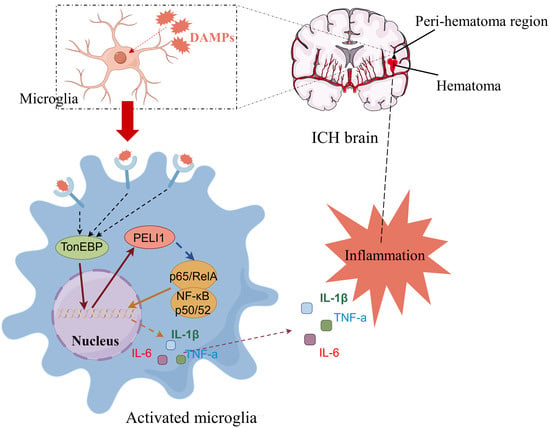
TonEBP: A Key Transcription Factor in Microglia Following Intracerebral Hemorrhage Induced-Neuroinflammation
Abstract
1. Introduction
2. Results
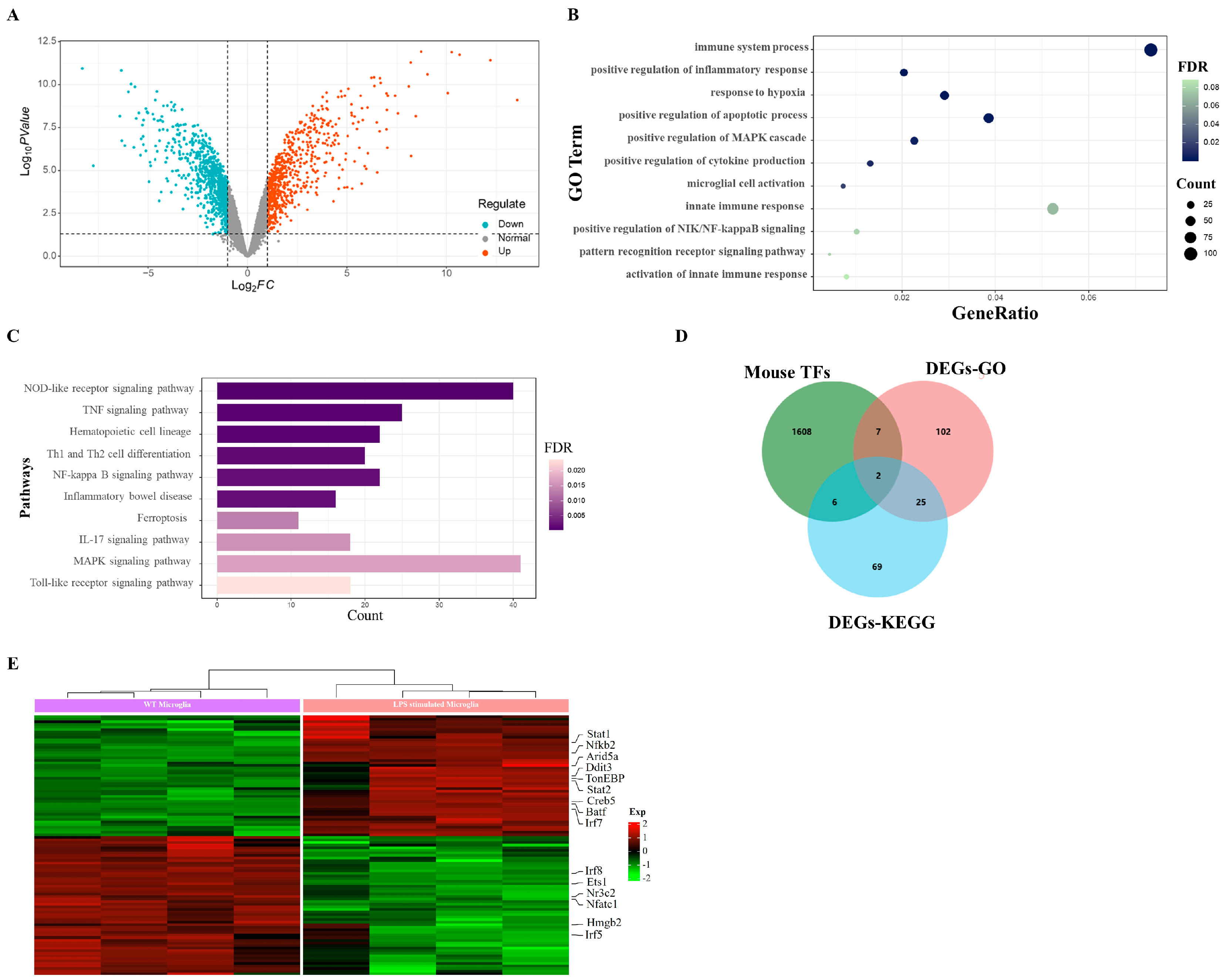
2.1. LPS Evokes Upregulated Transcription Factors in Microglia
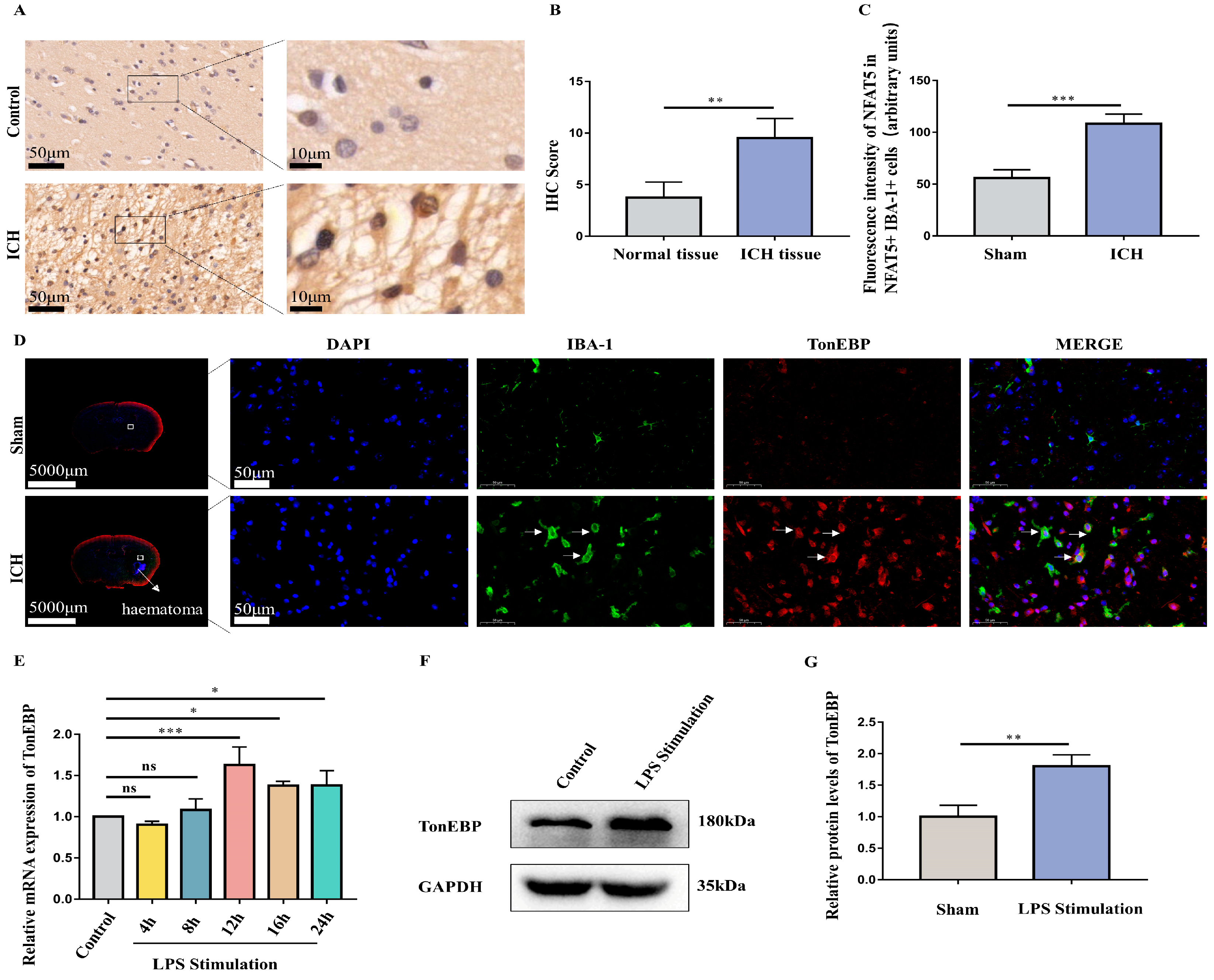
2.2. ICH and LPS Cause the Upregulation of Microglial TonEBP
