Abstract
The hippocampal formation, particularly the CA2 subregion, is critical for social memory formation and memory processing, relying on synaptic plasticity—a fundamental mechanism by which synapses strengthen. Given the role of the ubiquitin–proteasome system (UPS) in various nervous system processes, including learning and memory, we were particularly interested in exploring the involvement of RING-type ubiquitin E3 ligases, such as UHRF2 (NIRF), in social behavior and synaptic plasticity. Our results revealed altered social behavior in mice with systemic Uhrf2 knockout, including changes in nest building, tube dominance, and the three-chamber social novelty test. In Uhrf2 knockout mice, the entorhinal cortex-CA2 circuit showed significant reductions in synaptic plasticity during paired-pulse facilitation and long-term potentiation, while the inability to evoke synaptic plasticity in the Schaffer-collateral CA2 synapses remained unaffected. These changes in synaptic plasticity correlated with significant changes in gene expression including genes related to vesicle trafficking and transcriptional regulation. The effects of Uhrf2 knockout on synaptic plasticity and the observed gene expression changes highlight UHRF2 as a regulator of learning and memory processes at both the cellular and systemic levels. Targeting E3 ubiquitin ligases, such as UHRF2, may hold therapeutic potential for memory-related disorders, warranting further investigation.
1. Introduction
The hippocampus, a crucial brain region for memory formation, consists of distinct subregions with unique physiological properties that regulate different types of memory [1]. Among these subregions, Cornu Ammonis 1 (CA1) occupies a significant portion of the hippocampus and receives inputs from CA3 pyramidal neurons, as well as direct input from CA2, the medial entorhinal cortex (MEC), and the lateral entorhinal cortex (LEC) [2,3,4,5]. Cornu Ammonis 2 (CA2), located between CA1 and CA3, exhibits distinct molecular characteristics, including increased expression of specific genes and proteins such as RGS14, TCP4, ACTN2, and PTPN5 [6,7].
Recent studies have emphasized the critical role of the CA2 subregion in social memory formation and socio-cognitive information processing [8,9,10,11,12,13]. Notably, impairments in social memory have been observed when hippocampal CA2 neurons are inactivated, leading to the inability of animals to recognize familiar individuals [10,13]. Interestingly, studies have demonstrated that the inhibition of CA2 pyramidal neurons disrupts social memory, whereas their activation promotes both social memory and social aggression [10,13,14]. Moreover, the distinctive electrophysiological properties of CA2 excitatory neurons, characterized by their elevated negative resting membrane potentials, underscored the involvement of this region in social behavior [15]. Importantly, in addition to inputs from CA3, CA2 neurons receive afferent projections from layer II/III of the entorhinal cortex (EC) that synapse in the stratum lacunosum–moleculare. In this distal dendritic region, EC-CA2 synapses exhibit activity-dependent long-term potentiation (LTP) [16,17]. In contrast, proximal Schaffer collateral inputs (SC) from CA3 to CA2 do not support activity-dependent LTP [15,18], highlighting regional differences in the modulation of synaptic transmission by neural activity patterns [12].
While the morphological and physiological specializations underlying the role of CA2 role are becoming clearer, the specific molecular factors regulating social memory formation in this region remain less understood. One potential modulatory mechanism is protein turnover, which plays a pivotal role in neural development and plasticity [19,20,21]. The ubiquitin–proteasome system (UPS) is responsible for regulated protein degradation and is essential for synaptic plasticity and memory formation [22,23,24,25,26]. Dysregulation of the UPS contributes to synaptic dysfunction and memory impairments in Alzheimer’s disease (AD) and Parkinson’s disease (PD). Interestingly, targeted modulation of the UPS has shown promise in restoring impaired synaptic plasticity and memory in animal models of AD [27,28]. For example, overexpression of the ubiquitin E3 ligase Parkin, despite its association with PD pathogenesis, can counteract AD-related deficits in mice by ubiquitinating substrates [29], highlighting the potential of strategic UPS regulation as a therapeutic approach.
Given the central influence of protein regulation on cognition, elucidating the role of specific UPS components, such as the E3 ubiquitin ligase UHRF2, in hippocampus-dependent learning warrants exploration. UHRF2, encoded by the Uhrf2 gene [30], induces protein degradation through its RING finger domain and regulates gene transcription through histone modification [31,32]. Initial findings suggest that Uhrf2 deficiency may affect object recognition [33]. Intriguingly, while UPS components have been implicated in cognitive processes, UHRF2 remains uncharacterized in the context of social memory and CA2 synaptic plasticity. Thus, elucidating the influence of UHRF2-mediated ubiquitination on hippocampal circuitry may provide the first insights into how protein turnover mechanisms regulate this domain of memory formation.
To address these open questions, we conducted a study utilizing Uhrf2 knockout mice to evaluate the role of UHRF2 in modulating social behavior and synaptic plasticity specifically in the CA2 hippocampal subregion. By combining behavioral assays, electrophysiology, and unbiased RNA sequencing, we uncovered novel regulatory functions of UHRF2 in social behavior, entorhinal cortex–CA2 synaptic potentiation, and the modulation of a variety of gene expression networks. Thus, by evaluating the functions of UHRF2 in hippocampal social behavior and plasticity, our findings advance our understanding of how ubiquitin-dependent protein regulation shapes the properties of the CA2 memory circuit.
2. Results
2.1. Altered Social Interactions of Uhrf2 Knockout Mice
Social behavior constitutes a fundamental trait in numerous species, facilitating adaptation to environmental pressures, promoting community stability, and appropriate responses to external cues [34]. Perturbations in social behavior manifest prominently across neurological disorders, including depression, autism, bipolar disorder, and dementia, thereby underscoring its overarching importance in cognition [35,36]. In their natural habitats, mice exhibit a diverse array of social behaviors, such as nest construction, evaluating potential mates, mutual grooming, and hierarchy development, which rely extensively on proficient social cognition and communication abilities. More specifically, the skill to build nests constitutes an essential capability in mice, providing thermal regulation, shelter, and a venue for reproduction [37]. Therefore, it is important to consider nest-building skills as a potential marker for cognitive function in mice. Lack of nesting behavior has been associated with cognitive deficits after brain injury, pharmacological intervention, or in transgenic mouse strains [38]. Taken together, the assessment of nest-building capacity provides vital insights into general social competence and cognition in mice.
In our study, we evaluated the nesting behavior of Uhrf2 knockout and wild-type mice by distributing cotton balls in a 12-cell grid overlay and analyzing the arrangement and collection of the cotton balls at different time points (Figure 1A). Analysis of nest size revealed a significant difference between the two groups at multiple time points, 1, 3, 5, 10, and 24 h. Specifically, Uhrf2 knockout mice had a significantly larger nest size compared to wild-type mice (p < 0.0001). This finding was further supported by a two-way RM ANOVA, which revealed a significant effect of genotype (F (1, 16) = 41.25). Importantly, the lack of a significant interaction between time and genotype (F (4, 64) = 1.35, p = 0.3) indicated that the effect of genotype on nest size remained consistent across time points (Figure 1B).
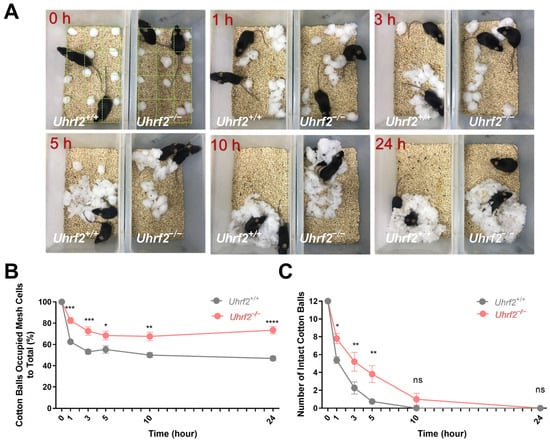
Figure 1.
Quantification of nest size and cotton ball use in Uhrf2 knockout and littermates. (A) Observational comparison of behavioral patterns between wild-type mice (littermates; left box) and Uhrf2 knockout mice (adjacent box) in the nest-building experiment. To calculate the percentage of cotton balls in the nest, we divided the number of grid cells containing cotton balls by the total number of cotton balls (12) and multiplied the result by 100. (B) Nest size of Uhrf2 knockout mice (red, n = 10) was significantly larger than that of wild-type mice (gray, n = 8) at 1, 3, 5, 10, and 24 h time points. Statistical significance of the differences was determined using Sidak’s multiple comparisons test, with p-values for each time point as follows: at 1 h, *** p = 0.0004; at 3 h, *** p = 0.0006; at 5 h, * p = 0.036; at 10 h, ** p = 0.002; at 24 h, **** p < 0.0001. (C) The number of intact cotton balls remaining in the cages of Uhrf2 knockout mice was significantly higher than in the cages of wild-type mice at 1, 3, and 5 h. The p-values obtained from the post hoc test were as follows: at 1 h, * p = 0.025; at 3 h, ** p = 0.003; at 5 h, ** p = 0.002; at 10 and 24 h, ns (not significant).
In addition, our study examined the disassembly of cotton balls, providing valuable insight into the preparation of nest-building material. Analysis of the data in Figure 1C showed that Uhrf2 knockout mice had a significantly higher number of intact cotton balls at multiple time points, indicating a decreased ability to disassemble them. A two-way RM ANOVA confirmed a significant genotype effect on the number of intact cotton balls (F (1, 16) = 8.54, p = 0.01). Sidak’s multiple comparisons tests also revealed significant group differences at 1, 3, and 5 h (see Figure 1 legend for specific values). These results provide valuable insight into the effects of Uhrf2 gene deletion on nesting behavior in mice. In addition, they suggest adverse effects on temperature regulation and imply an overall decrease in nesting ability in Uhrf2 knockout mice.
In addition to evaluating nest-building ability, we investigated the impact of Uhrf2 knockout on broader social behaviors, particularly the establishment of dominance hierarchies. For this purpose, we conducted the tube test and observed that Uhrf2 knockout mice exhibited a significant decrease in tube dominance compared to wild-type controls (Figure 2A,B). The outcomes of a two-way RM ANOVA elucidated a notable effect of genotype on winning times in the dominance tube test (F (1, 12) = 5.543, p = 0.036). However, the day of testing did not have a significant impact on the observed results (F (1.88, 22.56) = 0.87, not significant). Remarkably, Uhrf2 knockout mice consistently displayed lower winning scores on the first day of testing and throughout the two consecutive days (Figure 2C). These findings indicate that Uhrf2 knockout leads to diminished social dominance in comparison to littermates, potentially attributed to reduced aggression or heightened social anxiety.
