Stable Housekeeping Genes in Bone Marrow, Adipose Tissue, and Amniotic Membrane-Derived Mesenchymal Stromal Cells for Orthopedic Regenerative Medicine Approaches
Abstract
1. Introduction
2. Results
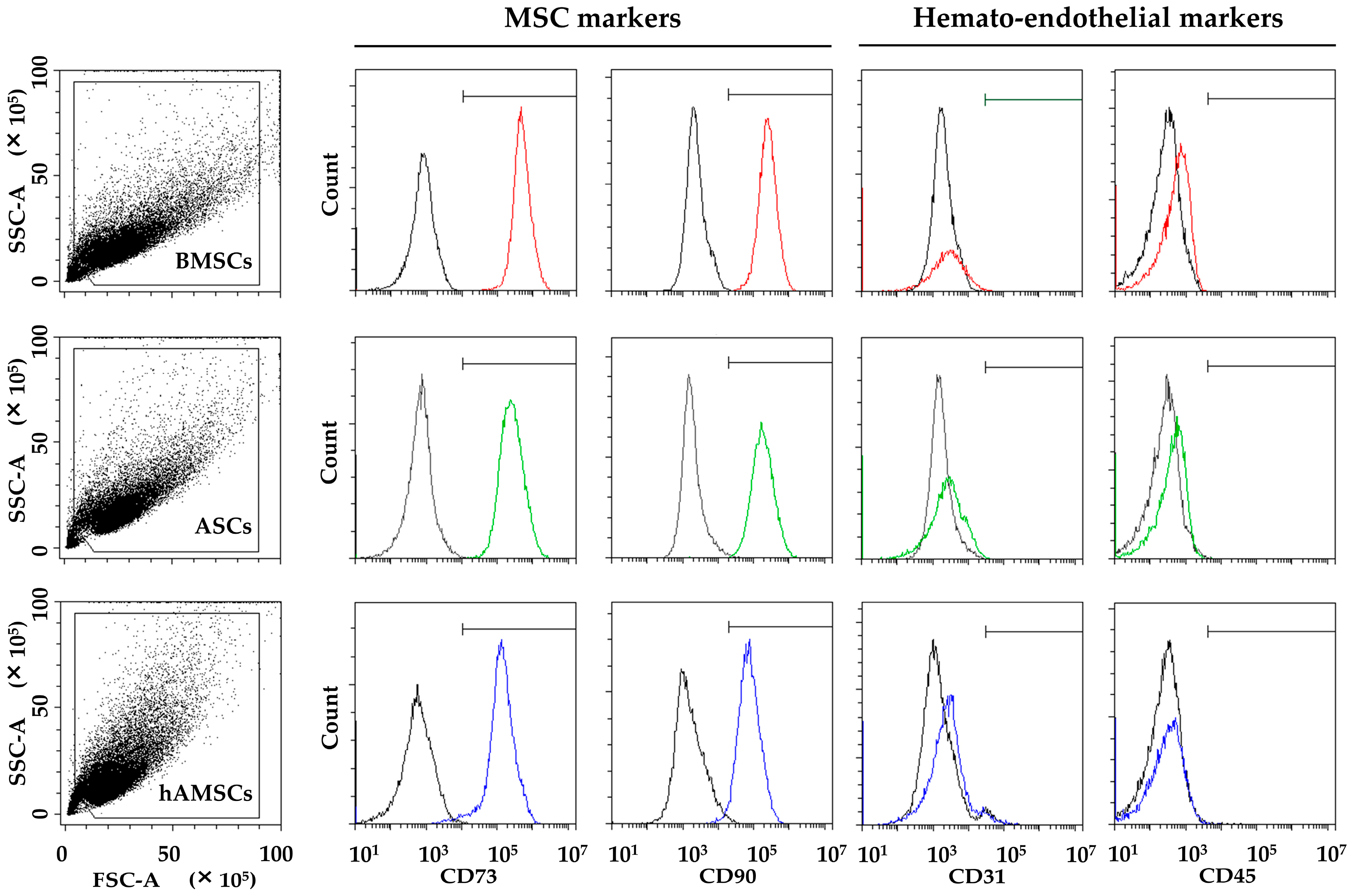
2.1. MSC Characterization
2.2. Candidate HKGs Expression
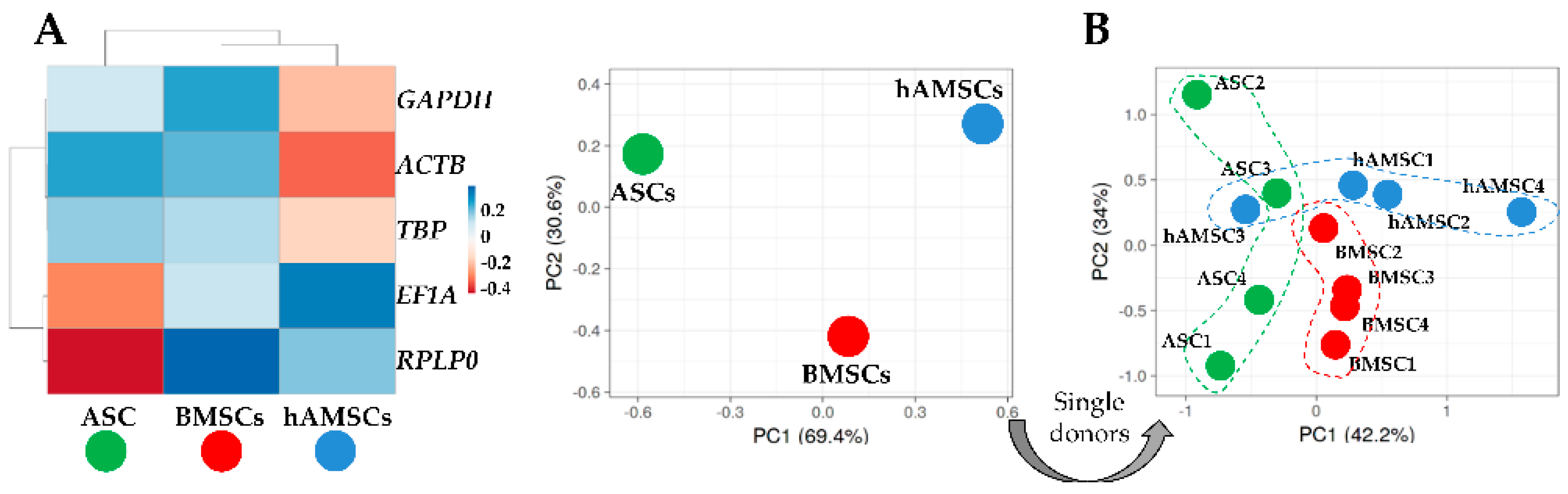
2.3. HKGs Stability
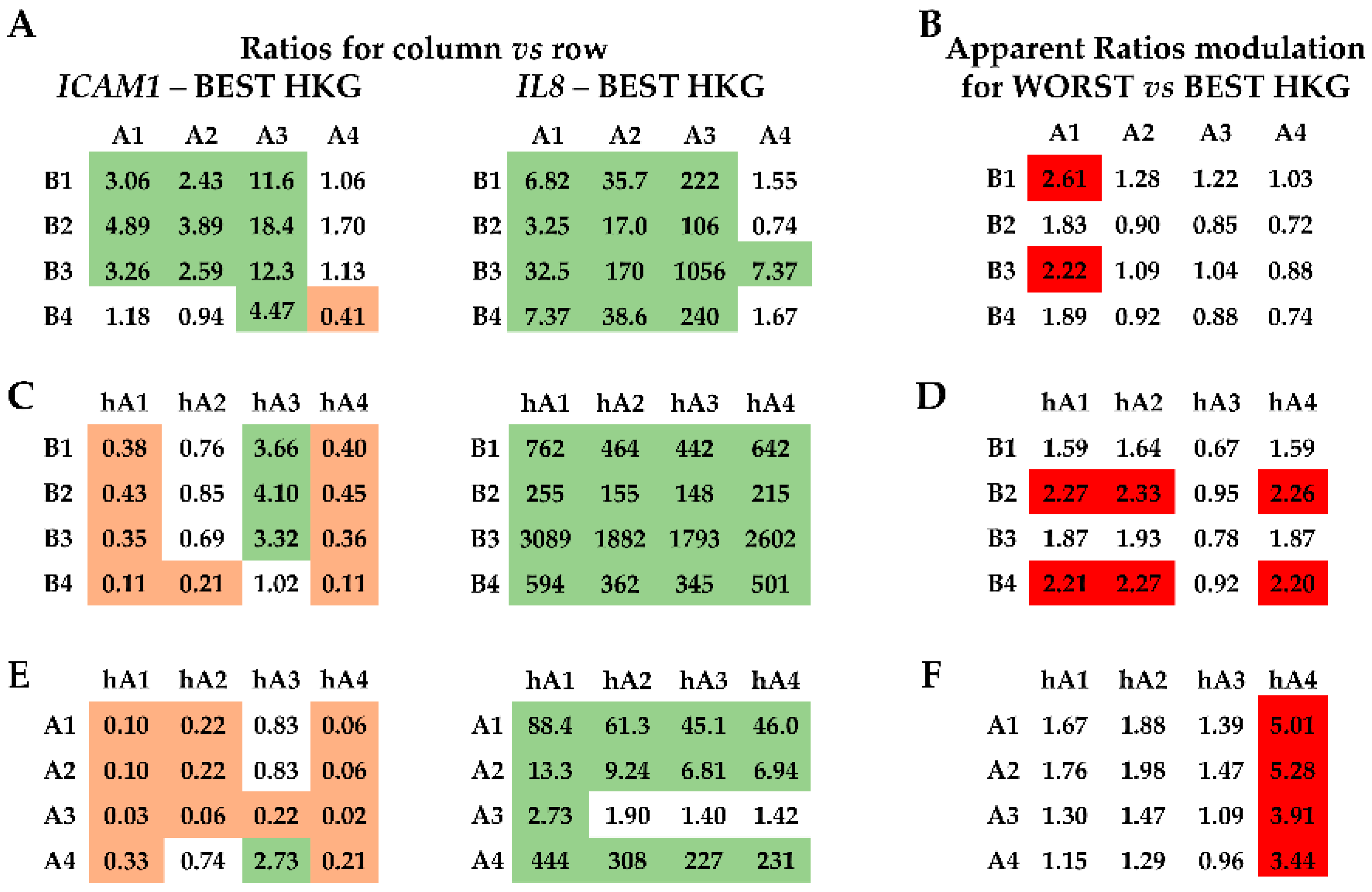
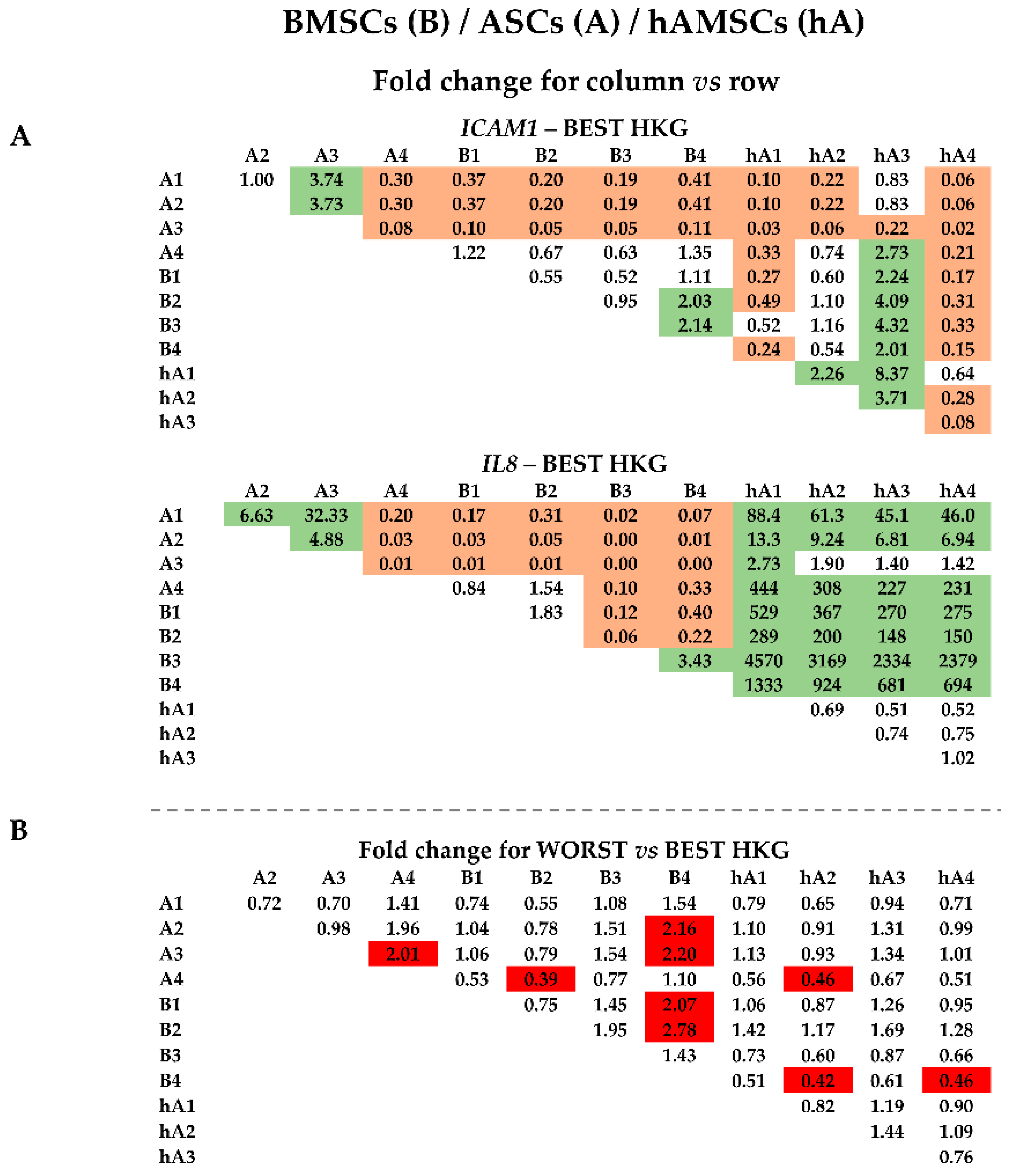
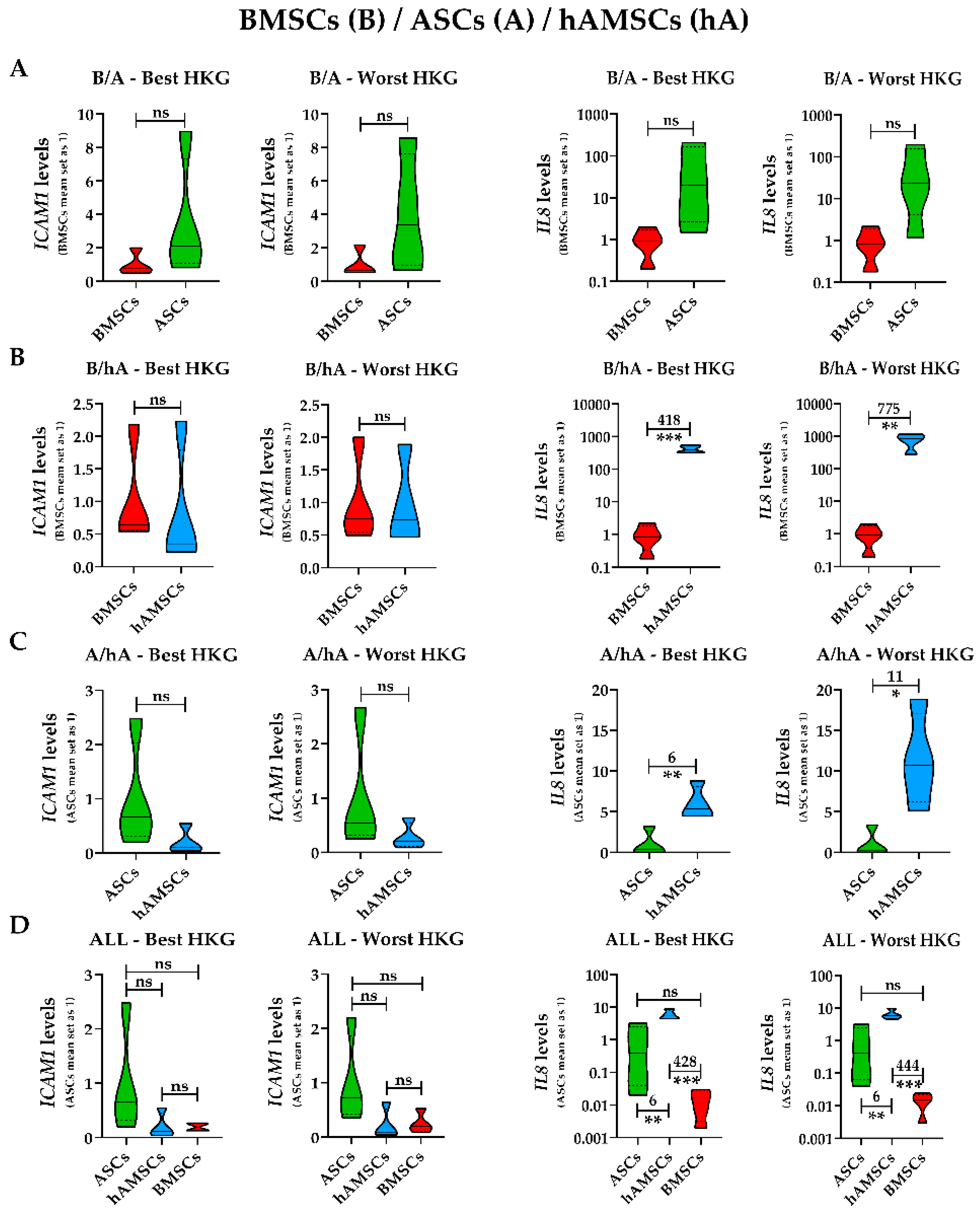
2.4. Effect of HKG Choice
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Adipose (ASCs), Bone Marrow (BMSCs), and Human Amniotic Membrane (hAMSCs) MSC Isolation and Culture
4.3. ASC, BMSC, and hAMSC Immunophenotype by Flow Cytometry
4.4. RNA Extraction and mRNA Profiling
4.5. Data Analysis
4.6. Hierarchical Clustering and Principal Component Analysis
4.7. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef] [PubMed]
- Gerami, M.H.; Khorram, R.; Rasoolzadegan, S.; Mardpour, S.; Nakhaei, P.; Hashemi, S.; Al-Naqeeb, B.Z.T.; Aminian, A.; Samimi, S. Emerging role of mesenchymal stem/stromal cells (MSCs) and MSCs-derived exosomes in bone- and joint-associated musculoskeletal disorders: A new frontier. Eur. J. Med. Res. 2023, 28, 86. [Google Scholar] [CrossRef]
- Caplan, A.I. Mesenchymal Stem Cells: Time to Change the Name! Stem Cells Transl. Med. 2017, 6, 1445–1451. [Google Scholar] [CrossRef]
- de Girolamo, L.; Filardo, G.; Laver, L. Disease-modifying effects of orthobiologics in the treatment of knee osteoarthritis: The lesson learned from preclinical research models. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 5286–5290. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho Carneiro, D.; de Araújo, L.T.; Santos, G.C.; Damasceno, P.K.F.; Vieira, J.L.; Santos, R.R.D.; Barbosa, J.D.V.; Soares, M.B.P. Clinical Trials with Mesenchymal Stem Cell Therapies for Osteoarthritis: Challenges in the Regeneration of Articular Cartilage. Int. J. Mol. Sci. 2023, 24, 9939. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, H.P.; Cohn, M.R.; Haunschild, E.D.; Wong, S.E.; Farr, J.; Yanke, A.B. Amniotic Product Treatments: Clinical and Basic Science Evidence. Curr. Rev. Musculoskelet. Med. 2020, 13, 148–154. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Lattanzi, W.; Ripoli, C.; Greco, V.; Barba, M.; Iavarone, F.; Minucci, A.; Urbani, A.; Grassi, C.; Parolini, O. Basic and Preclinical Research for Personalized Medicine. J. Pers. Med. 2021, 11, 354. [Google Scholar] [CrossRef]
