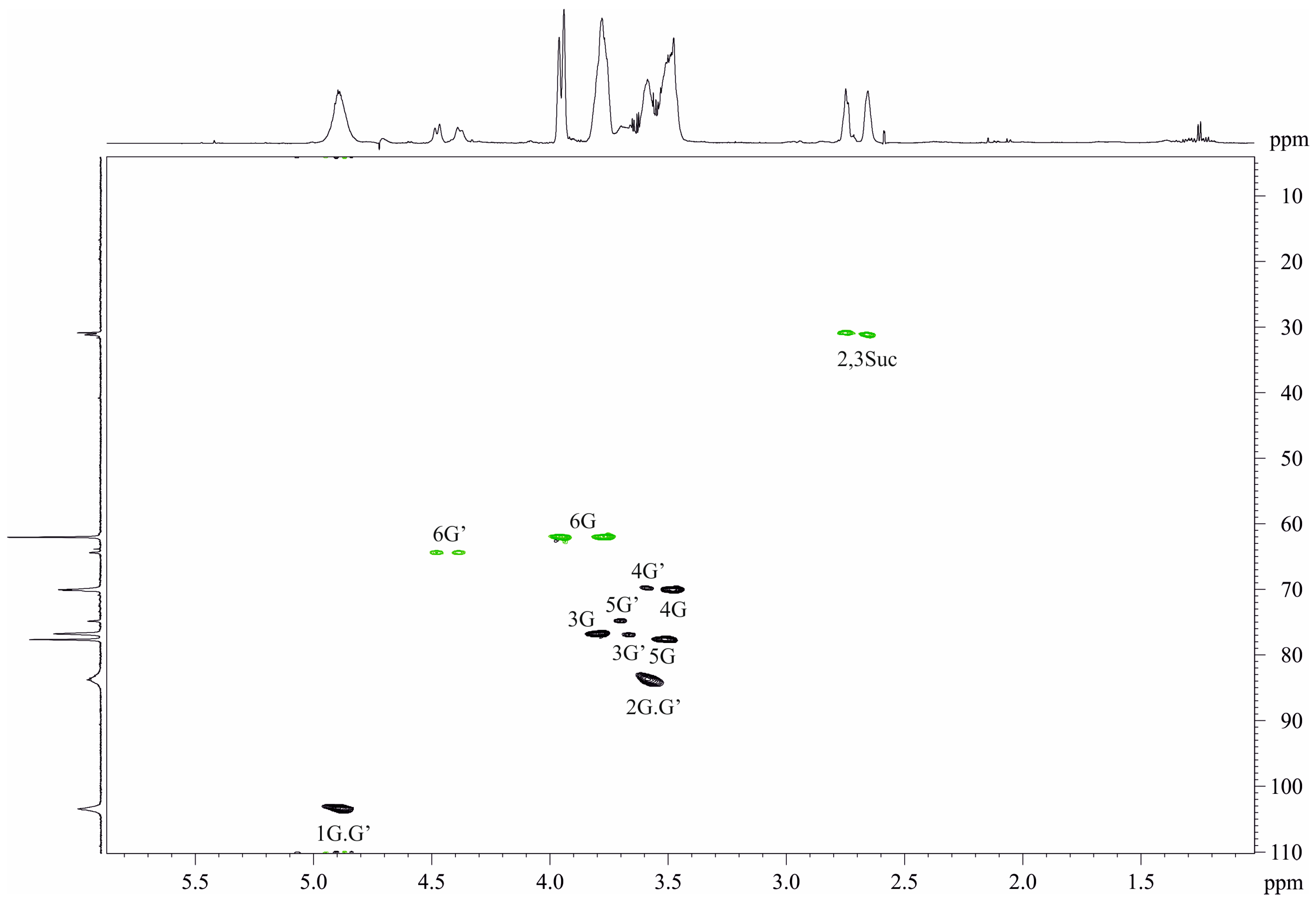



| Residue | C-1 H-1 | C-2 H-2 | C-3 H-3 | C-4 H-4 | C-5 H-5 | C-6 H-6 |
|---|---|---|---|---|---|---|
| DPS | ||||||
| →2)-β-d-Fucf-(1→ (A) | 109.0 5.43 | 90.5 4.34 | 77.9 4.10 | 88.4 3.88 | 68.6 3.94 | 19.5 1.26 |
| →3)-β-d-Fucp-(1→ (B) | 103.3 4.60 | 70.8 3.60 | 81.4 3.74 | 72.5 3.92 | 72.1 3.83 | 16.7 1.26 |
| PS-1 | ||||||
| →2)-β-d-Fucf-(1→ (A) | 108.8 5.43 | 90.5 4.35 | 77.7 4.11 | 88.3 3.88 | 68.6 3.89 | 19.5 1.26 |
| →3)-β-d-Fucp-(1→ (B′) | 102.9 4.67 | 70.9 3.60 | 81.0 3.77 | 72.3 3.90 | 72.2 3.80 | 16.6 1.26 |
| →2)-β-d-Fucf-(1→ (A′) 3) | OAc | 109.0 5.49 174.6 | 87.0 4.46 21.6 2.16 | 79.3 5.04 | 87.3 4.14 | 68.3 4.02 | 19.5 1.23 |
| →3)-β-d-Fucp-(1→ (B) | 103.4 4.60 | 70.9 3.62 | 81.4 3.74 | 72.3 3.90 | 72.2 3.83 | 16.6 1.27 |
| PS-2 | ||||||
| →2)-α-d-Glcp-(1→ (G) | 103.5 4.90 | 83.8 3.59 | 76.8 3.79 | 70.1 3.48 | 77.8 3.51 | 62.1 3.95, 3.77 |
| →2)-α-d-Glcp-(1→ (G′) 6) | 1)-Suc | 103.5 4.90 176.2 | 83.8 3.59 30.9 * 2.75 | 76.9 3.67 31.2 * 2.66 | 69.9 3.59 179.4 | 74.8 3.70 | 64.5 4.48, 4.39 |
| Anomeric Atom in Sugar Residue (δ) | Correlation(s) to Atoms in Sugar Residue(s) (δ) | |
|---|---|---|
| ROESY | HMBC | |
| DPS | ||
| A H-1 (5.43) | A H-2 (4.34), B H-3 (3.74), B H-2 (3.60) | A C-3 (77.6), B C-3 (814), A C-4 (88.4), A C-2 (90.5) |
| A C-1 (109.0) | A H-2 (4.34), A H-3 (4.10), B H-3 (3.74) | |
| B H-1 (4.60) | A H-2 (4.34), A H-3 (4.10), B H-5 (3.83), B H-3 (3.74), B H-2 (3.60) | B C-2 (90.5) |
| B C-1 (103.3) | A H-2 (4.34), B H-5 (3.83), B H-2 (3.60) | |
| PS-1 | ||
| A H-1 (5.43) | A H-2 (4.34), B′ H-4 (3.90), B′ H-3 (3.77), B′ H-2 (3.60) | A C-3 (77.7), B′ C-3 (81.4), A C-4 (88.3) |
| A C-1 (108.8) | A H-2 (4.34), B′ H-3 (3.77) | |
| B′ H-1 (4.67) | A′ H-2 (4.46), B′ H-3 (3.77), B′ H-5 (3.80), B′ H-2 (3.60) | A′ C-2 (87.0) |
| B′ C-1 (102.9) | A′ H-2 (4.45), B′ H-5 (3.80), B’ H-2 (3.60) | |
| A′ H-1 (5.49) | A′ H-2 (4.46), B H-4 (3.90), B H-3 (3,74), B H-2 (3.60) | A′ C-3 (79.3), A′ C-4 (87.3), B C-3 (81.4) |
| A′ C-1 (109.0) | A′ H-2 (4.46), B H-3 (3.77) | |
| B H-1 (4.60) | A H-2 (4.34), A H-3 (4.11), B H-5 (3.83), B H-3 (3.74) | A C-2 (90.5) |
| B C-1 (103.4) | A H-2 (4.35), B H-5 (3.83), B H-2 (3.62) | |
| Variable | Control | Experiment (+LPS) |
|---|---|---|
| Shoot length, mm | 59.0 ± 3.9 * | 79.8 ± 6.8 ** |
