Clinical Application of Liquid Biopsy in Pancreatic Cancer: A Narrative Review
Abstract
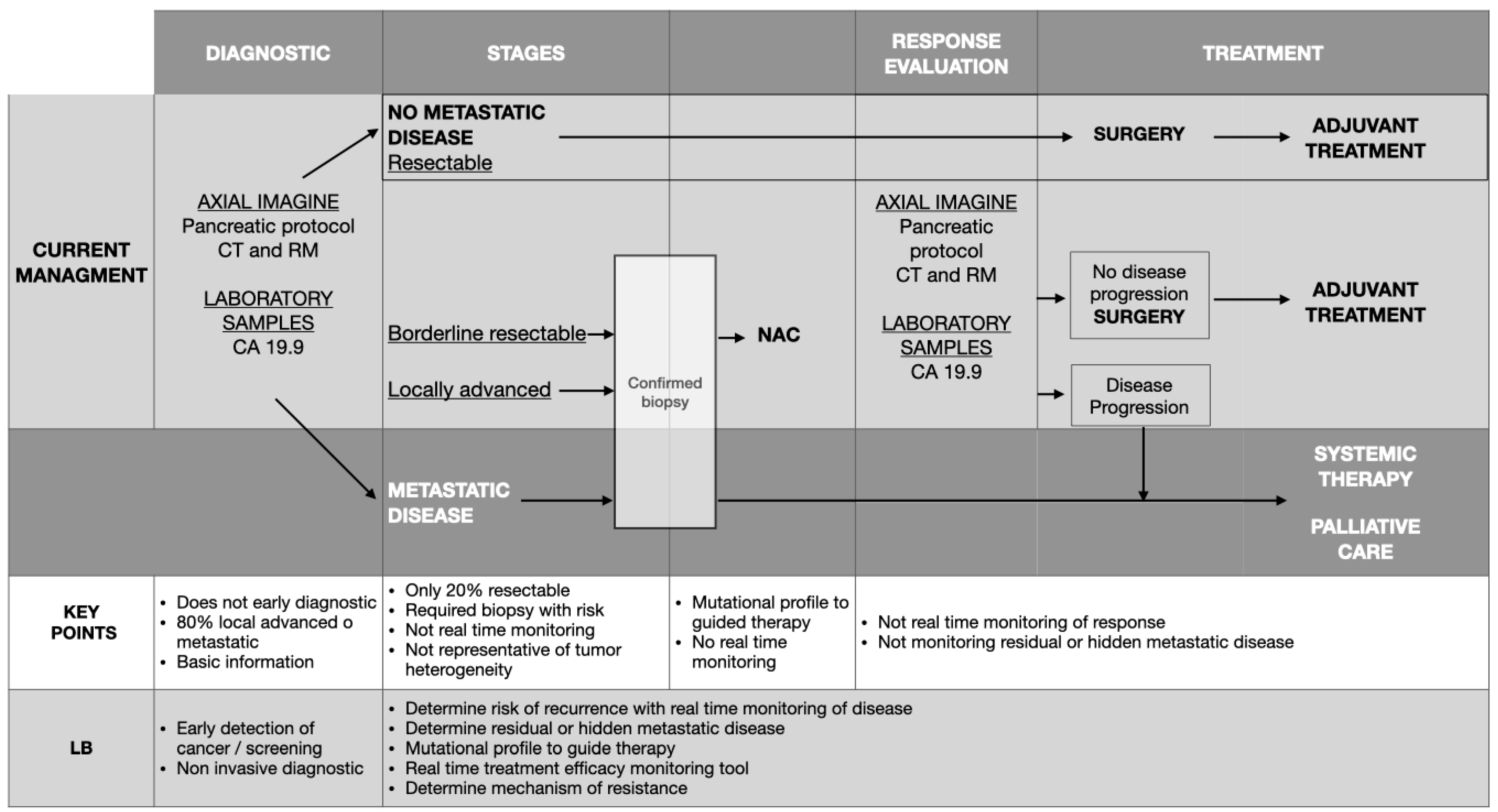
1. Introduction
2. Methodologies and Technological Approaches for LB
2.1. ctDNA
2.2. CTCs
2.3. EVs
3. Impact of LB in Early and Differential Diagnosis of PDAC
3.1. ctDNA
3.2. CTCs
3.3. EVs
3.4. miRNAs
4. Role of LB after Resection of PDAC
4.1. ctDNA
4.2. CTCs
4.3. EVs
5. Assessment of NAC in PDAC
6. The Use of LB and in Recurrence and Prognosis of PDAC
6.1. ctDNA
6.2. CTCs
6.3. EVs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AUC | area under curve |
| BEAMing | beads, emulsion, amplification, magnetics |
| CA 19-9 | carbohydrate antigen |
| CDKN2A | cyclin dependent kinase inhibitor 2A |
| cfDNA | circulating free DNA |
| CK | cytokeratin |
| CP | chronic pancreatitis |
| CTCs | circulating tumor cells |
| CTEC | tumor vascular endothelial cells |
| ctDNA | circulating tumor DNA |
| DFS | disease free survival |
| DPC4 | delete in Pancreatic Cancer-4 |
| dPCR | digital PCR |
| ddPCR | droplet digital PCR |
| ECM | extracellular matrix |
| EMT | epithelial–mesenchymal transition |
| EpCAM | epithelial cell adhesion |
| EV | extracellular vesicles |
| FDA | Food and Drug Administration |
| GPC1 | Glypican-1 |
| KRAS | Kristen rat sarcoma |
| LB | liquid biopsy |
| MIF | macrophage migration inhibitory factor |
| miRNA | micro-RNA |
| MST | median survival time |
| NAC | Neoadjuvant chemotherapy |
| NGS | next-generation sequencing |
| OS | overall survival |
| PCR | allele-specific polymerase chain reaction |
| PDAC | pancreatic ductal adenocarcinoma |
| SMAD4 | SMAD family member4 |
| TEP | tumor-educated platelet |
| TP53 | tumor protein 53 |
References
- Kamarajah, S.K.; Burns, W.R.; Frankel, T.L.; Cho, C.S.; Nathan, H. Validation of the American Joint Commission on Cancer (AJCC) 8th Edition Staging System for Patients with Pancreatic Adenocarcinoma: A Surveillance, Epidemiology and End Results (SEER) Analysis. Ann. Surg. Oncol. 2017, 24, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Allen, P.J.; Kuk, D.; Castillo, C.F.D.; Basturk, O.; Wolfgang, C.L.; Cameron, J.L.; Lillemoe, K.D.; Ferrone, C.R.; Morales-Oyarvide, V.; He, Y.; et al. Multi-institutional validation study of the American joint commission on cancer (8th edition) changes for Tand N staging in patients with pancreatic adenocarcinoma. Ann. Surg. 2017, 265, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Zhang, H.; Chen, N.; Hao, J.; Jin, H.; Ma, X. Diagnostic value of various liquid biopsy methods for pancreatic cancer: A systematic review and meta-analysis. Medicine 2020, 99, e18581. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.; Suzuki, K.; Noda, H.; Rikiyama, T. Liquid biopsy leads to a paradigm shift in the treatment of pancreatic cancer. World J. Gastroenterol. 2022, 28, 6478–6496. [Google Scholar] [CrossRef] [PubMed]
- Heredia-Soto, V.; Rodríguez-Salas, N.; Feliu, J. Liquid biopsy in pancreatic cancer: Are we ready to apply it in the clinical practice? Cancers 2021, 13, 1986. [Google Scholar] [CrossRef]