- Costela-Ruiz, V.J.; Melguizo-Rodríguez, L.; Bellotti, C.; Illescas-Montes, R.; Stanco, D.; Arciola, C.R.; Lucarelli, E. Different Sources of Mesenchymal Stem Cells for Tissue Regeneration: A Guide to Identifying the Most Favorable One in Orthopedics and Dentistry Applications. Int. J. Mol. Sci. 2022, 23, 6356. [Google Scholar] [CrossRef]
- Zucherato, V.S.; Penariol, L.B.C.; Silva, L.E.C.M.; Padovan, C.C.; Poli-Neto, O.B.; Rosa-E-Silva, J.C.; Ferriani, R.A.; Meola, J. Identification of suitable reference genes for mesenchymal stem cells from menstrual blood of women with endometriosis. Sci. Rep. 2021, 11, 5422. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Bai, J.; Yang, Y.; Zhong, L.; Wang, Y. Identification of optimal reference genes for quantitative PCR studies on human mesenchymal stem cells. Mol. Med. Rep. 2015, 11, 1304–1311. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ragni, E.; Viganò, M.; Rebulla, P.; Giordano, R.; Lazzari, L. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: How to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013, 17, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Viganò, M.; Perucca Orfei, C.; de Girolamo, L.; Pearson, J.R.; Ragni, E.; De Luca, P.; Colombini, A. Housekeeping Gene Stability in Human Mesenchymal Stem and Tendon Cells Exposed to Tenogenic Factors. Tissue Eng. Part C Methods 2018, 24, 360–367. [Google Scholar] [CrossRef]
- Tratwal, J.; Follin, B.; Ekblond, A.; Kastrup, J.; Haack-Sørensen, M. Identification of a common reference gene pair for qPCR in human mesenchymal stromal cells from different tissue sources treated with VEGF. BMC Mol. Biol. 2014, 15, 11. [Google Scholar] [CrossRef] [PubMed]
- Pannier, L.; Merino, E.; Marchal, K.; Collado-Vides, J. Effect of genomic distance on coexpression of coregulated genes in E. coli. PLoS ONE 2017, 12, e0174887. [Google Scholar] [CrossRef]