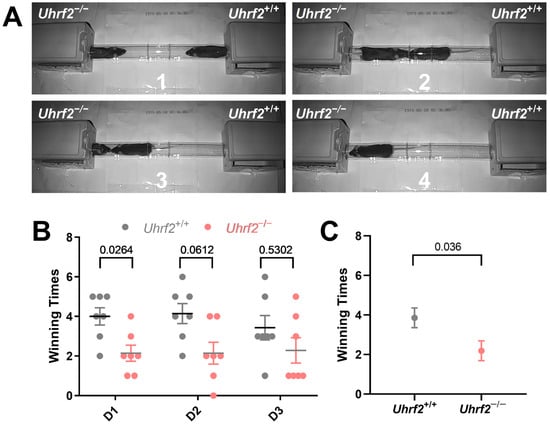
Figure 2.
Uhrf2 gene deletion impairs tube dominance behavior. (A) Observational comparison of behavioral patterns between Uhrf2 knockout mice (left panel) and wild-type mice (right panel) in the tube dominance test (1 to 4: time sequence of wild-type mice winning). (B) The bar graph presents individual winning times of mice per day, as well as the average and standard error of the mean (SEM), illustrating the tube dominance test results for wild-type mice (gray, n = 7) and Uhrf2 knockout mice (red, n = 7). On day one, Uhrf2 knockout mice had significantly lower winning times than the wild-type mice. The values of Sidak’s test for multiple comparisons are shown above the brackets in the graph. (C) Uhrf2 knockout mice exhibited significantly reduced average winning times across all days compared to their wild-type littermates (unpaired t-test, t = 2.35, df = 12, p < 0.05).
In addition to investigating social dominance, we explored the effects of Uhrf2 knockout on various components of social communication, including PPI. Impaired PPI has been implicated in several neurodegenerative and psychiatric disorders, highlighting its importance in cognitive and behavioral domains [39]. PPI is based on the principle that the startle response to a loud acoustic stimulus (e.g., 120 dB) is modulated by the presence of a preceding moderately intense acoustic stimulus (Figure 3A). Although the one-way RM ANOVA analysis did not reveal any significant effect of the three different prepulse acoustic stimuli within each of the three groups on the startle response (Uhrf2+/+: F (1.96, 21.52) = 0.69, p = 0.51; Uhrf2+/−: F (1.99, 21.92) = 0.075, p = 0.93; Uhrf2−/−: F (1.85, 16.63) = 0.77, p = 0.47), the consistent variation observed across subjects suggests that the experimental conditions had a consistent effect across stimuli, regardless of the specific prepulse stimulus intensity used (Figure 3B). In addition, our results showed a significant effect of genotype on sensorimotor gating, as revealed by a one-way ANOVA analysis (F (2, 6) = 68.58, p < 0.0001). Specifically, Uhrf2−/− mice displayed a deficit in PPI compared to their heterozygous and wild-type littermates, as evidenced by a significantly lower percentage of observed PPIs (Figure 3C). These results provide further insight into the multiple roles of Uhrf2 in regulating different facets of social communication.
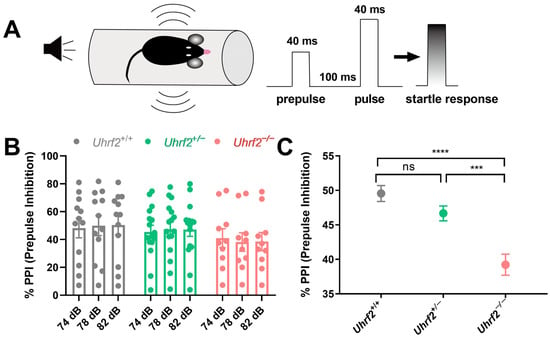
Figure 3.
Uhrf2 gene deletion results in impaired sensorimotor gating as measured by prepulse inhibition test (PPI). (A) A schematic of the PPI setup, the sequence of acoustic stimuli (S1 and S2), and the startle response measurement are presented. (B) Results of the PPI are shown for wild-type mice (gray, n = 12), heterozygous mice (green, n = 12), and Uhrf2 knockout mice (red, n = 10) at sound pressure levels of 74, 78, and 82 dB. (C) The results show a significant difference in PPI between Uhrf2+/+ and Uhrf2+/− mice compared to Uhrf2−/− mice. Sidak’s multiple comparisons yielded the following values: **** p < 0.0001, *** p = 0.0004, and ns (0.07), respectively.
In summary, the results revealed significant behavioral differences between Uhrf2 knockout mice and their wild-type littermates, suggesting a critical role for Uhrf2 in the regulation of social behaviors. The knockout mice exhibited impairments in nest-building ability, reduced dominance levels in the dominance tube test, and suppressed pre-pulse inhibition. These findings suggest that Uhrf2 and the gene expression network under its control are involved in the regulation of social behaviors, although the exact mechanisms and brain areas underlying these complex behaviors are not yet fully understood. Nonetheless, our results demonstrate that a deficiency in Uhrf2 leads to changes in social behaviors, highlighting its importance in maintaining normal social interactions.
2.2. Social Memory of Uhrf2 Knockout Mice
In addition to assessing general social behaviors, we aimed to evaluate the impact of Uhrf2 deficiency on short-term social memory, which is critically dependent on the CA2 region of the hippocampus. To achieve this, we conducted the three-chamber social interaction test, which enables the assessment of sociability and social memory in rodents [40].
During the three-day habituation phase, we measured the total distance traveled and velocity within the chambers. No significant differences were observed between wild-type littermates (n = 8) and Uhrf2 knockout mice (n = 7) on the first day. However, on the second day of habituation, homozygous knockouts exhibited increased distance traveled compared to heterozygotes (n = 6, p = 0.016), along with higher velocity compared to wild types (p = 0.032). Furthermore, on the last day of habituation, Uhrf2−/− mice exhibited greater distance and velocity compared to both heterozygous and wild-type littermates (p = 0.0011 and p = 0.018 for distance; p = 0.0003 for velocity) (see Figure 4B,C). These findings suggest that Uhrf2−/− mice display a level of hyperactivity in the habituated context.
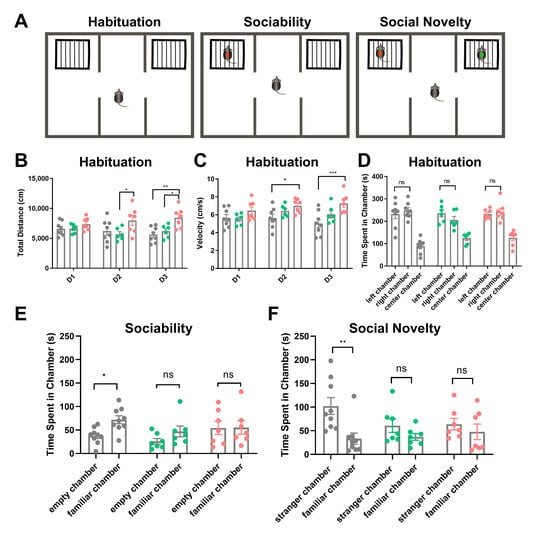
Figure 4.
Analysis of social behavior in wild-type, heterozygous, and homozygous Uhrf2 knockout mice using the three-chamber sociability and social novelty test. (A) The figure illustrates a schematic representation of the three-chamber social test. (B) The total distance traveled by mice with different genotypes was measured during the habituation period. A two-way RM ANOVA revealed a significant effect of genotype (F (2, 18) = 6.33, ** p = 0.008), but no significant effect of test day (F (2, 36) = 0.21, p = 0.82, ns). On the first day (D1), there was no significant difference in distance traveled among the three genotypes: wild-type mice (Uhrf2+/+, gray, n = 8), heterozygous mice (Uhrf2+/−, green, n = 6), and knockout mice (Uhrf2−/−, red, n = 7). However, on day 2 (D2), the heterozygous mice moved significantly shorter distances compared to the knockout mice (* p = 0.017). On day 3 (D3), the homozygous knockout mice traveled significantly longer distances than both the wild-type and heterozygous mice (** p = 0.001, * p = 0.02). (C) The velocity of mice with different genotypes was measured during the habituation period. Statistical analysis using a two-way RM ANOVA revealed a significant effect of genotype (F (2, 18) = 7.45, p = 0.004), while no significant effect of test days was observed (F (2, 36) = 1.59, p = 0.21; ns). On days 2 and 3, knockout mice traveled at a significantly higher velocity than wild-type mice (* p = 0.036, *** p < 0.0003). (D) The exploration preference of the mice during the habituation period on the test day (D4) was evaluated. A two-way RM ANOVA showed no significant effect of genotype (F (2, 18) = 3.37, p = 0.06, ns) and no significant effect of chamber (F (1, 18) = 0.01, ns) on the outcome of the test. There were no significant differences in exploration time between wild-type, heterozygous, and knockout mice in the left and right chambers (ns, not significant). (E) Sociability was assessed by measuring the amount of time mice spent exploring the familiar mouse compared to the empty cage. Wild-type mice spent significantly more time exploring the familiar mouse than the empty cage (* p = 0.043). However, no significant difference was observed between the exploration of the familiar mouse and the empty chamber in heterozygous and knockout mice (ns, not significant). A two-way repeated measures ANOVA revealed that the effect of genotype was not significant (F (2, 18) = 2.07, p = 0.16, ns), but the effect of chamber was significant (F (1, 18) = 5.16, p = 0.0357). (F) In terms of social novelty in mice, wild-type mice exhibited significantly higher levels of exploration towards the unfamiliar mouse compared to the familiar mouse (** p = 0.0025). Furthermore, no significant difference in exploration time between the unfamiliar and familiar mice was observed in heterozygous and knockout mice. A two-way repeated measures ANOVA revealed an effect of the chamber on the variations (F (1, 18) = 11.68, p = 0.003), but no significant effect of genotype (F (2, 18) = 0.94, p = 0.41). The p-values presented in this legend represent the results of comparisons analyzed using Sidak’s multiple comparisons.
