Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy
Abstract
1. Introduction
2. Results
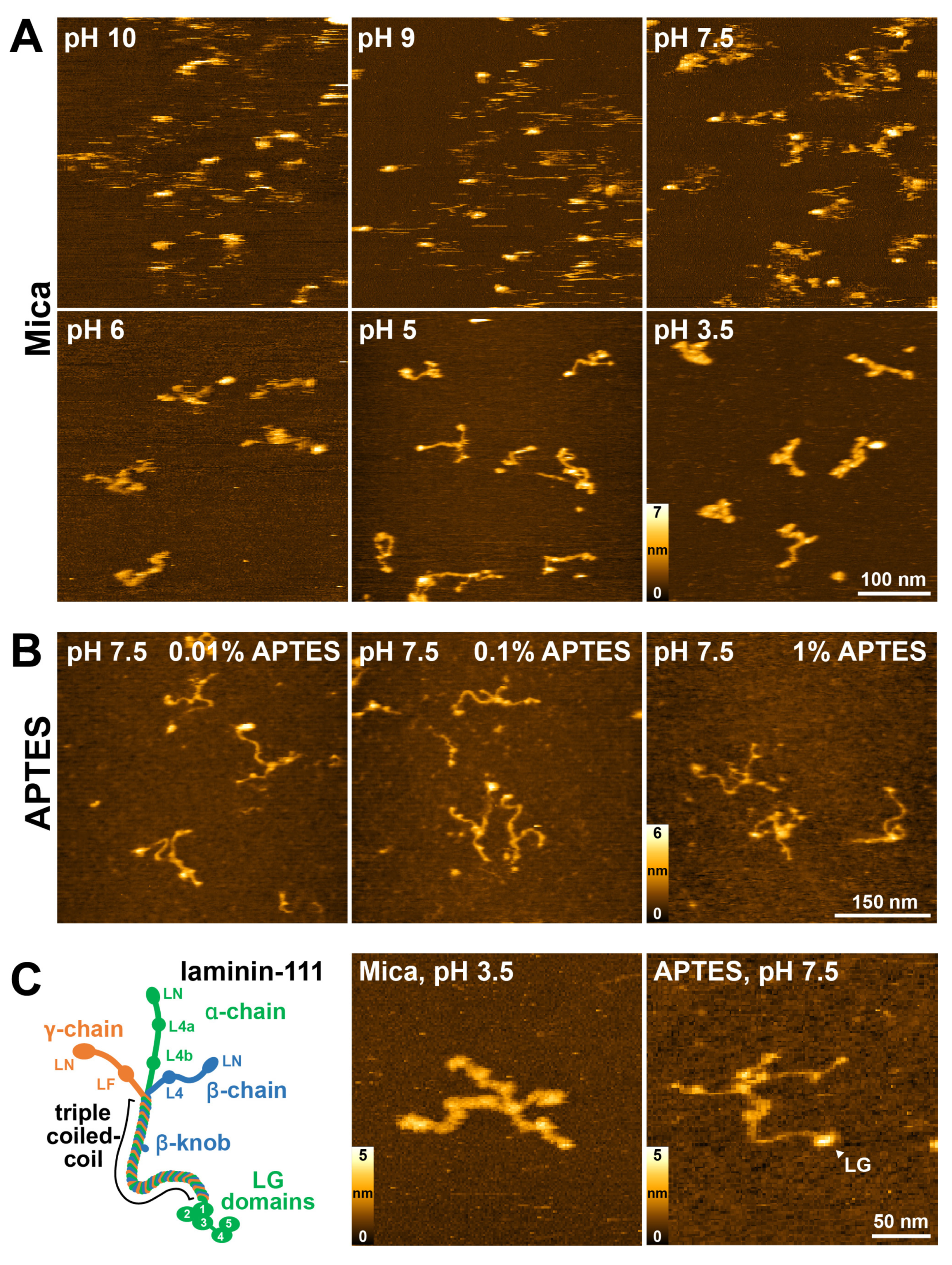
2.1. Optimization Surface Immobilization of Laminin-111 for HS-AFM Imaging
2.2. Investigating the Coiled-Coil Domain Structure of Laminin-111
2.3. HS-AFM Imaging of Laminin-3A32 and Laminin-3B32
2.4. Identifying the Hinge Region in the Laminin-332 Coiled-Coil Domain
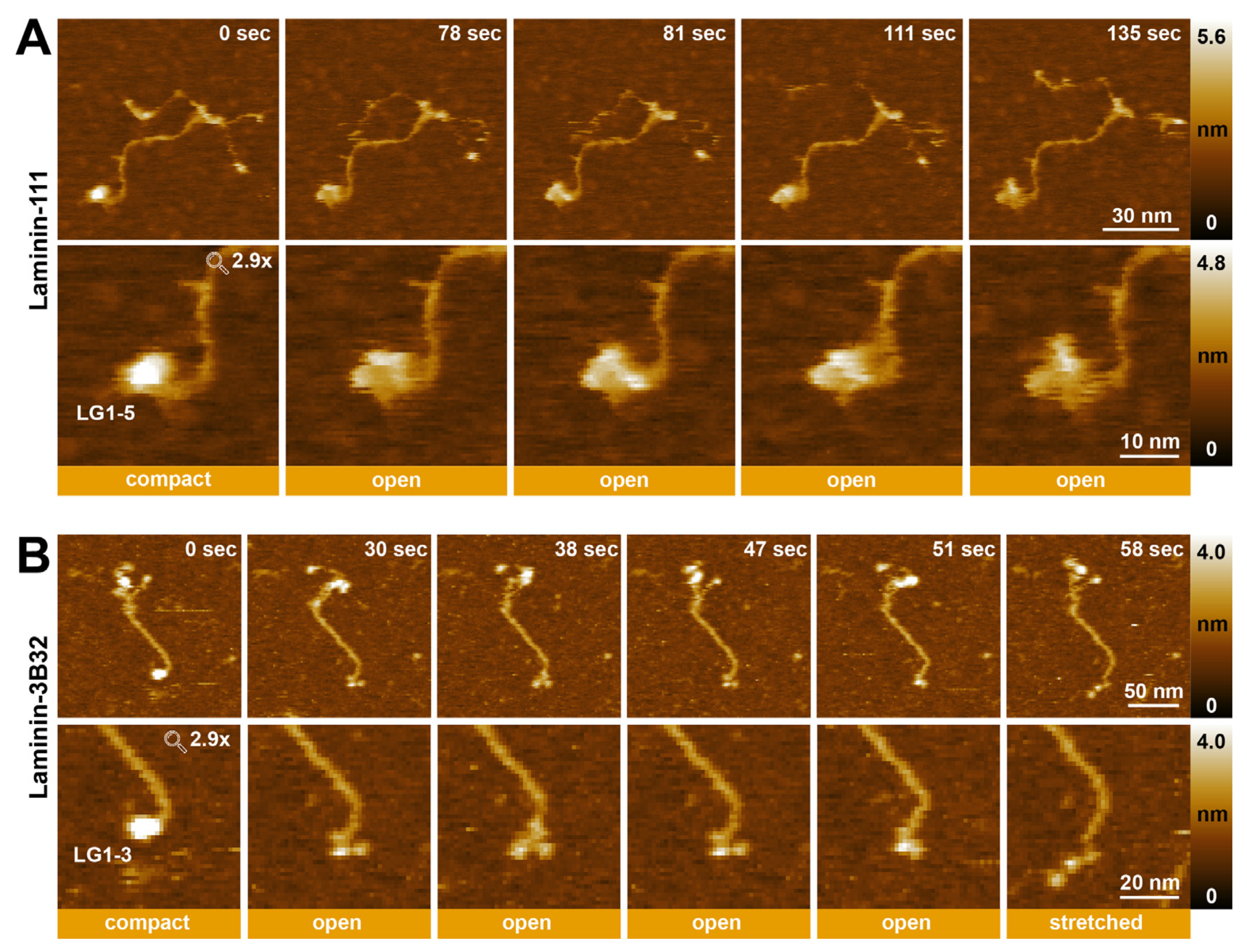
2.5. Visualizing the Dynamic Rearrangement of Laminin LG Domains Using HS-AFM
3. Discussion
4. Materials and Methods
4.1. Sample Preparation
4.2. Cantilever Preparation
4.3. HS-AFM Observation
4.4. HS-AFM Image Processing
4.5. Data Analysis
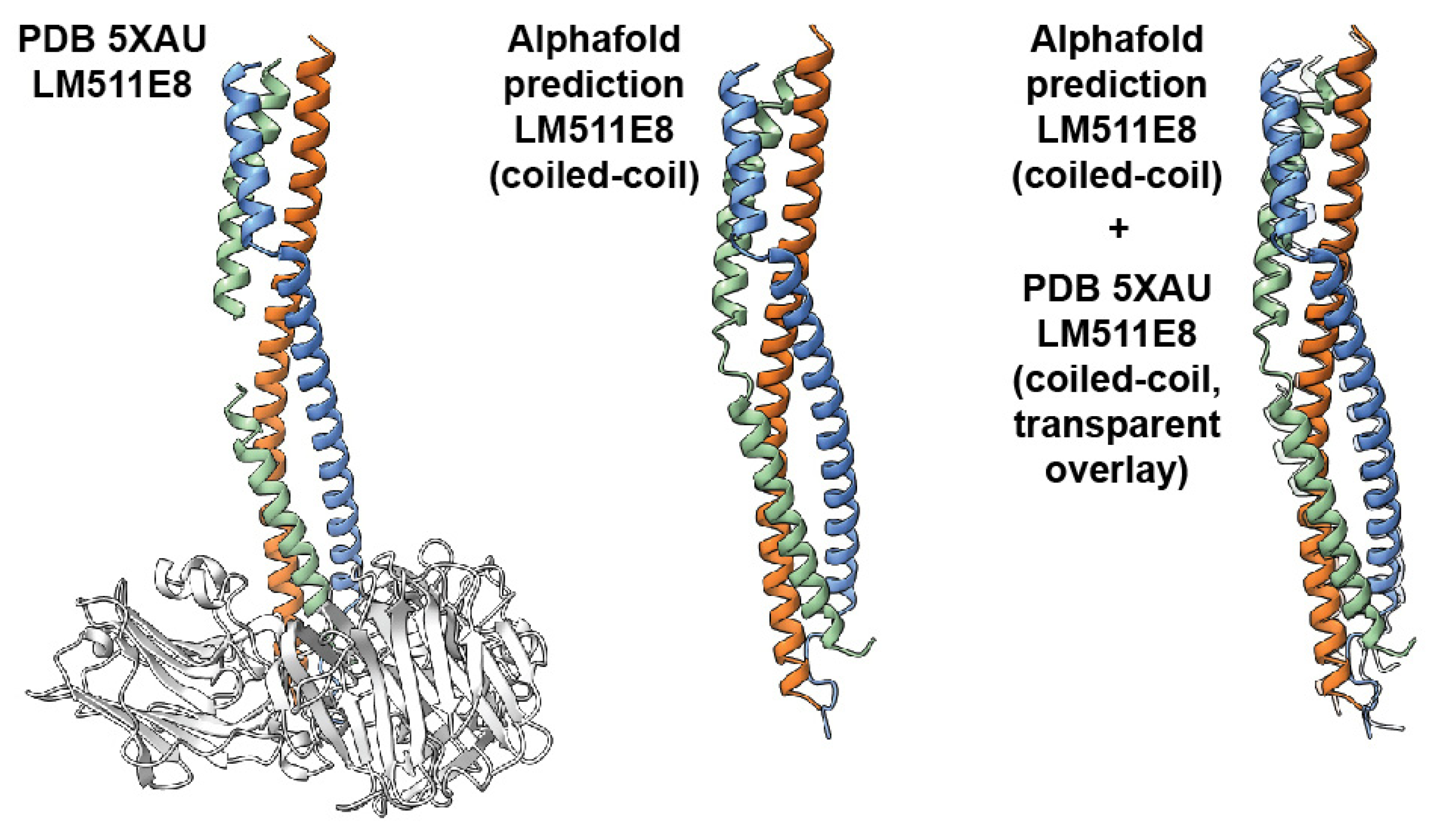
4.6. Structure Prediction of Laminin Coiled-Coiled Region
4.7. Simulated AFM Images of Laminin Structures
4.8. Molecular Dynamics Simulation of Structural Dynamics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakashita, S.; Engvall, E.; Ruoslahti, E. Basement membrane glycoprotein laminin binds to heparin. FEBS Lett. 1980, 116, 243–246. [Google Scholar] [CrossRef]
- Miner, J.H.; Yurchenco, P.D. Laminin functions in tissue morphogenesis. Annu. Rev. Cell Dev. Biol. 2004, 20, 255–284. [Google Scholar] [CrossRef]
- Hallmann, R.; Horn, N.; Selg, M.; Wendler, O.; Pausch, F.; Sorokin, L.M. Expression and function of laminins in the embryonic and mature vasculature. Physiol. Rev. 2005, 85, 979–1000. [Google Scholar] [CrossRef]
- Colognato, H.; Yurchenco, P.D. Form and function: The laminin family of heterotrimers. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2000, 218, 213–234. [Google Scholar] [CrossRef]
- Miner, J.H. Laminins and their roles in mammals. Microsc. Res. Tech. 2008, 71, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Miner, J.H.; Li, C.; Mudd, J.L.; Go, G.; Sutherland, A.E. Compositional and structural requirements for laminin and basement membranes during mouse embryo implantation and gastrulation. Development 2004, 131, 2247–2256. [Google Scholar] [CrossRef] [PubMed]