- Schroeder, A.; Rubin, J.P.; Kokai, L.; Sowa, G.; Chen, J.; Onishi, K. Use of Adipose-Derived Orthobiologics for Musculoskeletal Injuries: A Narrative Review. PM&R 2020, 12, 805–816. [Google Scholar] [CrossRef]
- Pasculli, R.M.; Kenyon, C.D.; Berrigan, W.A.; Mautner, K.; Hammond, K.; Jayaram, P. Mesenchymal stem cells for subchondral bone marrow lesions: From bench to bedside. Bone Rep. 2022, 17, 101630. [Google Scholar] [CrossRef]
- Kon, E.; Boffa, A.; Andriolo, L.; Di Martino, A.; Di Matteo, B.; Magarelli, N.; Trenti, N.; Zaffagnini, S.; Filardo, G. Combined subchondral and intra-articular injections of bone marrow aspirate concentrate provide stable results up to 24 months. Knee Surg. Sports Traumatol. Arthrosc. 2023, 31, 2511–2517. [Google Scholar] [CrossRef]
- Tabet, S.K.; Kimmerling, K.A.; Hale, G.J.; Munson, N.R.; Mowry, K.C. Hypothermically Stored Amniotic Membrane for the Treatment of Cartilage Lesions: A Single-Arm Prospective Study with 2-Year Follow-Up. Cartilage 2022, 13, 19476035211072213. [Google Scholar] [CrossRef]
- Schierenbeck, M.; Grözinger, M.; Reichardt, B.; Jansen, O.; Kauczor, H.; Campbell, G.M.; Sedaghat, S. Detecting Bone Marrow Edema of the Extremities on Spectral Computed Tomography Using a Three-Material Decomposition. Diagnostics 2023, 13, 2745. [Google Scholar] [CrossRef]
- Obana, K.K.; Schallmo, M.S.; Hong, I.S.; Ahmad, C.S.; Moorman, C.T., 3rd; Trofa, D.P.; Saltzman, B.M. Current Trends in Orthobiologics: An 11-Year Review of the Orthopaedic Literature. Am. J. Sports Med. 2022, 50, 3121–3129. [Google Scholar] [CrossRef]
- Dhillon, M.S.; Patel, S. Why OrthoBiologics? J. Clin. Orthop. Trauma 2022, 28, 101850. [Google Scholar] [CrossRef]
- Torres-Torrillas, M.; Rubio, M.; Damia, E.; Cuervo, B.; Del Romero, A.; Peláez, P.; Chicharro, D.; Miguel, L.; Sopena, J.J. Adipose-Derived Mesenchymal Stem Cells: A Promising Tool in the Treatment of Musculoskeletal Diseases. Int. J. Mol. Sci. 2019, 20, 3105. [Google Scholar] [CrossRef]
- Eder, C.; Schmidt-Bleek, K.; Geissler, S.; Sass, F.A.; Maleitzke, T.; Pumberger, M.; Perka, C.; Duda, G.N.; Winkler, T. Mesenchymal stromal cell and bone marrow concentrate therapies for musculoskeletal indications: A concise review of current literature. Mol. Biol. Rep. 2020, 47, 4789–4814. [Google Scholar] [CrossRef]