On the fourth day, we performed sociability and social novelty tests after 10 min of habituation, now with empty cages placed in the side chambers. No group differences arose in the exploration of the novel empty cages (Figure 4D). For sociability assessments, a familiar mouse was introduced into one cage and the exploration time was quantified. The wild-type mice exhibited greater exploration of the mouse cage compared to the empty cage (p = 0.043). However, Uhrf2 heterozygous and homozygous knockouts showed no preference between the two cages (Figure 4E). Notably, our social approach assay differed from the standard protocol by utilizing a familiar mouse rather than an unfamiliar stranger mouse, thereby assessing not just sociability but also the integration of social memory.
In the subsequent social novelty test, wild-type mice explored the cage with the unfamiliar mouse significantly more than the cage with the familiar mouse. Heterozygotes showed a non-significant increase in exploration of the novel mouse. Notably, Uhrf2−/− mice exhibited no difference in exploration between the unfamiliar and familiar cages (see Figure 4F).
In summary, the adapted protocol provides insight into social memory by quantifying interaction with a familiar mouse, while traditional sociability assesses preference for a stranger mouse. The results of tests assessing social memory showed that knockout mice performed significantly worse than wild-type littermates. Additionally, Uhrf2 knockout mice exhibited impaired habituation over repeated exposures. After the initial habituation period, the knockouts displayed hyperactive locomotion compared to the wild types.
2.3. Characterization of Afferent Pathway-Specific Synaptic Plasticity in Mice with Different Genotypes
The hippocampal CA2 region has emerged as an integral locus for social cognition, including roles in social memory, recognition, and preference. Using immunofluorescence against RGS14, a marker protein for CA2 [7,41], the relative position within a transversal hippocampal slice could be depicted. To ascertain if Uhrf2 knockout could perturb CA2-dependent processes, we performed in vitro electrophysiology to evaluate pathway-specific synaptic plasticity.
First, we examined basal synaptic transmission and input–output relationship at Schaffer collateral (SC) to CA2 synapses across wild-type, heterozygous, and Uhrf2 knockout mice. No significant differences arose in SC-CA2 synaptic transmission efficiency or field postsynaptic potential (fEPSP) input–output curves between groups (Figure 5A,B). Moreover, SC-CA2 paired-pulse facilitation was unaffected by Uhrf2 deletion (Figure 5C,D).
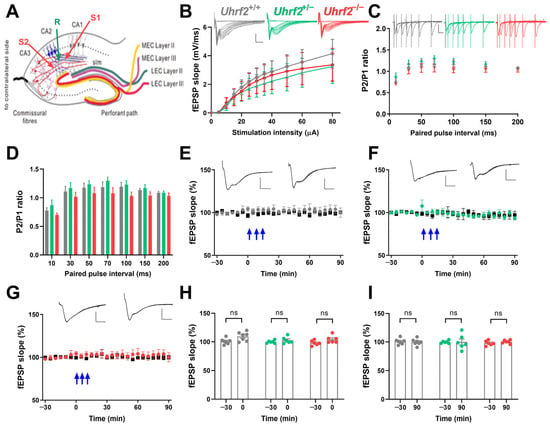
Figure 5.
Uhrf2 knockout does not affect input–output characteristics, paired-pulse facilitation, and synaptic plasticity in the SC-CA2 pathway. (A) This is a schematic diagram that illustrates the placement of electrodes in the SC-CA2 region. (B) Input–output curves were generated for the SC-CA2 synaptic transmission in Uhrf2+/+ (gray, n = 7), Uhrf2+/− (green, n = 6), and Uhrf2−/− mice (red, n = 7). fEPSP sample traces were presented with scale bars indicating an amplitude of 2 mV and a time interval of 5 ms. Statistical analysis using a two-way RM ANOVA revealed no significant differences in input–output curves among the different groups of mice (F (2, 17) = 0.13, p = 0.88). Post hoc comparisons using Sidak’s test also did not show any significant differences between groups per stimulation intensity (all p-values > 0.7). (C) Paired pulse facilitation fEPSP traces and ratios of P2/P1 fEPSP slopes were recorded for Uhrf2+/+ (gray, n = 8), Uhrf2+/− (green, n = 6), and Uhrf2−/− (red, n = 7) mice. Scale bars are 1 mV/10 ms. (D) No significant differences in paired-pulse facilitation were observed between Uhrf2+/+, Uhrf2+/−, and Uhrf2−/− mice (two-way RM ANOVA: F (2, 18) = 1.78, p = 0.12). Post hoc group comparisons per stimulus interval also revealed no significant differences (all p-values > 0.1). (E) LTP induction was observed in wild-type Uhrf2+/+ mice (gray, n = 7) following three tetanic stimulations (3 × 100 Hz, indicated by blue arrows). (F) LTP induction was performed in Uhrf2+/− mice (green, n = 6) after three tetanic stimulations. (G) LTP induction was also performed in Uhrf2−/− mice (red, n = 6). (H) No detectable effect of genotype on fEPSP slope potentiation immediately after tetanization was observed in Uhrf2+/+, Uhrf2+/−, and Uhrf2−/− mice (two-way RM ANOVA: F (2, 16) = 2.29, p = 0.14). Post hoc comparisons showed a significant difference between baseline and time 0 in wild-type mice (p = 0.044), while no significant differences were detectable in heterozygous and homozygous mice. (I) At the 90 min time point, there was no genotype effect on fEPSPs compared to baseline in Uhrf2+/+, Uhrf2+/−, and Uhrf2−/− mice (two-way RM ANOVA: F (2, 16) = 0.12, p = 0.89). Post hoc comparisons also showed no significant differences between baseline and 90 min time points for any group (all p-values > 0.9). Representative fEPSP traces at −30 and 90 min are shown with 2 mV/5 ms scale bars.
We examined the induction of long-term potentiation (LTP) using 3 × 100 Hz stimulus trains and found no differences in SC-CA2 fEPSP potentiation between genotypes (Figure 5E–I). In addition, we performed a two-way RM ANOVA to directly compare the time course of the fEPSP slope after tetanization among the three groups (Figure 5E–G). The analysis revealed no significant effect of genotype on fEPSP slope over time (F (2, 16) = 0.61, p = 0.56), and no significant effect of time (F (2.94, 47.04) = 2.63, p = 0.062) on fEPSP values. These results support previous studies indicating that SC-CA2 synapses lack activity-dependent plasticity [15,16,18].
Previous studies have shown that synapses form between entorhinal cortex (EC) afferents and CA2 neurons exhibit long-term potentiation (LTP) similar to Schaffer collateral–CA1 synapses [15]. Furthermore, reducing the activity of the entorhinal cortex afferents to the hippocampus decreased the extent of memory formation for new social encounters, highlighting their significance in enduring social interactions.
Therefore, we investigated the impact of Uhrf2 knockout on EC-CA2 plasticity. Although input–output curves were similar across all groups (Figure 6A,B), EC-CA2 paired-pulse facilitation was significantly reduced in Uhrf2−/− mice (Figure 6C,D). This implies that Uhrf2 selectively influences short-term plasticity dynamics at EC-CA2 synapses.
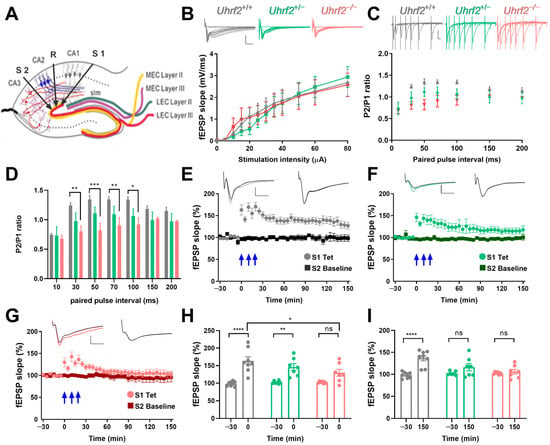
Figure 6.
Uhrf2 knockout impairs paired-pulse facilitation and LTP at EC-CA2 synapses. (A) Schematic showing electrode placement in the EC-CA2 pathway. (B) Input–output curves show the dynamics of the fEPSP slope in response to stimulation intensity (0–80 µA) for Uhrf2+/+ (gray, n = 10), Uhrf2+/− (green, n = 6), and Uhrf2−/− mice (red, n = 7). The input–output curves show no significant differences among the three groups of mice (two-way RM ANOVA: F (2, 20) = 0.025, p = 0.97). Scale bars: 2 mV/5 ms. (C) fEPSP traces recorded in response to paired-pulse facilitation stimulation using 40% of the maximum slope as the stimulation intensity in Uhrf2+/+ (gray, n = 8), Uhrf2+/− (green, n = 6), and Uhrf2−/− mice (red, n = 7). The scale bar is 1 mV/10 ms. (D) At 30, 50, 70, and 100 ms intervals, paired-pulse facilitation was significantly lower in Uhrf2 knockout mice compared to wild-type mice. A two-way RM ANOVA revealed a significant effect of genotype (F (2, 18) = 4.70, p = 0.021). Post hoc Sidak’s tests for paired-pulse intervals of 30 ms (** p = 0.0090), 50 ms (*** p = 0.0007), 70 ms (** p = 0.008), and 100 ms (* p = 0.015) showed significant differences between knockout and wild-type mice. (E) Induction of LTP was also recorded in hippocampal slices from Uhrf2+/+ mice (gray, n = 8) using three tetanic stimulations (3 × 100 Hz, indicated by blue arrows). (F) Induction of LTP was recorded in Uhrf2+/− mice (green, n = 7). (G) The induction of LTP was also observed in Uhrf2 knockout mice (red, n = 7). Scale bars: 1 mV/5 ms. (H) The fEPSP slope in EC-CA2 increased significantly after tetanization (0 min) in wild-type and heterozygous mice, whereas no significant difference was observed in knockout mice compared to baseline. The effect of genotype at this time point was not significant (F (2, 19) = 1.77, p = 0.20, ns). However, baseline to first value after tetanization was significant for wild-type mice (**** p < 0.0001) and heterozygous mice (** p = 0.002) but not for homozygous knockout mice (ns, p = 0.06). Additionally, the first value after tetanization in wild-type mice differed significantly from that in knockout mice (* p = 0.011). (I) There was a significant effect of genotype on fEPSP potentiation at 150 min after tetanization, based on a two-way RM ANOVA (F (2, 19) = 4.5, p = 0.025). In addition, fEPSP potentiation was significantly higher than baseline only in wild-type mice (**** p < 0.0001) but not in recordings from heterozygous or homozygous knockout mice. There were also significant differences between wild-type and heterozygous mice (*** p = 0.0002) and between wild-type and homozygous knockout mice (* p = 0.016). Brackets and asterisks in the graphs represent Sidak’s multiple comparison p-values: **** p < 0.0001, ** p < 0.002, and ns (not significant). The figure legend also provides the specific p-values of the post hoc tests for comparisons.