- Rofi, E.; Vivaldi, C.; Del Re, M.; Arrigoni, E.; Crucitta, S.; Fuel, N.; Fogli, S.; Vasile, E.; Musettini, G.; Fornaro, L.; et al. The emerginn role of liquid biopsy in diagnosis, prognosis, an treatment monitoring of pancreatic cancer. Pharmacogenomics 2019, 20, 49–68. [Google Scholar] [CrossRef]
- Grunvald, M.W.; Jacobson, R.A.; Kuzel, T.M.; Pappas, S.G.; Masood, A. Current status of circulating tumor DNA liquid biopsy in pancreatic cancer. Int. J. Mol. Sci. 2020, 21, 7651. [Google Scholar] [CrossRef]
- Raufi, A.G.; May, M.S.; Hadfield, M.J.; Seyhan, A.A.; El-Deiry, W.S. Advances in liquid biopsy technology and implications for pancreatic cancer. Int. J. Mol. Sci. 2023, 24, 4238. [Google Scholar] [CrossRef]
- Tempero, M.A.; Malafa, M.P.; Al-Hawari, M.; Behrman, S.W.; Benson, A.B.; Cardin, D.B.; Chiorean, E.G.; Chung, V.; Czito, B.; Del Chiaro, M.; et al. Pancreatic adenocarcinoma, Version 1.2024, NCCN Clinical practice guidelines in oncology. J. Natl. Cancer Netw. 2021, 19, 430–457. [Google Scholar] [CrossRef]
- Nagai, M.; Sho, M.; Akahori, T.; Nakagawa, K.; Nakamura, K. Application of liquid biopsy for surgical management of pancreatic cancer. Ann. Gastroenterol. Surg. 2020, 4, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.; Pando, E.; Blanco, L.; Fabregat-Franco, C.; Castet, F.; Sierra, A.; Macarulla, T.; Balsells, J.; Charco, R.; Vivancos, A. Liquid biopsy after resection of pancreatic adenocarcinoma and its relation to oncological outcomes. Systematic review and meta-analysis. Cancer Treat. Rev. 2023, 120, 102604. [Google Scholar] [CrossRef]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Mimeault, M.; Brand, R.E.; Sasson, A.A.; Batra, S.K. Recent advances on the molecular mechanisms involved in pancreatic cancer progression and therapies. Pancreas 2005, 31, 301–316. [Google Scholar] [CrossRef]
- Maitra, A.; Hruban, R.H. Pancreatic Cancer. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 157–188. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear Acids in Human Blood Plasma. Comptes Rendus Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Ashworth, T.R. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med. J. Aust. 1869, 14, 146–147. [Google Scholar]
- Yang, J.; Li, S.; Li, J.; Wang, F.; Chen, K.; Zheng, Y.; Wang, J.; Lu, W.; Zhou, Y.; Yin, Q.; et al. A meta-analysis of the diagnostic value of detecting K-ras mutation in pancreatic juice as a molecular marker for pancreatic cancer. Pancratology 2016, 16, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Fukushima, N.; Abe, T.; Bian, Y.; Hua, L.; Wendelburg, B.J.; Yeo, C.J.; Hruban, R.H.; Goggins, M.G.; Eshleman, J.R. Sensitive and quantitative detection of KRAS2 gene mutations in pancreatic duct juice differentiates patients with pancreatic cancer from chronic pancreatitis, potential for early detection. Cancer Biol. Ther. 2008, 7, 353–360. [Google Scholar] [CrossRef]
- Hata, T.; Ishida, M.; Motoi, F.; Yamaguchi, T.; Naitoh, T.; Katayose, Y.; Egawa, S.; Unno, M. Telomerase activity in pancreatic juice differentiates pancreatic cancer from chronic pancreatitis: A meta-analysis. Pancreatology 2016, 16, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Farrell, J.J.; Zhou, H.; Elashowff, D.; Akin, D.; Park, N.H.; Chia, D.; Wong, D.T. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010, 138, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419. [Google Scholar] [CrossRef]