- Kleinman, H.K.; Cannon, F.B.; Laurie, G.W.; Hassell, J.R.; Aumailley, M.; Terranova, V.P.; Martin, G.R.; DuBois-Dalcq, M. Biological activities of laminin. J. Cell. Biochem. 1985, 27, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Aumailley, M.; Brucknertuderman, L.; Carter, W.G.; Deutzmann, R.; Edgar, D.; Ekblom, P.; Engel, J.; Engvall, E.; Hohenester, E.; Jones, J.C.; et al. A simplified laminin nomenclature. Matrix Biol. 2005, 24, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Paulsson, M.; Deutzmann, R.; Timpl, R.; Dalzoppo, D.; Odermatt, E.; Engel, J. Evidence for coiled-coil alpha-helical regions in the long arm of laminin. EMBO J. 1985, 4, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Hohenester, E. Structural biology of laminins. Essays Biochem. 2019, 63, 285–295. [Google Scholar] [PubMed]
- Beck, K.; Dixon, T.W.; Engel, J.; Parry, D.A. Ionic interactions in the coiled-coil domain of laminin determine the specificity of chain assembly. J. Mol. Biol. 1993, 231, 311–323. [Google Scholar] [CrossRef]
- Tunggal, P.; Smyth, N.; Paulsson, M.; Ott, M.-C. Laminins: Structure and genetic regulation. Microsc. Res. Tech. 2000, 51, 214–227. [Google Scholar] [CrossRef]
- Yamada, M.; Sekiguchi, K. Molecular Basis of Laminin-Integrin Interactions. Curr. Top. Membr. 2015, 76, 197–229. [Google Scholar]
- Henry, M.D.; Campbell, K.P. A role for dystroglycan in basement membrane assembly. Cell 1998, 95, 859–870. [Google Scholar] [CrossRef]
- Yurchenco, P.D.; Cheng, Y.-S.; Colognato, H. Laminin forms an independent network in basement membranes. J. Cell Biol. 1992, 117, 1119–1133. [Google Scholar] [CrossRef]
- Kulczyk, A.W.; McKee, K.K.; Zhang, X.; Bizukojc, I.; Yu, Y.Q.; Yurchenco, P.D. Cryo-EM reveals the molecular basis of laminin polymerization and LN-lamininopathies. Nat. Commun. 2023, 14, 317. [Google Scholar] [CrossRef]
- Kleinman, H.K.; McGarvey, M.L.; Hassell, J.R.; Star, V.L.; Cannon, F.B.; Laurie, G.W.; Martin, G.R. Basement membrane complexes with biological activity. Biochemistry 1986, 25, 312–318. [Google Scholar] [CrossRef]
- Goletz, S.; Zillikens, D.; Schmidt, E. Structural proteins of the dermal-epidermal junction targeted by autoantibodies in pemphigoid diseases. Exp. Dermatol. 2017, 26, 1154–1162. [Google Scholar] [CrossRef]