Next, we investigated the long-term potentiation (LTP) in the EC-CA2 pathway. To directly compare the time course of the fEPSP slope after tetanization over 150 min among the three groups, we examined the data as shown in Figure 6E–G. The comparison was made using a two-way RM ANOVA, which revealed a significant effect of genotype on the fEPSP slope over time (F (2, 19) = 6.377, p = 0.0076).
A statistical comparison of the fEPSP slopes at different time points revealed that immediately after tetanization, both wild-type and heterozygous mice showed a significant increase in the slope compared to the baseline. However, in knockout mice, there was no significant difference between the fEPSP slopes of the tetanized synaptic input and the control input (Figure 6H). At the 150 min mark, the fEPSP slope in wild-type mice remained significantly higher than the baseline. In contrast, the slopes of heterozygous and knockout mice returned to baseline levels (Figure 6I).
The different response profiles of the fEPSP slope over time demonstrate how genetic variations influence synaptic plasticity. These results, depicted in Figure 6H,I, underscore the significance of genotype in shaping the dynamics of fEPSP slope potentiation and provide insights into the physiological implications of the observed genetic variation.
Taken together, these results suggest Uhrf2 knockout spares SC-CA2 synaptic transmission and plasticity, while selectively disrupting plasticity processes localized to the EC-CA2 pathway. These results shed light on the role of Uhrf2 in the hippocampus and suggest that Uhrf2 may be involved in the regulation of synaptic plasticity and information processing in the brain. Further elucidation of the molecular underpinnings of this effect could have important implications for comprehension of Uhrf2-mediated regulation of hippocampal learning and memory.
2.4. Transcriptomic Analysis Reveals Altered Gene Expression in the Hippocampus of Uhrf2 Knockout Mice
To gain insight into potential molecular mechanisms underlying the influence of Uhrf2 on cognition and synaptic function, we performed RNA sequencing on hippocampal tissue from Uhrf2 knockouts and wild-type littermates. Subsequent bioinformatic analysis identified differentially expressed genes (DEGs) and affected pathways resulting from Uhrf2 deletion. As expected, Uhrf2 exon 1 expression was absent in knockouts, as confirmed by analysis of reads from the RNA-seq data (Supplemental Figure S2). Expression of the housekeeping genes Gapdh and Actb remained unchanged (Figure 7A,B). Numerous DEGs arose from Uhrf2 knockout, including 421 upregulated and 129 downregulated genes (Figure 7C). Further examination highlighted DEGs involved in synaptic transmission and plasticity, such as Gnb3, Hrh1, Gabra2, and Gabra3 (Figure 8A,B).
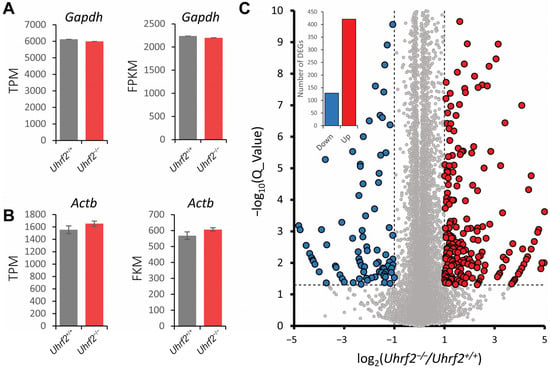
Figure 7.
Identification of differentially expressed genes (DEGs) in the hippocampus of Uhrf2 knockout mice using RNA sequencing. (A) The TPM (transcripts per million) and FPKM (fragments per kilobase million) expression of the housekeeping gene Gapdh in Uhrf2+/+ (gray, n = 3) and Uhrf2−/− (red, n = 3) mice did not show a significant difference. (B) Similarly, the TPM and FPKM expression of the housekeeping gene Actb in Uhrf2+/+ (gray, n = 3) and Uhrf2−/− mice (red, n = 3) show no significant difference. (C) The volcano plot displays the distribution of DEGs caused by Uhrf2 knockout, with upregulated genes in red and downregulated genes in blue. Only genes with Q values < 0.05 (−log10(0.05) = 1.3, horizontal dashed line) and log2(Uhrf2−/−/Uhrf2+/+) < −1 or > 1 (vertical dashed line) were considered DEGs. Genes that did not meet these criteria are shown as gray circles. The inset bar graph shows the total number of downregulated and upregulated DEGs associated with Uhrf2 knockout. Notably, some of these DEGs have established links to cognition. For instance, Gnb3 encodes a G protein subunit implicated in neurotransmitter signaling and risk for neurological disorders. Studies have shown that the Gnb3 gene is associated with the onset of neurological disorders such as depression and dementia.
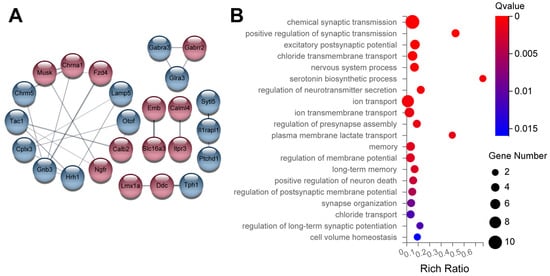
Figure 8.
Examples of DEGs related to synaptic transmission and regulation of synaptic plasticity. (A) The diagram shows genes that are DEGs and associated with synaptic transmission and regulation of synaptic plasticity. Red circles represent upregulated genes, while blue circles indicate downregulated genes. Notable highly integrated genes include Gnb3 and Hrh1. The numbers in (B) represent the number of DEGs in each enriched category, as represented by the size of the Gene Ontology pathway (GO pathway) circle. The Q values are color-coded to display the ratio of identified DEGs to the total number of genes in the category index. The DEGs are primarily associated with chemical synaptic transmission, excitatory postsynaptic potential, chloride ion transport, vesicle transport, and regulation of membrane potential.
Some of the DEGs have established links to cognition. For example, Gnb3 encodes a G protein subunit involved in neurotransmitter signaling and has been associated with an increased risk of neurological disorders [42]. Studies have shown that the Gnb3 gene is related to the occurrence of neurological diseases, such as depression and dementia [43].
The histamine receptor Hrh1 plays a role in memory, and its deletion disrupts hippocampal plasticity [44,45]. Gabra2 and Gabra3 encode subunits of the GABAA receptor, which is the primary receptor for the GABA neurotransmitter in the brain. GABA is the main inhibitory neurotransmitter in the central nervous system of mammals. GABAA receptors, along with excitatory glutamate, play a crucial role in maintaining the balance between inhibitory and excitatory responses, which is essential for the normal functioning of the brain. When activated, GABAA receptors selectively transmit chloride ions, leading to the hyperpolarization of neurons and regulating synaptic plasticity [46,47].
RNA-sequencing analysis provides novel insights into the molecular mechanisms underlying the effect of Uhrf2 knockout on synaptic transmission and plasticity. The identification of specific DEGs related to synaptic plasticity and neurotransmission opens new avenues for further investigation into the role of Uhrf2 in hippocampal function and its potential relevance to neurological diseases.
3. Discussion
Our findings elucidate the multiple roles of Uhrf2 in modulating social behavior and memory processes in mice. Behavioral assays revealed that Uhrf2 deficiency results in deficits in multiple domains of social interaction, including nest-building ability, dominance hierarchy establishment, and sensorimotor gating as measured by PPI. In addition, assessments relying on intact short-term social memory, mediated in part by entorhinal cortex-to-hippocampal CA2 neuronal circuits, revealed additional deficits resulting from Uhrf2 deletion. Electrophysiological analyses supported aberrant synaptic plasticity specific to the entorhinal cortex–CA2 pathway as a potential mechanism linking Uhrf2 to the regulation of social cognition. However, RNA sequencing revealed that Uhrf2 knockout also affected broad gene expression networks involving vesicle trafficking, transcriptional regulation, and neuroepithelial cell differentiation pathways. This suggests that the collective effects of Uhrf2 loss on social behavior, memory, and synaptic plasticity likely result from multiple effects on molecular processes beyond a single mechanism.
3.1. Uhrf2 Deletion Results in Reduced Nest Building Speed, Tube Dominance, and Prepulse Inhibition
Our studies revealed that Uhrf2 deletion results in reduced nest-building ability, social dominance, and sensorimotor gating quantified by the prepulse inhibition test in mice. Nest building facilitates thermoregulation, reproduction, and shelter construction in mammals, and deficiencies are associated with brain dysfunction.
Specifically, we found that Uhrf2-deficient mice had a reduced ability to disassemble cotton balls at different intervals and built a larger nest with the same number of cotton balls, suggesting reduced nesting behavior. While some research suggests that disruption of the UPS could disrupt nesting behavior, the links between Uhrf2 and nesting ability remain unexplored. Similarly, despite reduced dominance in the tube test in Uhrf2 knockouts, clear links between Uhrf2 and dominance have yet to emerge. Of note, proper cognitive and behavioral function relies on complex, interconnected brain networks rather than isolated regions. The broad behavioral perturbations observed following Uhrf2 knockout may result from diverse effects on neural circuits rather than singular mechanisms. Uhrf2 deletion also attenuated prepulse inhibition, which has been associated with neuropsychiatric conditions [48,49]. This is consistent with the spectrum of social behavioral deficits seen with Uhrf2 loss. Taken together, our behavioral findings demonstrate that Uhrf2 contributes broadly to the orchestration of social functioning in mice, potentially through effects on multiple neural pathways involved in these diverse behaviors. Elucidating the precise brain regions and molecular interactions that govern Uhrf2′s role is a goal for future mechanistic dissection.