- Xie, Z.; Yin, X.; Gong, B.; Nice, W.; Wu, B.; Zhang, X.; Huang, J.; Zhang, P.; Zhou, Z.; Li, Z. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev. Res. 2015, 8, 165–173. [Google Scholar] [CrossRef]
- Yoshizawa, N.; Sugimoto, K.; Tameda, M.; Inagaki, Y.; Ikejiri, M.; Inoue, H.; Usui, M.; Takei, Y. miR-3940-5p/miR-8069 ratio in urine exosomes is a novel diagnostic biomarker for pancreatic ductal adenocarcinoma. Once Lett. 2020, 19, 2677–2684. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, H.; Kinugasa, H.; Ako, S.; Hirai, M.; Matsushita, H.; Uchida, D.; Tomoda, T.; Matsumoto, K.; Horiguchi, S.; Kato, H.; et al. Utility of liquid biopsy using urine in patients with pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2019, 20, 1348–1353. [Google Scholar] [CrossRef] [PubMed]
- Adamo, P.; Cowley, C.M.; Neal, C.P.; Mistry, V.; Page, K.; Denninson, A.R.; Isherwood, J.; Hastings, R.; Luo, J.; Moore, D.A.; et al. Profiling tumour heterogeneity through circulating tumour DNA in patients with pancreatic cancer. Oncotarget 2017, 8, 87221–87233. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, S.D.; Bibbo, M.; Loren, D.E.; Siddiqui, A.A.; Solomides, C.; Kowalski, T.E.; Ellsworth, E. Molecular analysis of centrifugation supernatant fluid from pancreaticobiliary duct samples can improve cancer detection. Acta Cytol. 2012, 56, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Sikora, K.; Bedin, C.; Vicentini, C.; Malpeli, G.; D’Angelo, E.; Sperandio, N.; Lawlor, R.T.; Bassi, C.; Tortora, G.; Nitti, D.; et al. Evaluation of cell-free DNA as a biomarker for pancreatic malignancies. Int. J. Biol. Markers 2015, 30, e136–e141. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Thege, F.I.; Santana, S.M.; Lannin, T.B.; Saha, T.N.; Tsai, S.; Maggs, L.R.; Kochman, M.L.; Gingsberg, G.G.; Lieb, J.G.; et al. Detection of circulating pancreas epithelial cells in patients with pancreatic cystic lesions. Gastroenterology 2014, 146, 647–651. [Google Scholar] [CrossRef] [PubMed]
- Kulemann, B.; Pitman, M.B.; Liss, A.S.; Valsangkar, N.; Fernández-Del Castillo, C.; Lillemoe, K.D.; Hoeppner, J.; Mino-Kenudson, M.; Warshaw, A.L.; Thayer, S.P. Circulating tumor cells found in patients with localized and advanced pancreatic cancer. Pancreas 2015, 44, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Cauley, C.E.; Pitman, M.B.; Zhou, J.; Perkins, J.; Coleman, B.; Liss, A.S.; Fernández-Del Castillo, C.; Warshaw, A.L.; Lillemoe, K.D.; Ayer, S.P. Circulating Epithelial Cells in Patients with Pancreatic Lesions: Clinical and Pathologic Findings. J. Am. Coll. Surg. 2015, 221, 699–707. [Google Scholar] [CrossRef]
- Qi, Z.H.; Xu, H.X.; Zhang, S.R.; Xu, J.Z.; Li, S.; Gao, H.L.; Jin, W.; Wang, W.Q.; Wu, C.T.; Ni, Q.X.; et al. The Significance of Liquid Biopsy in Pancreatic Cancer. J. Cancer 2018, 9, 3417–3426. [Google Scholar] [CrossRef]
- Ankeny, J.S.; Court, C.M.; Hou, S.; Li, Q.; Song, M.; Wu, D.; Chen, J.F.; Lee, T.; Lin, M.; Sho, S.; et al. Circulating tumour cells as a biomarker for diagnosis and staging in pancreatic cancer. Br. J. Cancer 2016, 114, 1367–1375. [Google Scholar] [CrossRef]