| Node number, pcs | 7.85 ± 0.39 | 6.90 ± 0.55 |
| Root number, pcs | 10.0 ± 0.66 | 10.4 ± 0.53 |
| Sum of root length, cm | 240.6 ± 33.1 | 304.7 ± 33.9 |
| Wet weight of shoots, mg | 304.7 ± 23.4 | 334.3 ± 8.1 |
| Dry weight of shoots, mg | 31.5 ± 2.1 | 55.3 ± 11.4 ** |
| Wet weight of roots, mg | 89.2 ± 15.3 | 168.7 ± 23.6 *** |
| Dry weight of roots, mg | 9.76 ± 0.92 | 15.33 ± 2.33 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivoruchko, A.A.; Zdorovenko, E.L.; Ivanova, M.F.; Kostina, E.E.; Fedonenko, Y.P.; Shashkov, A.S.; Dmitrenok, A.S.; Ul’chenko, E.A.; Tkachenko, O.V.; Astankova, A.S.; et al. Structure, Physicochemical Properties and Biological Activity of Lipopolysaccharide from the Rhizospheric Bacterium Ochrobactrum quorumnocens T1Kr02, Containing d-Fucose Residues. Int. J. Mol. Sci. 2024, 25, 1970. https://doi.org/10.3390/ijms25041970
Krivoruchko AA, Zdorovenko EL, Ivanova MF, Kostina EE, Fedonenko YP, Shashkov AS, Dmitrenok AS, Ul’chenko EA, Tkachenko OV, Astankova AS, et al. Structure, Physicochemical Properties and Biological Activity of Lipopolysaccharide from the Rhizospheric Bacterium Ochrobactrum quorumnocens T1Kr02, Containing d-Fucose Residues. International Journal of Molecular Sciences. 2024; 25(4):1970. https://doi.org/10.3390/ijms25041970
Chicago/Turabian StyleKrivoruchko, Aleksandra A., Evelina L. Zdorovenko, Maria F. Ivanova, Ekaterina E. Kostina, Yulia P. Fedonenko, Alexander S. Shashkov, Andrey S. Dmitrenok, Elizaveta A. Ul’chenko, Oksana V. Tkachenko, Anastasia S. Astankova, and et al. 2024. "Structure, Physicochemical Properties and Biological Activity of Lipopolysaccharide from the Rhizospheric Bacterium Ochrobactrum quorumnocens T1Kr02, Containing d-Fucose Residues" International Journal of Molecular Sciences 25, no. 4: 1970. https://doi.org/10.3390/ijms25041970
APA StyleKrivoruchko, A. A., Zdorovenko, E. L., Ivanova, M. F., Kostina, E. E., Fedonenko, Y. P., Shashkov, A. S., Dmitrenok, A. S., Ul’chenko, E. A., Tkachenko, O. V., Astankova, A. S., & Burygin, G. L. (2024). Structure, Physicochemical Properties and Biological Activity of Lipopolysaccharide from the Rhizospheric Bacterium Ochrobactrum quorumnocens T1Kr02, Containing d-Fucose Residues. International Journal of Molecular Sciences, 25(4), 1970. https://doi.org/10.3390/ijms25041970