3.2. Uhrf2 Deletion Impairs Social Novelty
In addition to general social behaviors, our studies revealed that Uhrf2 knockout mice failed to habituate to the test apparatus over three repeated exposures. The knockouts also exhibited increased locomotor activity compared to wild-type controls during the habituation phase. Concerning short-term social memory, Uhrf2 deletion significantly impaired performance compared to wild-type littermates. Interestingly, we did not find any previous publications explicitly linking Uhrf2 loss to habituation, hyperlocomotion, or social memory deficits in mice. However, accumulating evidence highlights the hippocampal CA2 subregion as a critical site for social novelty and memory consolidation, with dysfunction potentially contributing to social impairments in several brain disorders [10]. In particular, lateral projections from the entorhinal cortex to CA2 represent a central circuit for stabilizing social memories, with plasticity in this pathway regulating social cognition [17,50]. Therefore, the potential changes in synaptic plasticity in Uhrf2 knockout mice within the EC-CA2 circuit may be involved in the regulation of social memory.
3.3. Uhrf2 Deletion Attenuates LTP in EC-CA2 Synapse
While Uhrf2 deletion spares SC-CA2 synaptic transmission and plasticity, our studies revealed a selective disruption of plasticity mechanisms localized to the entorhinal cortex–CA2 pathway. Specifically, electrophysiological studies revealed a selective disruption of LTP at EC-CA2 synapses in Uhrf2 knockouts, while LTP at SC-CA2 synapses remained absent. We found that Uhrf2 knockout did not alter SC-CA2 input–output relationships or paired-pulse facilitation, indicating that basal synaptic transmission was unaffected. However, Uhrf2 loss significantly suppressed short-term plasticity dynamics measured by paired-pulse responses at EC-CA2 synapses, suggesting a deficit in presynaptic function. Critically, we observed a robust inhibition of both the induction and maintenance phases of long-term potentiation exclusively at EC-CA2 synapses in Uhrf2 knockouts. This suggests a critical role for Uhrf2 in enabling long-term synaptic strengthening, which is thought to underlie memory formation within the EC-CA2 circuit. While studies directly examining the involvement of ubiquitin pathways in EC-CA2 plasticity are limited, more general evidence suggests that UPS components can modulate synaptic plasticity and cognition through transcriptional and protein turnover mechanisms [51]. Therefore, our findings provide the first evidence implicating Uhrf2 in the coordination of EC-CA2 plasticity and warrant further mechanistic studies to elucidate its potential role in enabling the long-lasting synaptic strengthening hypothesized to underlie memory formation within this circuit.
3.4. Changes of Gene Expression by Uhrf2 Knockout
Uhrf2 knockout resulted in the upregulation of 421 genes and the downregulation of 129 genes. The results of the study, which used RNA sequencing technology, indicate that Uhrf2 knockout leads to DEGs related to vesicle trafficking and transcription factors. Concerning vesicle trafficking, Uhrf2 downregulation leads to a decrease in Actc1 and Myl2, which encode actin and myosin, respectively. Actin is an essential structural protein that plays a critical role in cell functions such as division, migration, and vesicle trafficking [52]. Similarly, ACTC1 is one of the isoforms of myosin involved in early mammalian neurodevelopment, particularly in brain and glial cells (Goggolidou [53,54,55]. In addition, ACTC1 regulates vesicle trafficking and interacts with calcium–phospholipid-binding proteins [56,57]. The Myl2 gene encodes myosin regulatory light chain, which specifically binds to myosin heavy chain, an essential component of myosin filaments. MYL2 is also involved in the AMPK pathway, which regulates energy balance in eukaryotes, and abnormal expression of MYL2 is associated with schizophrenia [58,59]. The study suggests that UHRF2 may regulate the transport and release of presynaptic vesicles by modulating ACTC1 and MYL2. The deletion of Uhrf2 also affects the expression of genes related to neuroepithelium development. During early brain development, neuroepithelial cells interact with cerebrospinal fluid, which promotes brain expansion and development. The flow of cerebrospinal fluid is crucial for clearing metabolic waste from the brain, eliminating inflammatory factors, and regulating the physiological status of mice through neuroimmune responses [60,61,62,63,64]. Six3, which regulates neural development by interacting with signaling factors, is significant in mRNA sequencing results and has a significant increase in the content of SIX3 protein in Uhrf2 knockout mice. Although there is no direct evidence that Six3 is involved in learning and memory, studies have shown that Six3 deficiency hinders the formation of dopamine receptor type 2-expressing striatal medium spiny neurons (D2-MSNs) that express dopamine receptor 2 (DRD2) in the striatum. Activation of D2-MSNs in the nucleus accumbens reduces cocaine-induced conditioned place preference memory [65,66,67,68].
We acknowledge that our study has several limitations. In particular, we did not examine the downstream functional consequences of differential expression of genes related to vesicle trafficking, transcription factors, or neuroepithelial cell differentiation. In addition, we did not directly examine the role of UHRF2-mediated transcriptional changes in memory processes, particularly in the CA2 area of the hippocampus. However, our integrative approach combining behavioral, electrophysiological, and genomic data provides novel evidence for the potential involvement of UHRF2 in modulating learning and memory capacities that support social cognition. Notably, UHRF2 appears to play a larger role in regulating hippocampal function through intricate effects on gene expression networks related to synaptic plasticity than previously recognized.
Moreover, the observed behavioral impairments in Uhrf2 gene-deleted mice may involve contributions from motor and sensory alterations. Our RNA sequencing analysis revealed potential mechanisms related to GABA synthesis that affect the excitatory–inhibitory balance in neuronal inputs. Perturbations in this balance are associated with neurological disorders and changes in social behavior, such as those seen in schizophrenia. In addition, we found increased hyperactivity, suggesting motor dysfunction. However, pinpointing specific physiological mechanisms at the systemic level is challenging due to substantial changes in overall gene expression. Further studies are needed to understand the intricate relationship between gene expression changes, motor and sensory alterations, and the resulting behavioral phenotypes in Uhrf2 knockout mice. Overall, our study highlights the multidimensional nature of the observed behavioral impairments in Uhrf2 knockout mice.
Increased trimethylation of histone H3 lysine 9 in hematopoietic stem/progenitor cells in Uhrf2−/− mice has been reported, indicating impaired repopulating ability and functioning of hematopoietic progenitors [69]. Additionally, Uhrf2 accumulation has been observed in retinal progenitor cells (RPCs), and its conditional deletion resulted in reduced 5hmC levels, altered gene expressions, and disruption of retinal cell proliferation and differentiation [70]. Furthermore, Uhrf2 has been implicated in the ubiquitination and degradation of nuclear aggregates containing polyglutamine repeats, such as seen in Huntington’s disease and related polyglutamine diseases [71]. These findings highlight the potential effects of Uhrf2 beyond the hippocampus, encompassing alterations in hematopoietic cells, retinal progenitor cells, protein degradation processes, synaptic transmission, and transcriptional regulation.
4. Materials and Methods
4.1. Ethics Statement
We made efforts to minimize animal usage. All experiments adhered to the guidelines of the Institutes of Brain Science and State Key Laboratory of Medical Neurobiology of Fudan University, Shanghai, China, and were approved by the Institutional Animal Care and Use Committee of Fudan University, Shanghai Medical College (approval Nr. 31320103906).
4.2. Animals
To characterize the effects of genome-wide Uhrf2 knockout, we utilized a mutant mouse strain with Uhrf2 deletion generated via CRISPR-Cas9 targeting the start codon in exon 1. This mouse strain was previously created and validated by Dr. Dali Li’s group at East China Normal University [72,73]. Founder mice were kindly provided by Dr. Dali Li. Breeding colonies were maintained at the Institutes of Brain Science, Fudan University. Experimental animals included Uhrf2−/− knockouts, as well as Uhrf2+/− and Uhrf2+/+ littermate controls derived from Uhrf2+/− x Uhrf2+/− crosses. Genotyping utilized PCR with the forward primer CCATTTCTTGCTCACGCCAG and the reverse primer TCGGGCCTTACATCGAAGAG (Chen et al., 2017). Mice aged 6–8 months were group housed, with four mice per cage. They were kept in a 12 h reversed light/dark cycle at ambient temperature, with ad libitum access to food and water.
4.3. Observation of Nest-Building Behavior
To evaluate nest-building behavior, we distributed twelve 1 cm cotton balls evenly across the cage floor. The arrangement of the cotton balls within a 12-cell grid overlay was analyzed at different time points, following the methods described by Deacon, Jirk of et al., and Li et al. [37,74,75]. The analysis included assessing the amount of biting and shredding of the cotton balls, as well as their collection in a corner, within the 12-cell grid (Figure 1). To calculate the percentage of cotton balls in the nest, we divided the number of grid cells containing cotton balls by the total number of cotton balls (12) and multiplied the result by 100.
4.4. Social Tube Dominance Test
The tube test, also known as the social tube dominance test, is a widely used experimental paradigm to assess social hierarchies in mice [76,77]. A transparent tube was selected with a diameter that allowed forward and backward movement, enabling the evaluation of social hierarchies. During the habituation period, mice were given three consecutive days to freely explore the tube. On one of the competition days, when both mice entered the tube from opposite entrances, the dominant mouse displacing the other mouse scored one winning time (point), while the displaced mouse scored 0 points. To ensure fairness, we employed a round-robin design to semi-randomize pairings, guaranteeing that each wild-type (WT) mouse would compete against each knockout (KO) mouse. As a result, every mouse participated in a total of seven runs, with a new run beginning against a randomly selected competitor once all other mice in the group had finished their contests. Therefore, a maximum of seven wins per mouse per day could be achieved. However, there were instances where both mice remained in the tube, resulting in neither mouse receiving a victory point. This process was repeated over the next two days.