- Buscail, E.; Alix-Panabieres, C.; Quincy, P.; Cauvin, T.; Chauvet, A.; Degrandi, O.; Caumont, C.; Verdom, S.; Lamrissi, I.; Moranvillier, I.; et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers 2019, 11, 1656. [Google Scholar] [CrossRef]
- Allenson, K.; Castillo, J.; San Lucas, F.A.; Scale, G.; Kim, D.U.; Bernard, V.; Davis, G.; Kumar, T.; Katz, M.; Overman, M.J.; et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann. Oncol. 2017, 28, 741–747. [Google Scholar] [CrossRef] [PubMed]
- Maire, F.; Micard, S.; Hammel, P.; Voitot, H.; Lévy, P.; Cugnenc, P.H.; Ruszniewski, P.; Puig, P.L. Differential diagnosis between chronic pancreatitis and pancreatic cancer: Value of the detection of KRAS2 mutations in circulating DNA. Br. J. Cancer 2002, 87, 551–554. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gaming, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Y.; Shi, J.; Zhang, K.; Zhang, Z.; Zhang, H. Sensitive Signal Amplifying a Diagnostic Biochip Based on a Biomimetic Periodic Nanostructure for Detecting Cancer Exosomes. ACS Appl. Mater. Interfaces 2020, 12, 33473–33482. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.M.; Vyas, A.D.; Qiu, Y.; Messer, K.S.; White, R.; Heller, M.J. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano 2018, 12, 3311–3320. [Google Scholar] [CrossRef] [PubMed]
- Lux, A.; Kahlert, C.; Grützmann, R.; Pilarsky, C. c-Met and PD-l1 on circulating exosomes as diagnostic and prognostic markers for pancreatic cancer. Int. J. Mol. Sci. 2019, 20, 3305. [Google Scholar] [CrossRef]
- Yan, T.B.; Huang, J.Q.; Huang, S.Y.; Ahir, B.K.; Li, L.M.; Mo, Z.N.; Zhong, J.H. Advances in the Detection of Pancreatic Cancer Through Liquid Biopsy. Front. Oncol. 2021, 11, 801173. [Google Scholar] [CrossRef]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.P.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef]
- Yan, Q.; Hu, D.; Li, M.; Chen, Y.; Wu, X.; Ye, Q.; Wang, Z.; He, L.; Zhu, J. The Serum MicroRNA Signatures for Pancreatic Cancer Detection and Operability Evaluation. Front. Bioeng. Biotechnol. 2020, 8, 379. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Miyamae, M.; Kawaguchi, T.; Morimura, R.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; et al. Malignant potential in pancreatic neoplasm; New insights provided by circulating miR-223 in plasma. Expert Opin. Biol. Ther. 2015, 15, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, D.K.; Gercke, N.; Strauch, K.; Wieboldt, R.; Matthäi, E.; Wagner, V.; Rospleszcz, S.; Schäfer, A.; Franke, F.S.; Mintziras, I.; et al. The combination of miRNA-196b, LCN2, and TIMP1 is a potential set of circulating biomarkers for screening individuals at risk for familial pancreatic cancer. J. Clin. Med. 2018, 7, 295. [Google Scholar] [CrossRef] [PubMed]