- Song, I.; Rim, J.; Lee, J.; Jang, I.; Jung, B.; Kim, K.; Lee, S. Therapeutic Potential of Human Fetal Mesenchymal Stem Cells in Musculoskeletal Disorders: A Narrative Review. Int. J. Mol. Sci. 2022, 23, 1439. [Google Scholar] [CrossRef]
- Hasler, J.; Hatt, L.P.; Stoddart, M.J.; Armiento, A.R. Stable Reference Genes for qPCR Analysis in BM-MSCs Undergoing Osteogenic Differentiation within 3D Hyaluronan-Based Hydrogels. Int. J. Mol. Sci. 2020, 21, 9195. [Google Scholar] [CrossRef]
- Amable, P.R.; Teixeira, M.V.T.; Carias, R.B.V.; Granjeiro, J.M.; Borojevic, R. Identification of appropriate reference genes for human mesenchymal cells during expansion and differentiation. PLoS ONE 2013, 8, e73792. [Google Scholar] [CrossRef]
- Curtis, K.M.; Gomez, L.A.; Rios, C.; Garbayo, E.; Raval, A.P.; Perez-Pinzon, M.A.; Schiller, P.C. EF1alpha and RPL13a represent normalization genes suitable for RT-qPCR analysis of bone marrow derived mesenchymal stem cells. BMC Mol. Biol. 2010, 11, 61. [Google Scholar] [CrossRef]
- Roson-Burgo, B.; Sanchez-Guijo, F.; Del Cañizo, C.; De Las Rivas, J. Transcriptomic portrait of human Mesenchymal Stromal/Stem Cells isolated from bone marrow and placenta. BMC Genom. 2014, 15, 910. [Google Scholar] [CrossRef]
- Bui, T.M.; Wiesolek, H.L.; Sumagin, R. ICAM-1: A master regulator of cellular responses in inflammation, injury resolution, and tumorigenesis. J. Leukoc. Biol. 2020, 108, 787–799. [Google Scholar] [CrossRef]
- Tang, B.; Li, X.; Liu, Y.; Chen, X.; Li, X.; Chu, Y.; Zhu, H.; Liu, W.; Xu, F.; Zhou, F.; et al. The Therapeutic Effect of ICAM-1-Overexpressing Mesenchymal Stem Cells on Acute Graft-Versus-Host Disease. Cell Physiol. Biochem. 2018, 46, 2624–2635. [Google Scholar] [CrossRef]
- Woodell-May, J.E.; Sommerfeld, S.D. Role of Inflammation and the Immune System in the Progression of Osteoarthritis. J. Orthop. Res. 2020, 38, 253–257. [Google Scholar] [CrossRef]
- Kim, M.; Erickson, I.E.; Huang, A.H.; Garrity, S.T.; Mauck, R.L.; Steinberg, D.R. Donor Variation and Optimization of Human Mesenchymal Stem Cell Chondrogenesis in Hyaluronic Acid. Tissue Eng. Part A 2018, 24, 1693–1703. [Google Scholar] [CrossRef]