4.5. Prepulse Inhibition Test
Prepulse inhibition (PPI) is a well-established experimental paradigm used to study sensorimotor gating in rodents [49,78,79]. In this experiment, we induced and quantified PPI using acoustic stimuli and a mouse startle response recording system. The experiment included two types of acoustic stimuli, referred to as S1 and S2, which had different sound intensities. The PPI experiment consisted of eight trials, with each trial involving the presentation of S1 and S2 stimuli at a specific dB combination. Trial 2 served as a baseline measurement with S1 at 0 dB and S2 at 120 dB. Trials 4, 6, and 8 induced prepulse inhibition with S1 levels of 74, 78, and 82 dB, respectively, while S2 remained at a fixed intensity of 120 dB. In trials 3, 5, and 7, S2 was set to 0 dB and S1 was set to 74, 78, and 78 dB, respectively. These trials were designed to evaluate prepulse inhibition at different sound intensities by measuring the startle response of the mice. Each trial was randomly presented six times throughout the experiment. The PPI was calculated by comparing the mean startle response values of S2 in trials 4, 6, and 8 (averaging the values from the six repetitions of each trial) with the baseline value from trial 2. The percentage PPI for a given S1–S2 combination in trials 4, 6, and 8 was determined using the formula: %PPI = 100 − (mean of S2/mean of baseline) × 100%.
4.6. Three-Chamber Tasks: Sociability and Social Memory
We followed the previously published methods with minor modifications [40,80]. Briefly, we habituated the mice for three consecutive days by allowing them to move freely in the 3-chamber box for 20 min. The testing phase consisted of a final 10 min of habituation, a 10 min sociability test with a familiar mouse in one chamber, and a 10 min social novelty test with the familiar mouse in a cage of one chamber and an unfamiliar mouse in another cage in the opposite chamber. This study measured the extent of exploration and interaction with cages through video analysis using a tracking software package (EthoVision 14, Noldus (Beijing) Information Technology Co. Ltd., Beijing, China). The total time spent by the animals in the vicinity (within 2 cm) of the cages in either the left or right chamber was calculated by summing the respective time periods during which the animal’s nose tip was detected within the specified distance range.
4.7. Preparation of Acute Hippocampal Slices
The preparation of acute hippocampal slices followed previously published methodologies [26,75]. After anesthetizing the animal using isoflurane, the brain was removed immediately and immersed in ice-cold artificial cerebrospinal fluid (ACSF) composed of the following (in mM): 124 NaCl, 4.9 KCl, 1.2 KH2PO4, 25.6 NaHCO3, 1.3 MgSO4, 2.5 CaCl2, 10 d-glucose, and previously bubbled with 95% O2 and 5% CO2. Transverse slices (350 μm thick) were cut perpendicularly to the long axis of the hippocampus with a vibratome (VT-1200, Leica, Germany) and incubated for at least 2 h after slicing in a custom-made interface-type recording chamber at 32.5 °C under the constant perfusion with carbogenated ACSF at a flow rate of 4 mL/min.
4.8. Field Excitatory Postsynaptic Potentials (fEPSPs) Recording
The field excitatory postsynaptic potentials (fEPSPs) were evoked by stimulation with biphasic rectangular current pulses (100 μs/polarity) in a range of 15–25 μA through tungsten electrodes (A-M Systems) [18,75]. Stimulation was applied either at Schaffer collateral fibers for SC-CA2 recording or at axon terminals from the entorhinal cortex for EC-CA2 recording. The fEPSPs were recorded from the CA2 region using stainless steel electrodes (5 MΩ, A-M Systems) placed either in the stratum radiatum or stratum lacunosum–moleculare. The recording electrode (Rec) was positioned between the two independent stimulation inputs (S1 and S2) along the two different layers (Supplementary Figure S1A,B for location of CA2). The strength of synaptic transmission was determined by measuring the slope of the fEPSP. The stimulation intensity was set to 40–50% of the maximum fEPSP response. The size of fEPPSs was monitored by stimulation every 5 min as an average of four sweeps, with 10 s intervals between each sweep, throughout the experiment. Field potentials were recorded and digitized at a sampling frequency of 10 kHz using a CED 1401 plus AD/DA converter (Cambridge Electronics Design, Cambridge, UK). LTP was induced by a tetanization (TET) protocol consisting of three trains of 100 stimuli at 100 Hz separated by 10 min.
4.9. Immunofluorescence
For immunofluorescence, animals were anesthetized with 4% isoflurane and acute hippocampal slices were prepared as described in the section on slice preparation. The slices were then placed in 4% paraformaldehyde for 1 h at room temperature. After washing with PBS, the slice was transferred to a 30% sucrose solution in PBS and left overnight at 4 °C. Sections (40 μm) were cut with a cryotome (Leica Biosystems, Wetzlar, Germany). The sections were then mounted and permeabilized with 0.3% Triton X-100 in PBS for 30 min, and nonspecific binding sites were blocked with 5% goat serum in PBS for two hours at room temperature. Sections were then incubated with primary antibody (anti-RGS14 1:200; Abcam, Waltham, MA, USA) overnight at 4 °C. After three washes with PBS, sections were incubated with a species-specific secondary antibody (goat anti-rabbit Alexa 546, 1:400; Abcam) in 5% goat serum for 1 h. After application of DAPI in 0.01 M PBS (100 ng/mL, 28718-90-3, Roche, Switzerland) for 5 min and three additional washes with PBS, the sections were mounted onto slides (Fluoromount Aqueous Mounting Medium, Sigma, Livonia, MI, USA), and fluorescence images were captured using a fluorescence microscope system.
4.10. RNA Sequencing and Analysis
Total RNA was isolated from the hippocampi of Uhrf2 knockout and wild-type littermate mice using TRIzol reagent (ThermoScientific, Shanghai, China) according to the manufacturer’s instructions. RNA sequencing was performed by the Beijing Genomics Institute. RNA sequencing libraries were prepared according to the Illumina TruSeq protocol v2 (Illumina, San Diego, CA, USA). Gene expression levels were determined using edgeR v3.32.1 software (https://bioconductor.org/) with detection rate adjustment and reported as fragments per kilobase of transcript per million mapped reads. A gene was considered to be expressed if it had fragments per kilobase per million greater than one. For a gene to be considered differentially expressed, it had to meet two criteria: a greater than 2.0-fold change in expression level and a p-value < 0.05 [81,82].
4.11. Statistical Analysis
Data are presented as mean ± standard error of the mean (SEM). Statistical analysis was performed using GraphPad Prism 8. For comparisons between more than two groups with a single independent variable, ANOVA was used. In the case of two independent variables, two-way RM ANOVA was used. Any additional statistical methods used are mentioned in the text. A p-value less than 0.05 was considered to indicate a statistically significant difference between groups or an effect on independent variables. The significance values of the tests performed are given in the text of the figure legends and/or in the graphs themselves.
5. Conclusions
In summary, our behavioral, electrophysiological, and transcriptomic findings implicate UHRF2 in the regulation of social interaction, memory, and synaptic potentiation in the entorhinal cortex–CA2 pathway. While further research is imperative to elucidate the precise mechanisms involved, this study provides evidence that UHRF2 likely contributes to the modulation of learning, memory, and goal-directed behaviors related to social functioning through coordinated effects on multiple molecular pathways. Targeted investigation of these complex gene regulatory networks mediated by UHRF2 may provide novel insights into neurological disorders characterized by social dysfunction.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25031543/s1.
Author Contributions
Conceptualization, Y.Z., C.L. and T.B.; methodology, Y.Z., C.L., F.Z. and Y.Y.; validation, Y.Z., C.L., T.B. and J.Z.; formal analysis, Y.Z. and C.L.; investigation, Y.Z. and C.L.; resources, H.P., J.Z. and T.B.; data curation, Y.Z., C.L., F.Z. and Y.Y.; writing—original draft preparation, Y.Z., C.L. and T.B.; writing—review and editing, Y.Z., C.L., F.Z., Y.Y. and T.B.; visualization, Y.Z., C.L. and Y.Y.; supervision, J.Z. and T.B.; project administration, T.B.; funding acquisition, T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China: 31871076 to TB; Shanghai Municipal Science and Technology Major Project: 2018SHZDZX01; ZJ Lab., and Shanghai Center for Brain Science and Brain-Inspired Technology: 2023.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Animal Care and Use Committee of Fudan University, Shanghai Medical College of the Institutes of Brain Science and State Key Laboratory of Medical Neurobiology of Fudan University, Shanghai, China (approval Nr. 20211028-003, and date of approval: 28 October 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary files; further inquiries can be directed to the corresponding author. The original RNA-sequencing data are available in SRA and accessible under the link: https://dataview.ncbi.nlm.nih.gov/object/PRJNA1044912.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Henriksen, E.J.; Colgin, L.L.; Barnes, C.A.; Witter, M.P.; Moser, M.-B.; Moser, E.I. Spatial Representation along the Proximodistal Axis of CA1. Neuron 2010, 68, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Baltaci, S.B.; Mogulkoc, R.; Baltaci, A.K. Molecular Mechanisms of Early and Late LTP. Neurochem. Res. 2019, 44, 281–296. [Google Scholar] [CrossRef]