- Peng, C.; Wang, J.; Gao, W.; Huang, L.; Liu, Y.; Li, X.; Li, Z.; Yu, X. Meta-analysis of the diagnostic performance of circulating micrornas for pancreatic cancer. Int. J. Med. Sci. 2021, 18, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Raimondo, M.; Guha, S.; Chen, J.; Diao, L.; Dong, X.; Wallacew, M.B.; Killary, A.M.; Frazier, M.L.; Woodward, T.A.; et al. Circulating microRNAs in pancreatic juice as candidate biomarkers of pancreatic cancer. J. Cancer 2014, 5, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Burnt, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary microRNA in pancreatic cancer patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Dong, Z.; Zhen, L.; Xia, G.; Huang, X.; Wang, T.; Geo, H.; Yang, B.; Xu, C.; Wu, W.; et al. Combined exosomal GPC1, CD82, and serum CA19-9 as multiplex targets: A specific, sensitive, and reproducible detection panel for the diagnosis of pancreaticcancer. Mol. Cancer Res. 2020, 18, 1300–1310. [Google Scholar] [CrossRef]
- Pu, X.; Ding, G.; Wu, M.; Zhou, S.; Jia, S.; Cao, L. Elevated expression of exosomal microRNA–21 as a potential biomarker for the early diagnosis of pancreatic cancer using a tethered cationic lipoplex nanoparticle biochip. Oncol. Lett. 2020, 19, 2062–2070. [Google Scholar] [CrossRef]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdix, A.; Pares, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef]
- Wu, H.; Guo, S.; Liu, X.; Li, Y.; Su, Z.; He, Q.; Liu, X.; Zhang, Z.; Yu, L.; Shi, X.; et al. Noninvasive detection of pancreatic ductal adenocarcinoma using the methylation signature of circulating tumour DNA. BMC Med. 2022, 20, 458. [Google Scholar] [CrossRef]
- Xu, Y.; Qin, T.; Li, J.; Wang, X.; Gao, C.; Xu, C.; Hao, J.; Liu, J.; Gao, S.; Ren, H. Detection of circulating tumor cells using negative enrichment immunofluorescence and an in situ hybridization system in pancreatic cancer. Int. J. Mol. Sci. 2017, 18, 622. [Google Scholar] [CrossRef]
- Verstei Jne, E.; van Dam, J.L.; Suker, M.; Janssend, Q.P.; Groothuis, K.; Akkermans-Vogelaar, J.M.; Basselink, M.G.; Bonsing, B.A.; Buijsen, J.; Busch, O.R.; et al. Neoadjuvant chemoradiotherapy versus upfront surgery for resectable and borderline resectable pancreatic cancer: Long-term results of the Dutch randomized PREOPANC trial. J. Clin. Oncol. 2022, 40, 1220–1230. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analysis identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Kruger, S.; Heinemann, V.; Ross, C.; Diehl, F.; Nagel, D.; Ormanns, S.; Liebmann, S.; Prinz-Bravin, I.; Westphalen, C.B.; Haas, M.; et al. Repeated mutKRAS ctDNA measurements represent a novel and promising tool for early response prediction and therapy monitoring in advanced pancreatic cancer. Ann. Oncol. 2018, 29, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Gemenetzis, G.; Groot, V.P.; Yu, J.; Ding, D.; Tenor, J.A.; Javed, A.A.; Wood, L.D.; Burkhart, R.A.; Cameron, J.L.; Makary, M.; et al. Circulating tumor cells dynamics in pancreatic adenocarcinoma correlate with disease status: Results of the prospective CLUSTER study. Ann. Surf. 2018, 268, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Pu, N.; Thompson, E.; Miao, Y.; Wolfgang, C.; Yu, J. Improved assessment of response status in patients with pancreatic cancer treated with neoadjuvant therapy using somatic mutations and liquid biopsy analysis. Clin. Cancer Res. 2021, 27, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gupta, S.; Pandey, R.M.; Chauhan, S.S.; Saraya, A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Investig. 2015, 33, 78–85. [Google Scholar] [CrossRef]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Li, Q.K.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Berry, K.S.O.; Gilje, B.; Soreide, J.A.; Javle, M.; Nordgard, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2016, 10, 635–643. [Google Scholar] [CrossRef]