- Matsushima, K.; Yang, D.; Oppenheim, J.J. Interleukin-8: An evolving chemokine. Cytokine 2022, 153, 155828. [Google Scholar] [CrossRef]
- Molnar, V.; Matišić, V.; Kodvanj, I.; Bjelica, R.; Jeleč, Z.; Hudetz, D.; Rod, E.; Čukelj, F.; Vrdoljak, T.; Vidović, D.; et al. Cytokines and Chemokines Involved in Osteoarthritis Pathogenesis. Int. J. Mol. Sci. 2021, 22, 9208. [Google Scholar] [CrossRef]
- Jiang, F.; Zhang, W.; Zhou, M.; Zhou, Z.; Shen, M.; Chen, N.; Jiang, X. Human amniotic mesenchymal stromal cells promote bone regeneration via activating endogenous regeneration. Theranostics 2020, 10, 6216–6230. [Google Scholar] [CrossRef]
- Alonso-Goulart, V.; Braga Ferreira, L.; Angélico Duarte, C.; Lemos de Lima, I.; Ferreira, E.R.; Candido de Oliveira, B.; Nascimento Vargas, L.; Dotto de Moraes, D.; Borges Silva, I.B.; de Oliveira Faria, R.; et al. Mesenchymal stem cells from human adipose tissue and bone repair: A literature review. Biotechnol. Res. Innov. 2018, 2, 74–80. [Google Scholar] [CrossRef]
- Salehinejad, P.; Moshrefi, M.; Eslaminejad, T. An Overview on Mesenchymal Stem Cells Derived from Extraembryonic Tissues: Supplement Sources and Isolation Methods. Stem Cells Cloning 2020, 13, 57–65. [Google Scholar] [CrossRef]
- Murray, I.R.; Chahla, J.; Wordie, S.J.; Shapiro, S.A.; Piuzzi, N.S.; Frank, R.M.; Halbrecht, J.; Okada, K.; Nakamura, N.; Mandelbaum, B.; et al. Regulatory and Ethical Aspects of Orthobiologic Therapies. Orthop. J. Sports Med. 2022, 10, 23259671221101626. [Google Scholar] [CrossRef]
- Becherucci, V.; Piccini, L.; Casamassima, S.; Bisin, S.; Gori, V.; Gentile, F.; Ceccantini, R.; De Rienzo, E.; Bindi, B.; Pavan, P.; et al. Human platelet lysate in mesenchymal stromal cell expansion according to a GMP grade protocol: A cell factory experience. Stem Cell Res. Ther. 2018, 9, 124. [Google Scholar] [CrossRef]
- Bui, H.T.H.; Nguyen, L.T.; Than, U.T.T. Influences of Xeno-Free Media on Mesenchymal Stem Cell Expansion for Clinical Application. Tissue Eng. Regen. Med. 2021, 18, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tseng, P.; Lo, W.; Wang, F.; Lee, C. Comparing the impact of 3D bioreactor and 2D culture system on immunomodulation potency of Warton’s jelly derived-MSC. Cytotherapy 2017, 19, S186. [Google Scholar] [CrossRef]