- Goldfinger, L.E.; Stack, M.S.; Jones, J.C. Processing of laminin-5 and its functional consequences: Role of plasmin and tis-sue-type plasminogen activator. J. Cell Biol. 1998, 141, 255–265. [Google Scholar] [CrossRef]
- Amano, S.; Scott, I.C.; Takahara, K.; Koch, M.; Champliaud, M.-F.; Gerecke, D.R.; Keene, D.R.; Hudson, D.L.; Nishiyama, T.; Lee, S.; et al. Bone morphogenetic protein 1 is an extracellular processing enzyme of the laminin 5 γ2 chain. J. Biol. Chem. 2000, 275, 22728–22735. [Google Scholar] [CrossRef]
- Veitch, D.P.; Nokelainen, P.; McGowan, K.A.; Nguyen, T.-T.; Nguyen, N.E.; Stephenson, R.; Pappano, W.N.; Keene, D.R.; Spong, S.M.; Greenspan, D.S.; et al. Mammalian tolloid metalloproteinase, and not matrix metalloprotease 2 or membrane type 1 metalloprotease, processes laminin-5 in keratinocytes and skin. J. Biol. Chem. 2003, 278, 15661–15668. [Google Scholar] [CrossRef]
- Rousselle, P.; Beck, K. Laminin 332 processing impacts cellular behavior. Cell Adhes. Migr. 2013, 7, 122–134. [Google Scholar] [CrossRef]
- Tsubota, Y.; Yasuda, C.; Kariya, Y.; Ogawa, T.; Hirosaki, T.; Mizushima, H.; Miyazaki, K. Regulation of biological activity and matrix assembly of laminin-5 by COOH-terminal, lg4–5 domain of α3 chain. J. Biol. Chem. 2005, 280, 14370–14377. [Google Scholar] [CrossRef]
- Engel, J.; Odermatt, E.; Engel, A.; Madri, J.A.; Furthmayr, H.; Rohde, H.; Timpl, R. Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J. Mol. Biol. 1981, 150, 97–120. [Google Scholar] [CrossRef]
- Rousselle, P.; Lunstrum, G.P.; Keene, D.R.; E Burgeson, R. Kalinin: An epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J. Cell Biol. 1991, 114, 567–576. [Google Scholar] [CrossRef]
- Vailly, J.; Verrando, P.; Champliaud, M.; Gerecke, D.; Wagman, D.W.; Baudoin, C.; Aberdam, D.; Burgeson, R.; Bauer, E.; Ortonne, J. The 100-kDa chain of nicein/kalinin is a laminin B2 chain variant. Eur. J. Biochem. 1994, 219, 209–218. [Google Scholar] [CrossRef]