- Nicoll, R.A. A Brief History of Long-Term Potentiation. Neuron 2017, 93, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Zhang, Y.; Zhang, J.; Wang, H.; Ren, B. Glutamate receptors and signal transduction in learning and memory. Mol. Biol. Rep. 2011, 38, 453–460. [Google Scholar] [CrossRef]
- Tsien, J.Z.; Huerta, P.T.; Tonegawa, S. The Essential Role of Hippocampal CA1 NMDA Receptor–Dependent Synaptic Plasticity in Spatial Memory. Cell 1996, 87, 1327–1338. [Google Scholar] [CrossRef]
- Dudek, S.M.; Alexander, G.M.; Farris, S. Introduction to the special issue on: A new view of hippocampal area CA2. Hippocampus 2023, 33, 127–132. [Google Scholar] [CrossRef]
- Zhao, F.; Behnisch, T. The Enigmatic CA2: Exploring the Understudied Region of the Hippocampus and Its Involvement in Parkinson’s Disease. Biomedicines 2023, 11, 1996. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; He, L.; Huang, A.J.Y.; Boehringer, R.; Robert, V.; Wintzer, M.E.; Polygalov, D.; Weitemier, A.Z.; Tao, Y.; Gu, M.; et al. A hypothalamic novelty signal modulates hippocampal memory. Nature 2020, 586, 270–274. [Google Scholar] [CrossRef]
- Cui, Z.; Gerfen, C.R.; Young, W.S., 3rd. Hypothalamic and other connections with dorsal CA2 area of the mouse hippocampus. J. Comp. Neurol. 2013, 521, 1844–1866. [Google Scholar] [CrossRef] [PubMed]
- Hitti, F.L.; Siegelbaum, S.A. The hippocampal CA2 region is essential for social memory. Nature 2014, 508, 88–92. [Google Scholar] [CrossRef]
- Kohara, K.; Pignatelli, M.; Rivest, A.J.; Jung, H.-Y.; Kitamura, T.; Suh, J.; Frank, D.; Kajikawa, K.; Mise, N.; Obata, Y.; et al. Cell type–specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 2014, 17, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; Brann, D.H.; Meira, T.; Siegelbaum, S.A. Input-Timing-Dependent Plasticity in the Hippocampal CA2 Region and Its Potential Role in Social Memory. Neuron 2019, 102, 260–262. [Google Scholar] [CrossRef] [PubMed]
- Meira, T.; Leroy, F.; Buss, E.W.; Oliva, A.; Park, J.; Siegelbaum, S.A. A hippocampal circuit linking dorsal CA2 to ventral CA1 critical for social memory dynamics. Nat. Commun. 2018, 9, 4163. [Google Scholar] [CrossRef] [PubMed]
- Leroy, F.; Park, J.; Asok, A.; Brann, D.H.; Meira, T.; Boyle, L.M.; Buss, E.W.; Kandel, E.R.; Siegelbaum, S.A. A circuit from hippocampal CA2 to lateral septum disinhibits social aggression. Nature 2018, 564, 213–218. [Google Scholar] [CrossRef]
- Zhao, M.; Choi, Y.-S.; Obrietan, K.; Dudek, S.M. Synaptic Plasticity (and the Lack Thereof) in Hippocampal CA2 Neurons. J. Neurosci. 2007, 27, 12025–12032. [Google Scholar] [CrossRef]
- Chevaleyre, V.; Siegelbaum, S.A. Strong CA2 Pyramidal Neuron Synapses Define a Powerful Disynaptic Cortico-Hippocampal Loop. Neuron 2010, 66, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rojas, J.; de Solis, C.A.; Leroy, F.; Kandel, E.R.; Siegelbaum, S.A. A direct lateral entorhinal cortex to hippocampal CA2 circuit conveys social information required for social memory. Neuron 2022, 110, 1559–1572.e4. [Google Scholar] [CrossRef]
- Dasgupta, A.; Baby, N.; Krishna, K.; Hakim, M.; Wong, Y.P.; Behnisch, T.; Soong, T.W.; Sajikumar, S. Substance P induces plasticity and synaptic tagging/capture in rat hippocampal area CA2. Proc. Natl. Acad. Sci. USA 2017, 114, E8741–E8749. [Google Scholar] [CrossRef]
- Dubiel, W.; Dubiel, D.; Wolf, D.A.; Naumann, M. Cullin 3-Based Ubiquitin Ligases as Master Regulators of Mammalian Cell Differentiation. Trends Biochem. Sci. 2018, 43, 95–107. [Google Scholar] [CrossRef]
- Pinto, M.J.; Tomé, D.; Almeida, R.D. The Ubiquitinated Axon: Local Control of Axon Development and Function by Ubiquitin. J. Neurosci. 2021, 41, 2796–2813. [Google Scholar] [CrossRef]
- Vriend, J.; Ghavami, S.; Marzban, H. The role of the ubiquitin proteasome system in cerebellar development and medulloblastoma. Mol. Brain 2015, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Frey, J.; Sanna, P.; Behnisch, T. Protein degradation by the proteasome is required for synaptic tagging and the heterosynaptic stabilization of hippocampal late-phase long-term potentiation. Neuroscience 2010, 169, 1520–1526. [Google Scholar] [CrossRef]
- Chen, Y.; Yuanxiang, P.; Knöpfel, T.; Thomas, U.; Behnisch, T. Hippocampal LTP triggers proteasome-mediated SPAR degradation in CA1 neurons. Synapse 2012, 66, 142–150. [Google Scholar] [CrossRef]
- Ehlers, M.D. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 2003, 6, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Karpova, A.; Mikhaylova, M.; Thomas, U.; Knöpfel, T.; Behnisch, T. Involvement of Protein Synthesis and Degradation in Long-Term Potentiation of Schaffer Collateral CA1 Synapses. J. Neurosci. 2006, 26, 4949–4955. [Google Scholar] [CrossRef]
- Yun, D.; Zhuang, Y.; Kreutz, M.R.; Behnisch, T. The role of 19S proteasome associated deubiquitinases in activity-dependent hippocampal synaptic plasticity. Neuropharmacology 2018, 133, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Krishna, K.K.; Baby, N.; Raghuraman, R.; Navakkode, S.; Behnisch, T.; Sajikumar, S. Regulation of aberrant proteasome activity re-establishes plasticity and long-term memory in an animal model of Alzheimer’s disease. FASEB J. 2020, 34, 9466–9479. [Google Scholar] [CrossRef] [PubMed]
- Krishna, K.K.; Behnisch, T.; Sajikumar, S. Modulation of the Ubiquitin-Proteasome System Restores Plasticity in Hippocampal Pyramidal Neurons of the APP/PS1 Alzheimer’s Disease-Like Mice. J. Alzheimer’s Dis. 2022, 86, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Liu, J.; Zhu, G.; Zhuang, Y.; Suo, H.; Wang, P.; Huang, D.; Xu, J.; Huang, Y.; Yu, M.; et al. Parkin overexpression ameliorates hippocampal long-term potentiation and beta-amyloid load in an Alzheimer’s disease mouse model. Hum. Mol. Genet. 2014, 23, 1056–1072. [Google Scholar] [CrossRef]
- Lu, H.; Hallstrom, T.C. The Nuclear Protein UHRF2 Is a Direct Target of the Transcription Factor E2F1 in the Induction of Apoptosis. J. Biol. Chem. 2013, 288, 23833–23843. [Google Scholar] [CrossRef]
- Irwin, R.E.; Pentieva, K.; Cassidy, T.; Lees-Murdock, D.J.; McLaughlin, M.; Prasad, G.; McNulty, H.; Walsh, C.P. The interplay between DNA methylation, folate and neurocognitive development. Epigenomics 2016, 8, 863–879. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, L.; Zeng, S.; Bai, L.; Chen, J.; Zhang, Z.; Wang, Y.; Duan, C. UHRF2 decreases H3K9ac expression by interacting with it through the PHD and SRA/YDG domain in HepG2 hepatocellular carcinoma cells. Int. J. Mol. Med. 2017, 39, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.-R.; Sun, S.-C.; Teng, S.-W.; Li, L.; Bie, Y.-F.; Yu, H.; Li, D.-L.; Chen, Z.-Y.; Wang, Y. Uhrf2 deletion impairs the formation of hippocampus-dependent memory by changing the structure of the dentate gyrus. Anat. Embryol. 2018, 223, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Sapolsky, R.M. The Influence of Social Hierarchy on Primate Health. Science 2005, 308, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Kaidanovich-Beilin, O.; Lipina, T.; Vukobradovic, I.; Roder, J.; Woodgett, J.R. Assessment of social interaction behaviors. J. Vis. Exp. 2011, 48, e2473. [Google Scholar] [CrossRef]
- Takumi, T.; Tamada, K.; Hatanaka, F.; Nakai, N.; Bolton, P.F. Behavioral neuroscience of autism. Neurosci. Biobehav. Rev. 2020, 110, 60–76. [Google Scholar] [CrossRef] [PubMed]
- Deacon, R.M. Assessing nest building in mice. Nat. Protoc. 2006, 1, 1117–1119. [Google Scholar] [CrossRef] [PubMed]
- Neely, C.L.C.; Pedemonte, K.A.; Boggs, K.N.; Flinn, J.M. Nest Building Behavior as an Early Indicator of Behavioral Deficits in Mice. J. Vis. Exp. 2019, 152, e60139. [Google Scholar] [CrossRef]
- Ferguson, B.R.; Gao, W.-J. PV Interneurons: Critical Regulators of E/I Balance for Prefrontal Cortex-Dependent Behavior and Psychiatric Disorders. Front. Neural Circuits 2018, 12, 37. [Google Scholar] [CrossRef]
- Moy, S.S.; Nadler, J.J.; Perez, A.; Barbaro, R.P.; Johns, J.M.; Magnuson, T.R.; Piven, J.; Crawley, J.N. Sociability and preference for social novelty in five inbred strains: An approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004, 3, 287–302. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Parra-Bueno, P.; Smirnov, M.S.; Lustberg, D.J.; Dudek, S.M.; Hepler, J.R.; Yasuda, R. RGS14 Restricts Plasticity in Hippocampal CA2 by Limiting Postsynaptic Calcium Signaling. eNeuro 2018, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, C.; Serretti, A. Pharmacogenetics of Major Depressive Disorder: Top Genes and Pathways Toward Clinical Applications. Curr. Psychiatry Rep. 2015, 17, 50. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Cha, J.-H.; Ham, B.-J.; Han, C.-S.; Kim, Y.-K.; Lee, S.-H.; Ryu, S.-H.; Kang, R.-H.; Choi, M.-J.; Lee, M.-S. Association between a G-protein beta 3 subunit gene polymorphism and the symptomatology and treatment responses of major depressive disorders. Pharmacogenomics J. 2004, 4, 29–33. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Yan, X.; Zeng, S.; Zhang, T.; Cheng, F.; Chen, R.; Duan, C. UHRF2 promotes DNA damage response by decreasing p21 via RING finger domain. Biotechnol. Lett. 2018, 40, 1181–1188. [Google Scholar] [CrossRef]