- Shankar, P.; Jagtap, J.; Sharma, G.; Prasad Sharma, G.; Singh, J.; Parashar, M.; Kumar, G.; Mittal, S.; Sharma, M.K.; Jadhav, K.; et al. Chapter 12-A revolutionary breakthrough of bionanomaterials in tissue engineering and regenerative medicine. In Bionanotechnology: Emerging Applications of Bionanomaterials; Elsevier: Amsterdam, The Netherlands, 2022; pp. 399–441. [Google Scholar] [CrossRef]
- Dias, I.X.; Cordeiro, A.; Guimarães, J.a.M.; Silva, K.R. Potential and Limitations of Induced Pluripotent Stem Cells-Derived Mesenchymal Stem Cells in Musculoskeletal Disorders Treatment. Biomolecules 2023, 13, 1342. [Google Scholar] [CrossRef]
- Magatti, M.; Pianta, S.; Silini, A.; Parolini, O. Isolation, Culture, and Phenotypic Characterization of Mesenchymal Stromal Cells from the Amniotic Membrane of the Human Term Placenta. Methods Mol. Biol. 2016, 416, 233–244. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Parolini, O.; Alviano, F.; Bagnara, G.P.; Bilic, G.; Bühring, H.; Evangelista, M.; Hennerbichler, S.; Liu, B.; Magatti, M.; Mao, N.; et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells 2008, 26, 300–311. [Google Scholar] [CrossRef]
- Silini, A.R.; Di Pietro, R.; Lang-Olip, I.; Alviano, F.; Banerjee, A.; Basile, M.; Borutinskaite, V.; Eissner, G.; Gellhaus, A.; Giebel, B.; et al. Perinatal Derivatives: Where Do We Stand? A Roadmap of the Human Placenta and Consensus for Tissue and Cell Nomenclature. Front. Bioeng. Biotechnol. 2020, 8, 610544. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, I.; Lay, S. Thein Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
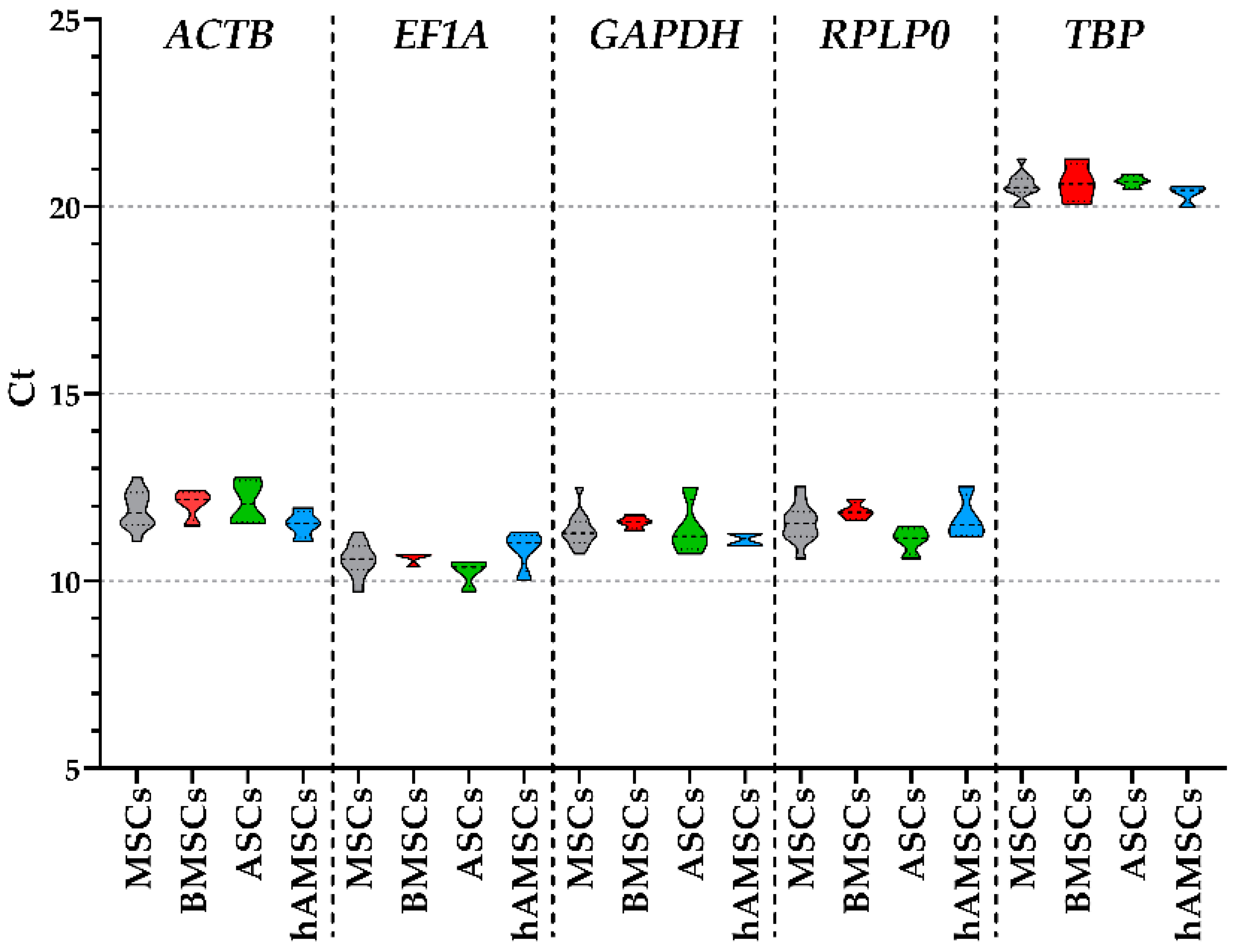

| ACTB | EF1A | GAPDH | RPLP0 | TBP | |
|---|---|---|---|---|---|
| MSCs | 11.90 ± 0.14 | 10.57 ± 0.12 | 11.37 ± 0.13 | 11.55 ± 0.14 | 20.55 ± 0.10 |
| BMSCs | 12.06 ± 0.17 | 10.63 ± 0.07 | 11.57 ± 0.07 | 11.87 ± 0.10 | 20.64 ± 0.23 |
| ASCs | 12.10 ± 0.25 | 10.24 ± 0.16 | 11.41 ± 0.33 | 11.09 ± 0.16 | 20.67 ± 0.07 |
| hAMSCs | 11.53 ± 0.16 | 10.84 ± 0.24 | 11.12 ± 0.08 | 11.68 ± 0.26 | 20.35 ± 0.11 |