- Baker, S.E.; Hopkinson, S.B.; Fitchmun, M.; Andreason, G.L.; Frasier, F.; Plopper, G.; Quaranta, V.; Jones, J.C.R. Laminin-5 and hemidesmosomes: Role of the α3 chain subunit in hemidesmosome stability and assembly. J. Cell Sci. 1996, 109, 2509–2520. [Google Scholar] [CrossRef]
- Hussain, S.; Carafoli, F.; Hohenester, E. Determinants of laminin polymerization revealed by the structure of the α5 chain amino-terminal region. Embo Rep. 2011, 12, 276–282. [Google Scholar] [CrossRef]
- Carafoli, F.; Hussain, S.-A.; Hohenester, E. Crystal Structures of the Network-Forming Short-Arm Tips of the Laminin β1 and γ1 Chains. PLoS ONE 2012, 7, e42473. [Google Scholar] [CrossRef]
- Stetefeld, J.; Mayer, U.; Timpl, R.; Huber, R. Crystal structure of three consecutive laminin-type epidermal growth factor-like (le) modules of laminin γ1 chain harboring the nidogen binding site. J. Mol. Biol. 1996, 257, 644–657. [Google Scholar] [CrossRef]
- Hohenester, E.; Tisi, D.; Talts, J.F.; Timpl, R. The crystal structure of a laminin G–like module reveals the molecular basis of α-dystroglycan binding to laminins, perlecan, and agrin. Mol. Cell 1999, 4, 783–792. [Google Scholar] [CrossRef]
- Tisi, D.; Talts, J.F.; Timpl, R.; Hohenester, E. Structure of the C-terminal laminin G-like domain pair of the laminin α2 chain harbouring binding sites for α-dystroglycan and heparin. EMBO J. 2000, 19, 1432–1440. [Google Scholar] [CrossRef]
- Pulido, D.; Hussain, S.-A.; Hohenester, E. Crystal structure of the heterotrimeric integrin-binding region of laminin-111. Structure 2017, 25, 530–535. [Google Scholar] [CrossRef][Green Version]
- Takizawa, M.; Arimori, T.; Taniguchi, Y.; Kitago, Y.; Yamashita, E.; Takagi, J.; Sekiguchi, K. Mechanistic basis for the recognition of laminin-511 by α6β1 integrin. Sci. Adv. 2017, 3, e1701497. [Google Scholar] [CrossRef]
- Chen, C.H.; Clegg, D.O.; Hansma, H.G. Structures and dynamic motion of laminin-1 as observed by atomic force micros-copy. Biochemistry 1998, 37, 8262–8267. [Google Scholar] [CrossRef][Green Version]