- Yang, A.; Daya, T.; Carlton, K.; Yan, J.H.; Schmid, S. Differential effect of clomipramine on habituation and prepulse inhibition in dominant versus subordinate rats. Eur. Neuropsychopharmacol. 2016, 26, 591–601. [Google Scholar] [CrossRef] [PubMed]
- Basavaraj, S.; Yan, J. Prepulse Inhibition of Acoustic Startle Reflex as a Function of the Frequency Difference between Prepulse and Background Sounds in Mice. PLoS ONE 2012, 7, e45123. [Google Scholar] [CrossRef] [PubMed]
- Kinkead, B.; Yan, F.; Owens, M.J.; Nemeroff, C.B. Endogenous neurotensin is involved in estrous cycle related alterations in prepulse inhibition of the acoustic startle reflex in female rats. Psychoneuroendocrinology 2008, 33, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Ayyar, P.; Ravinder, J.R. Animal models for the evaluation of antipsychotic agents. Fundam. Clin. Pharmacol. 2023, 37, 447–460. [Google Scholar] [CrossRef]
- Swerdlow, N.R.; Geyer, M.A.; Braff, D.L. Neural circuit regulation of prepulse inhibition of startle in the rat: Current knowledge and future challenges. Psychopharmacology 2001, 156, 194–215. [Google Scholar] [CrossRef]
- Pannoni, K.E.; Farris, S. LEC to CA2: A circuit to remember. Neuron 2022, 110, 1443–1445. [Google Scholar] [CrossRef]
- Tai, H.-C.; Schuman, E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008, 9, 826–838. [Google Scholar] [CrossRef] [PubMed]
- Guhathakurta, P.; Prochniewicz, E.; Thomas, D.D. Actin-Myosin Interaction: Structure, Function and Drug Discovery. Int. J. Mol. Sci. 2018, 19, 2628. [Google Scholar] [CrossRef]
- Goggolidou, P.; Soneji, S.; Powles-Glover, N.; Williams, D.; Sethi, S.; Baban, D.; Simon, M.M.; Ragoussis, I.; Norris, D.P. A chronological expression profile of gene activity during embryonic mouse brain development. Mamm. Genome 2013, 24, 459–472. [Google Scholar] [CrossRef]
- James, V.; Nizamudeen, Z.A.; Lea, D.; Dottorini, T.; Holmes, T.L.; Johnson, B.B.; Arkill, K.P.; Denning, C.; Smith, J.G.W. Transcriptomic Analysis of Cardiomyocyte Extracellular Vesicles in Hypertrophic Cardiomyopathy Reveals Differential snoRNA Cargo. Stem Cells Dev. 2021, 30, 1215–1227. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, S.; Wanibuchi, M.; Kataoka-Sasaki, Y.; Sasaki, M.; Oka, S.; Noshiro, S.; Akiyama, Y.; Mikami, T.; Mikuni, N.; Kocsis, J.D.; et al. ACTC1 as an invasion and prognosis marker in glioma. J. Neurosurg. 2017, 126, 467–475. [Google Scholar] [CrossRef]
- Gou, R.; Zhu, L.; Zheng, M.; Guo, Q.; Hu, Y.; Li, X.; Liu, J.; Lin, B. Annexin A8 can serve as potential prognostic biomarker and therapeutic target for ovarian cancer: Based on the comprehensive analysis of Annexins. J. Transl. Med. 2019, 17, 275. [Google Scholar] [CrossRef] [PubMed]
- Suresh, R.; Diaz, R.J. The remodelling of actin composition as a hallmark of cancer. Transl. Oncol. 2021, 14, 101051. [Google Scholar] [CrossRef]
- Thaiparambil, J.T.; Eggers, C.M.; Marcus, A.I. AMPK Regulates Mitotic Spindle Orientation through Phosphorylation of Myosin Regulatory Light Chain. Mol. Cell. Biol. 2012, 32, 3203–3217. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, C.; Xu, Y.; Qi, G.; Yuan, G.; Cheng, Z.; Wang, J.; Wang, G.; Wang, Z.; Zhu, W.; et al. A Two-Stage Association Study Suggests BRAP as a Susceptibility Gene for Schizophrenia. PLoS ONE 2014, 9, e86037. [Google Scholar] [CrossRef] [PubMed]
- Bothwell, S.W.; Janigro, D.; Patabendige, A. Cerebrospinal fluid dynamics and intracranial pressure elevation in neurological diseases. Fluids Barriers CNS 2019, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Cserr, H.F.; DePasquale, M.; Harling-Berg, C.J.; Park, J.T.; Knopf, P.M. Afferent and efferent arms of the humoral immune response to CSF-administered albumins in a rat model with normal blood-brain barrier permeability. J. Neuroimmunol. 1992, 41, 195–202. [Google Scholar] [CrossRef]
- Gato, A.; Desmond, M.E. Why the embryo still matters: CSF and the neuroepithelium as interdependent regulators of embryonic brain growth, morphogenesis and histiogenesis. Dev. Biol. 2009, 327, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Parada, C.; Martín, C.; Alonso, M.I.; Moro, J.A.; Bueno, D.; Gato, A. Embryonic cerebrospinal fluid collaborates with the isthmic organizer to regulate mesencephalic gene expression. J. Neurosci. Res. 2005, 82, 333–345. [Google Scholar] [CrossRef]
- Sakka, L.; Coll, G.; Chazal, J. Anatomy and physiology of cerebrospinal fluid. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2011, 128, 309–316. [Google Scholar] [CrossRef]
- Lobo, M.K.; Covington, H.E.; Chaudhury, D.; Friedman, A.K.; Sun, H.; Damez-Werno, D.; Dietz, D.M.; Zaman, S.; Koo, J.W.; Kennedy, P.J.; et al. Cell Type–Specific Loss of BDNF Signaling Mimics Optogenetic Control of Cocaine Reward. Science 2010, 330, 385–390. [Google Scholar] [CrossRef]
- Ma, P.; Song, N.-N.; Li, Y.; Zhang, Q.; Zhang, L.; Zhang, L.; Kong, Q.; Ma, L.; Yang, X.; Ren, B.; et al. Fine-Tuning of Shh/Gli Signaling Gradient by Non-proteolytic Ubiquitination during Neural Patterning. Cell Rep. 2019, 28, 541–553.e4. [Google Scholar] [CrossRef]
- Palma, V.; Ruiz i Altaba, A. Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 2004, 131, 337–345. [Google Scholar] [CrossRef]
- Xu, Z.; Liang, Q.; Song, X.; Zhang, Z.; Lindtner, S.; Li, Z.; Wen, Y.; Liu, G.; Guo, T.; Qi, D.; et al. Sp8 and Sp9 coordinately promote D2-type medium spiny neuron production by activating Six3 expression. Development 2018, 145, dev165456. [Google Scholar] [CrossRef] [PubMed]
- Sano, T.; Ueda, K.; Minakawa, K.; Mori, T.; Hashimoto, Y.; Koseki, H.; Takeishi, Y.; Ikeda, K.; Ikezoe, T. Impaired Repopulating Ability of Uhrf2−/− Hematopoietic Progenitor Cells in Mice. Genes 2023, 14, 1531. [Google Scholar] [CrossRef]
- Wang, X.; Sarver, A.L.; Han, Q.; Seiler, C.L.; Xie, C.; Lu, H.; Forster, C.L.; Tretyakova, N.Y.; Hallstrom, T.C. UHRF2 regulates cell cycle, epigenetics and gene expression to control the timing of retinal progenitor and ganglion cell differentiation. Development 2022, 149, dev195644. [Google Scholar] [CrossRef]
- Iwata, A.; Nagashima, Y.; Matsumoto, L.; Suzuki, T.; Yamanaka, T.; Date, H.; Deoka, K.; Nukina, N.; Tsuji, S. Intranuclear Degradation of Polyglutamine Aggregates by the Ubiquitin-Proteasome System. J. Biol. Chem. 2009, 284, 9796–9803. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, Q.; Duan, X.; York, P.; Chen, G.-D.; Yin, P.; Zhu, H.; Xu, M.; Chen, P.; Wu, Q.; et al. The 5-Hydroxymethylcytosine (5hmC) Reader UHRF2 Is Required for Normal Levels of 5hmC in Mouse Adult Brain and Spatial Learning and Memory. J. Biol. Chem. 2017, 292, 4533–4543. [Google Scholar] [CrossRef]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X.; et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Jirkof, P.; Fleischmann, T.; Cesarovic, N.; Rettich, A.; Vogel, J.; Arras, M. Assessment of postsurgical distress and pain in laboratory mice by nest complexity scoring. Lab Anim. 2013, 47, 153–161. [Google Scholar] [CrossRef]
- Li, D.; Jing, D.; Liu, Z.; Chen, Y.; Huang, F.; Behnisch, T. Enhanced Expression of Secreted α-Klotho in the Hippocampus Alters Nesting Behavior and Memory Formation in Mice. Front. Cell. Neurosci. 2019, 13, 133. [Google Scholar] [CrossRef]
- Fan, Z.; Zhu, H.; Zhou, T.; Wang, S.; Wu, Y.; Hu, H. Using the tube test to measure social hierarchy in mice. Nat. Protoc. 2019, 14, 819–831. [Google Scholar] [CrossRef]
- Zhou, T.; Zhu, H.; Fan, Z.; Wang, F.; Chen, Y.; Liang, H.; Yang, Z.; Zhang, L.; Lin, L.; Zhan, Y.; et al. History of winning remodels thalamo-PFC circuit to reinforce social dominance. Science 2017, 357, 162–168. [Google Scholar] [CrossRef]
- Kohl, S.; Heekeren, K.; Klosterkötter, J.; Kuhn, J. Prepulse inhibition in psychiatric disorders—Apart from schizophrenia. J. Psychiatr. Res. 2013, 47, 445–452. [Google Scholar] [CrossRef]
- Perry, W.; Minassian, A.; Lopez, B.; Maron, L.; Lincoln, A. Sensorimotor Gating Deficits in Adults with Autism. Biol. Psychiatry 2007, 61, 482–486. [Google Scholar] [CrossRef]
- Nakamoto, C.; Kawamura, M.; Nakatsukasa, E.; Natsume, R.; Takao, K.; Watanabe, M.; Abe, M.; Takeuchi, T.; Sakimura, K. GluD1 knockout mice with a pure C57BL/6N background show impaired fear memory, social interaction, and enhanced depressive-like behavior. PLoS ONE 2020, 15, e0229288. [Google Scholar] [CrossRef]
- Pan, H.; Jiang, N.; Sun, S.; Jiang, H.; Xu, J.; Jiang, X.; Gao, Q.; Li, L.; Wu, H.; Zheng, H.; et al. UHRF1-repressed 5′-hydroxymethylcytosine is essential for the male meiotic prophase I. Cell Death Dis. 2020, 11, 142. [Google Scholar] [CrossRef]
- Wang, C.; Pan, Y.; Zhang, W.; Chen, Y.; Li, C.; Zhao, F.; Behnisch, T. Positive Regulatory Domain I-binding Factor 1 Mediates Peripheral Nerve Injury-induced Nociception in Mice by Repressing Kv4.3 Channel Expression. Anesthesiology 2021, 134, 435–456. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).