- Nakano, Y.; Kitago, M.; Matsuda, S.; Nakamura, Y.; Fujita, Y.; Imai, S.; Shinoda, M.; Yagi, H.; Abe, Y.; Hibi, T.; et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: A retrospective study. Br. J. Cancer 2018, 118, 662–669. [Google Scholar] [CrossRef]
- Watanabe, F.; Suzuki, K.; Tamaki, S.; Abe, I.; Endo, Y.; Takayama, Y.; Ishikawa, H.; Kakizawa, N.; Saito, M.; Futsuhara, K.; et al. Sequential assessments of KRAS- mutated circulating tumor DNA in longitudinal monitoring enable the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. J. Clin. Oncol. 2019, 37, e15712. [Google Scholar] [CrossRef]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodríguez-Garrote, J.; Lisa, E.; Mendía, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, F.; Ning, N.; Chen, Q.; Yang, Z.; Guo, Y.; Xu, D.; Zhang, D.; Han, T.; Cui, W. Patterns of circulating tumor cells identified by CEP8, CK and CD45 in pancreatic cancer. Int. J. Cancer 2015, 136, 1228–1233. [Google Scholar] [CrossRef]
- Okubo, K.; Uenosono, Y.; Arigami, T.; Makati, Y.; Matsushita, D.; Yanagita, S.; Kuruhara, H.; Sakoda, M.; Kijima, Y.; Maemura, K.; et al. Clinical impact of circulating tumor cells and therapy response in pancreatic cancer. Eur. J. Surg. Oncol. 2017, 43, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tao, Y.; Wang, X.; Jiang, P.; Li, J.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Zhen, P.; et al. Tumor-secreted exosomal miR-222 promotes tumor progression via regulating P27 expression and re-localization in pancreatic cancer. Cell. Physiol. Biochem. 2018, 51, 610–629. [Google Scholar] [CrossRef]
- Frampton, A.E.; Prado, M.M.; López-Jiménez, E.; Fajardo-Puerta, A.B.; Jawad, Z.A.R.; Lawton, P.; Giovanetti, E.; Habib, N.A.; Castellano, L.; Stebbing, J.; et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget 2018, 9, 19006–19013. [Google Scholar] [CrossRef]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kg to promote pancreatic cancer metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed]
| ctDNA | CTCs | EVs | |
|---|---|---|---|
| Target | KRAS, TP53, CDKN2A, SMAD4, BRAF, PIK3CA, ADAMTS1, BNC1, 5MC, H2AZ, H2A1.1, H3K4me2, h2ak119ub | CD45, CEP8, CK, EpCAM | KRAS, TP53, RNA: miRNA, longRNA Proteins markers: EFGR, EPCAM, MUC-1, GPC-1, WNT2 |
| Isolation | Blood | Blood | Body fluids |
| Tumor information | Epigenetic information | DNA, RNA, Protein | DNA, RNA, Protein |
| Technological approaches | qPCR, dPCR, ddPCR, NGS, commercial kits | Immunoaffinity, Physical methods (size and density) | Density-based, size-based, affinity-based, commercial kits |
| Advantages | qPCR: Fast and low-cost dPCR: High sensitivity/Specificity NGS: capability to screen for a broad range of genetic variants using high DNA input | Immunoaffinity: Specific, label-free obtained Physical methods: Fast, simple, Low-cost, label-free obtained | Density-based: low cost. Independent of marker expression. Size-based: Low-cost, fast, Independent of marker expression. Affinity-based: Specificity. High purity. Commercial kits: Simple, fast. |