- Chen, C.H.; Hansma, H.G. Basement Membrane Macromolecules: Insights from Atomic Force Microscopy. J. Struct. Biol. 2000, 131, 44–55. [Google Scholar] [CrossRef][Green Version]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef]
- Ando, T. High-speed atomic force microscopy. Curr. Opin. Chem. Biol. 2019, 51, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Fukuda, S.; Ngo, K.X.; Flechsig, H. High-Speed Atomic Force Microscopy for Filming Protein Molecules in Dynamic Action. Annu. Rev. Biophys. 2024, 53, 1. [Google Scholar] [CrossRef]
- Marchesi, A.; Umeda, K.; Komekawa, T.; Matsubara, T.; Flechsig, H.; Ando, T.; Watanabe, S.; Kodera, N.; Franz, C.M. An ultra-wide scanner for large-area high-speed atomic force microscopy with megapixel resolution. Sci. Rep. 2021, 11, 13003. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Kodera, N.; Yamamoto, D.; Miyagi, A.; Taniguchi, M.; Yamashita, H. High-speed AFM and nano-visualization of biomolecular processes. Pflügers Arch. Eur. J. Physiol. 2008, 456, 211–225. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Scheuring, S. Filming Biomolecular Processes by High-Speed Atomic Force Microscopy. Chem. Rev. 2014, 114, 3120–3188. [Google Scholar] [CrossRef]
- Boyken, S.E.; Benhaim, M.A.; Busch, F.; Jia, M.; Bick, M.J.; Choi, H.; Klima, J.C.; Chen, Z.; Walkey, C.; Mileant, A. De novo design of tunable, pH-driven conformational changes. Science 2019, 364, 658–664. [Google Scholar] [CrossRef]
- Kozai, T.; Sekiguchi, T.; Satoh, T.; Yagi, H.; Kato, K.; Uchihashi, T. Two-step process for disassembly mechanism of proteasome α7 homo-tetradecamer by α6 revealed by high-speed atomic force microscopy. Sci. Rep. 2017, 7, 15373. [Google Scholar] [CrossRef]
- Beck, K.; Hunter, I.; Engel, J. Structure and function of laminin: Anatomy of a multidomain glycoprotein. FASEB J. 1990, 4, 148–160. [Google Scholar] [CrossRef]
- Kariya, Y.; Ishida, K.; Tsubota, Y.; Nakashima, Y.; Hirosaki, T.; Ogawa, T.; Miyazaki, K. Efficient Expression System of Human Recombinant Laminin-51. J. Biochem. 2002, 132, 607–612. [Google Scholar] [CrossRef]
- Ryan, M.; Tizard, R.; VanDevanter, D.; Carter, W. Cloning of the LamA3 gene encoding the alpha 3 chain of the adhesive ligand epiligrin. Expression in wound repair. J. Biol. Chem. 1994, 269, 22779–22787. [Google Scholar] [CrossRef]