| Source | Geomean | Delta CT | BestKeeper | NormFinder | Genorm | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| BMSCs | EF1A | 1.19 | RPLP0 | 0.30 | EF1A | 0.12 | EF1A | 0.08 | EF1A|RPLP0 | 0.17 |
| RPLP0 | 1.57 | EF1A | 0.31 | GAPDH | 0.12 | RPLP0 | 0.08 | |||
| ACTB | 3.22 | ACTB | 0.40 | RPLP0 | 0.16 | ACTB | 0.29 | ACTB | 0.22 | |
| GAPDH | 3.36 | GAPDH | 0.44 | ACTB | 0.29 | GAPDH | 0.36 | GAPDH | 0.29 | |
| TBP | 5.00 | TBP | 0.58 | TBP | 0.39 | TBP | 0.55 | TBP | 0.41 | |
| ASCs | EF1A | 1.32 | EF1A | 0.33 | TBP | 0.12 | EF1A | 0.08 | EF1A|RPLP0 | 0.17 |
| TBP | 1.86 | TBP | 0.40 | RPLP0 | 0.25 | TBP | 0.2 | |||
| RPLP0 | 2.06 | RPLP0 | 0.41 | EF1A | 0.26 | RPLP0 | 0.28 | TBP | 0.23 | |
| ACTB | 4.00 | ACTB | 0.48 | ACTB | 0.49 | ACTB | 0.36 | ACTB | 0.32 | |
| GAPDH | 5.00 | GAPDH | 0.66 | GAPDH | 0.55 | GAPDH | 0.62 | GAPDH | 0.46 | |
| hAMSCs | GAPDH | 1.32 | GAPDH | 0.50 | GAPDH | 0.16 | GAPDH | 0.19 | ACTB|TBP | 0.23 |
| TBP | 1.68 | TBP | 0.54 | TBP | 0.18 | TBP | 0.34 | |||
| ACTB | 2.45 | ACTB | 0.63 | ACTB | 0.26 | EF1A | 0.54 | GAPDH | 0.34 | |
| EF1A | 3.72 | EF1A | 0.67 | EF1A | 0.40 | ACTB | 0.55 | EF1A | 0.55 | |
| RPLP0 | 5.00 | RPLP0 | 0.70 | RPLP0 | 0.43 | RPLP0 | 0.59 | RPLP0 | 0.61 | |
| BMSCs/ASCs | EF1A | 1.00 | EF1A | 0.38 | EF1A | 0.23 | EF1A | 0.13 | EF1A|RPLP0 | 0.26 |
| RPLP0 | 2.59 | ACTB | 0.50 | TBP | 0.25 | ACTB | 0.35 | |||
| ACTB | 2.63 | RPLP0 | 0.50 | GAPDH | 0.37 | RPLP0 | 0.38 | ACTB | 0.39 | |
| TBP | 3.36 | TBP | 0.54 | ACTB | 0.39 | TBP | 0.42 | TBP | 0.45 | |
| GAPDH | 4.40 | GAPDH | 0.56 | RPLP0 | 0.39 | GAPDH | 0.46 | GAPDH | 0.49 | |
| BMSCs/hAMSCs | GAPDH | 1.00 | GAPDH | 0.49 | GAPDH | 0.22 | GAPDH | 0.26 | ACTB|GAPDH | 0.41 |
| TBP | 2.71 | RPLP0 | 0.53 | EF1A | 0.28 | RPLP0 | 0.36 | |||
| RPLP0 | 2.83 | TBP | 0.57 | TBP | 0.28 | TBP | 0.41 | TBP | 0.47 | |
| ACTB | 2.99 | ACTB | 0.57 | RPLP0 | 0.32 | ACTB | 0.45 | RPLP0 | 0.53 | |
| EF1A | 4.40 | EF1A | 0.58 | ACTB | 0.39 | EF1A | 0.46 | EF1A | 0.55 | |
| ASCs/hAMSCs | TBP | 1.00 | TBP | 0.57 | TBP | 0.18 | TBP | 0.30 | ACTB|TBP | 0.38 |
| GAPDH | 2.21 | GAPDH | 0.64 | GAPDH | 0.32 | GAPDH | 0.42 | |||