| Disadvantages | General: No early stages qPCR: Low sensitivity. Only points mutations. dPCR: High cost. Only points mutations. NGS: Variable sensitivity. High cost. | General: Isolation complex and expensive. Technical variability Immunoaffinity: capture only one subpopulation. Low purity. Physical methods: Needs immuno-labeling techniques to distinguish CTCs | General: Isolation complex by contamination and expensive Density-based: Time, high volume sample, can damage EVs. Size based: contamination. Affinity-based: low sample yield. Commercial kits: High cost. |
| Sensitivity (S) (%) | 34–71% KRAS mutations: codons 12, 13, 61, in different stages. | 73–76% CD45/CEP8 100% Mt, 58% resectable Anti-EpCAM portal vein Blood | 67% ES, 80% LA, 85% Mt KRAS mutations in exoDNA 50% ES GPC1 miRNAs Increased expression |
| Specificity (Sp) (%) | 75–81% Mutations KRAS exon 2 | 68% CD45/CEP8 | 90% ES GPC1 |
| Combined techniques (%) | S: 85–98%, Sp: 77–81% ctDNA (KRAS exon 2) with CA19.9 S: 47% ctDNA (KRAS MAFs) with CA19.9 | S: 100%, Sp: 80% CTCs.with EVs | NR |
| Application | No suitable for screening of PDAC Monitoring postoperative minimal residual disease Predictor of disease recurrence and prognosis | Not present in healthy controls Variable sensitivity in early diagnosis Excellent specificity. Follow-up of disease recurrence and prognosis Functional analysis drug resistance | The highest sensitivity and specificity in early detection Evaluated response of resection or any therapy Biotherapeutic application |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Maldonado, E.; López Gordo, S.; Major Branco, R.P.; Pavel, M.-C.; Estalella, L.; Llàcer-Millán, E.; Guerrero, M.A.; López-Gordo, E.; Memba, R.; Jorba, R. Clinical Application of Liquid Biopsy in Pancreatic Cancer: A Narrative Review. Int. J. Mol. Sci. 2024, 25, 1640. https://doi.org/10.3390/ijms25031640
Ramírez-Maldonado E, López Gordo S, Major Branco RP, Pavel M-C, Estalella L, Llàcer-Millán E, Guerrero MA, López-Gordo E, Memba R, Jorba R. Clinical Application of Liquid Biopsy in Pancreatic Cancer: A Narrative Review. International Journal of Molecular Sciences. 2024; 25(3):1640. https://doi.org/10.3390/ijms25031640
Chicago/Turabian StyleRamírez-Maldonado, Elena, Sandra López Gordo, Rui Pedro Major Branco, Mihai-Calin Pavel, Laia Estalella, Erik Llàcer-Millán, María Alejandra Guerrero, Estrella López-Gordo, Robert Memba, and Rosa Jorba. 2024. "Clinical Application of Liquid Biopsy in Pancreatic Cancer: A Narrative Review" International Journal of Molecular Sciences 25, no. 3: 1640. https://doi.org/10.3390/ijms25031640
APA StyleRamírez-Maldonado, E., López Gordo, S., Major Branco, R. P., Pavel, M.-C., Estalella, L., Llàcer-Millán, E., Guerrero, M. A., López-Gordo, E., Memba, R., & Jorba, R. (2024). Clinical Application of Liquid Biopsy in Pancreatic Cancer: A Narrative Review. International Journal of Molecular Sciences, 25(3), 1640. https://doi.org/10.3390/ijms25031640