- Galliano, M.F.; Aberdam, D.; Aguzzi, A.; Ortonne, J.P.; Meneguzzi, G. Cloning and Complete Primary Structure of the Mouse Laminin α3 Chain: DISTINCT EXPRESSION PATTERN OF THE LAMININ α3A AND α3B CHAIN ISOFORMS (∗). J. Biol. Chem. 1995, 270, 21820–21826. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kenzaki, H.; Koga, N.; Hori, N.; Kanada, R.; Li, W.; Okazaki, K.-I.; Yao, X.-Q.; Takada, S. CafeMol: A Coarse-Grained Biomolecular Simulator for Simulating Proteins at Work. J. Chem. Theory Comput. 2011, 7, 1979–1989. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Carafoli, F.; Clout, N.J.; Hohenester, E. Crystal structure of the LG1-3 region of the laminin alpha2 chain. J. Biol. Chem 2009, 284, 22786–22792. [Google Scholar] [CrossRef]
- McKee, K.K.; Hohenester, E.; Aleksandrova, M.; Yurchenco, P.D. Organization of the laminin polymer node. Matrix Biol. 2021, 98, 49–63. [Google Scholar] [CrossRef]
- Arimori, T.; Miyazaki, N.; Mihara, E.; Takizawa, M.; Taniguchi, Y.; Cabañas, C.; Sekiguchi, K.; Takagi, J. Structural mechanism of laminin recognition by integrin. Nat. Commun. 2021, 12, 4012. [Google Scholar] [CrossRef]
- Bruch, M.; Landwehr, R.; Engel, J. Dissection of laminin by cathepsin G into its long-arm and short-arm structures and localization of regions involved in calcium dependent stabilization and self-association. Eur. J. Biochem. 1989, 185, 271–279. [Google Scholar] [CrossRef]
- Kammerer, R.A.; Schulthess, T.; Landwehr, R.; Schumacher, B.; Lustig, A.; Yurchenco, P.D.; Ruegg, M.A.; Engel, J.; Denzer, A.J. Interaction of agrin with laminin requires a coiled-coil conformation of the agrin-binding site within the laminin gamma1 chain. EMBO J. 1999, 18, 6762–6770. [Google Scholar] [CrossRef]
- Mason, J.M.; Arndt, K.M. Coiled Coil Domains: Stability, Specificity, and Biological Implications. ChemBioChem 2004, 5, 170–176. [Google Scholar] [CrossRef]
- Elliott, A.; Offer, G. Shape and flexibility of the myosin molecule. J. Mol. Biol. 1978, 123, 505–519. [Google Scholar] [CrossRef]