| ACTB | 2.99 | EF1A | 0.66 | RPLP0 | 0.38 | EF1A | 0.51 | GAPDH | 0.47 | |
| EF1A | 3.46 | ACTB | 0.69 | EF1A | 0.42 | ACTB | 0.56 | EF1A | 0.62 | |
| RPLP0 | 4.40 | RPLP0 | 0.69 | ACTB | 0.43 | RPLP0 | 0.57 | RPLP0 | 0.65 | |
| ALL | TBP | 1.41 | TBP | 0.57 | TBP | 0.25 | TBP | 0.38 | EF1A|RPLP0 | 0.32 |
| EF1A | 2.06 | EF1A | 0.57 | GAPDH | 0.33 | GAPDH | 0.38 | |||
| GAPDH | 2.45 | GAPDH | 0.58 | EF1A | 0.34 | EF1A | 0.41 | GAPDH | 0.53 | |
| RPLP0 | 2.83 | RPLP0 | 0.60 | RPLP0 | 0.40 | RPLP0 | 0.47 | TBP | 0.57 | |
| ACTB | 5.00 | ACTB | 0.61 | ACTB | 0.42 | ACTB | 0.47 | ACTB | 0.59 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Piccolo, S.; Papait, A.; De Luca, P.; Taiana, M.; Grieco, G.; Silini, A.R.; Parolini, O.; de Girolamo, L. Stable Housekeeping Genes in Bone Marrow, Adipose Tissue, and Amniotic Membrane-Derived Mesenchymal Stromal Cells for Orthopedic Regenerative Medicine Approaches. Int. J. Mol. Sci. 2024, 25, 1461. https://doi.org/10.3390/ijms25031461
Ragni E, Piccolo S, Papait A, De Luca P, Taiana M, Grieco G, Silini AR, Parolini O, de Girolamo L. Stable Housekeeping Genes in Bone Marrow, Adipose Tissue, and Amniotic Membrane-Derived Mesenchymal Stromal Cells for Orthopedic Regenerative Medicine Approaches. International Journal of Molecular Sciences. 2024; 25(3):1461. https://doi.org/10.3390/ijms25031461
Chicago/Turabian StyleRagni, Enrico, Simona Piccolo, Andrea Papait, Paola De Luca, Michela Taiana, Giulio Grieco, Antonietta Rosa Silini, Ornella Parolini, and Laura de Girolamo. 2024. "Stable Housekeeping Genes in Bone Marrow, Adipose Tissue, and Amniotic Membrane-Derived Mesenchymal Stromal Cells for Orthopedic Regenerative Medicine Approaches" International Journal of Molecular Sciences 25, no. 3: 1461. https://doi.org/10.3390/ijms25031461
APA StyleRagni, E., Piccolo, S., Papait, A., De Luca, P., Taiana, M., Grieco, G., Silini, A. R., Parolini, O., & de Girolamo, L. (2024). Stable Housekeeping Genes in Bone Marrow, Adipose Tissue, and Amniotic Membrane-Derived Mesenchymal Stromal Cells for Orthopedic Regenerative Medicine Approaches. International Journal of Molecular Sciences, 25(3), 1461. https://doi.org/10.3390/ijms25031461