- Tanizawa, H.; Taniguchi, M.; Ghimire, G.D.; Mitaku, S. Prediction of fragile points of coiled coils. Chem-Bio Inform. J. 2009, 9, 12–29. [Google Scholar] [CrossRef]
- Lupas, A. Coiled coils: New structures and new functions. Trends Biochem. Sci. 1996, 21, 375–382. [Google Scholar] [CrossRef]
- Armony, G.; Jacob, E.; Moran, T.; Levin, Y.; Mehlman, T.; Levy, Y.; Fass, D. Cross-linking reveals laminin coiled-coil architecture. Proc. Natl. Acad. Sci. USA 2016, 113, 13384–13389. [Google Scholar] [CrossRef]
- Uchihashi, T.; Kodera, N.; Ando, T. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protoc. 2012, 7, 1193–1206. [Google Scholar] [CrossRef]
- Ye, Z.; Galvanetto, N.; Puppulin, L.; Pifferi, S.; Flechsig, H.; Arndt, M.; Triviño, C.A.S.; Di Palma, M.; Guo, S.; Vogel, H.; et al. Structural heterogeneity of the ion and lipid channel TMEM16F. Nat. Commun. 2024, 15, 110. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Amyot, R.; Flechsig, H. BioAFMviewer: An interactive interface for simulated AFM scanning of biomolecular structures and dynamics. PLoS Comput. Biol. 2020, 16, e1008444. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, L.; Flechsig, H.; Marchesi, A.; Franz, C.M. Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy. Int. J. Mol. Sci. 2024, 25, 1951. https://doi.org/10.3390/ijms25041951
Akter L, Flechsig H, Marchesi A, Franz CM. Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy. International Journal of Molecular Sciences. 2024; 25(4):1951. https://doi.org/10.3390/ijms25041951
Chicago/Turabian StyleAkter, Lucky, Holger Flechsig, Arin Marchesi, and Clemens M. Franz. 2024. "Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy" International Journal of Molecular Sciences 25, no. 4: 1951. https://doi.org/10.3390/ijms25041951
APA StyleAkter, L., Flechsig, H., Marchesi, A., & Franz, C. M. (2024). Observing Dynamic Conformational Changes within the Coiled-Coil Domain of Different Laminin Isoforms Using High-Speed Atomic Force Microscopy. International Journal of Molecular Sciences, 25(4), 1951. https://doi.org/10.3390/ijms25041951