2.3. TonEBP Exacerbates Neuroinflammation and Microglial Activation
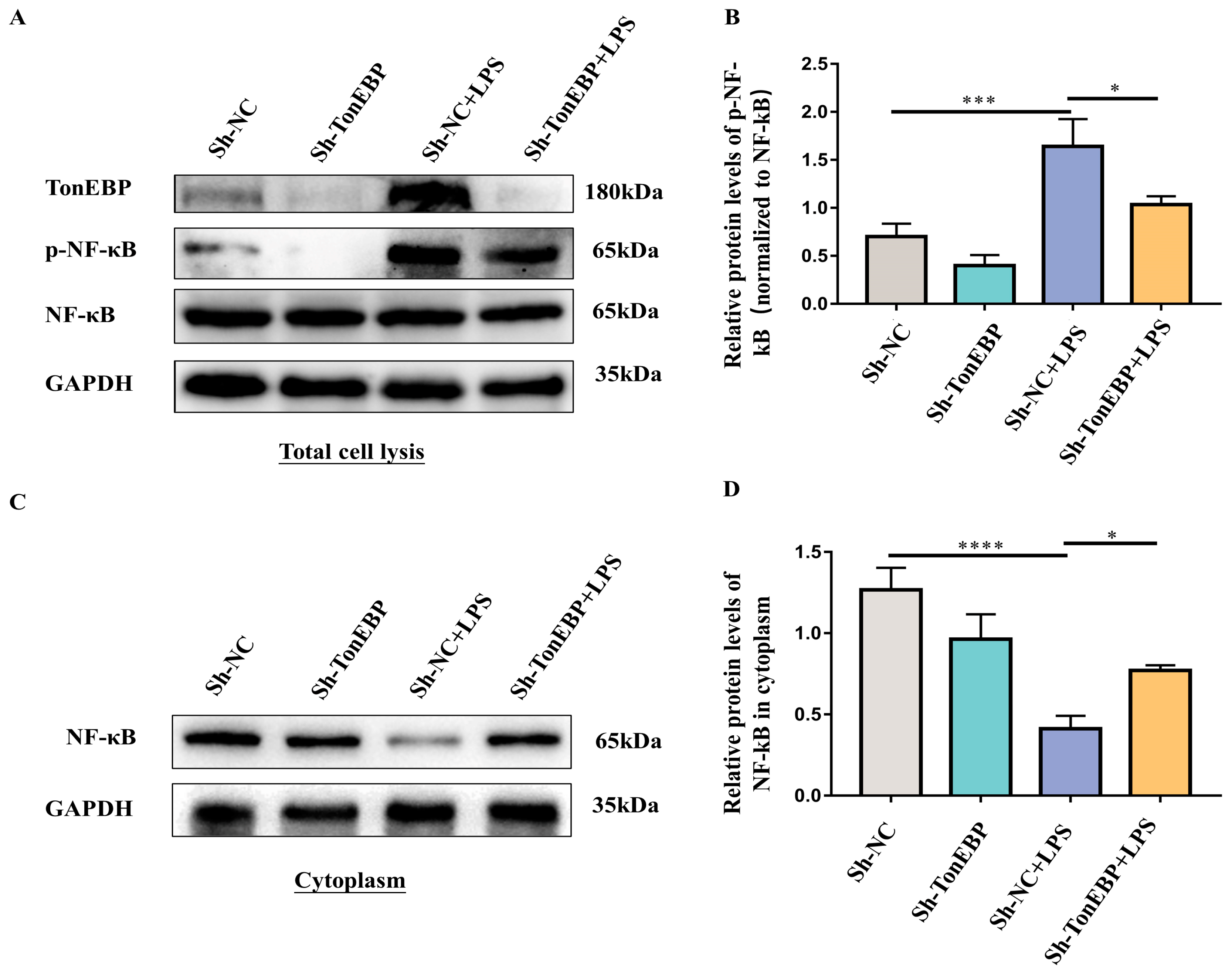
2.4. TonEBP Contributes to the Activation of the NF-κB Signaling Pathway
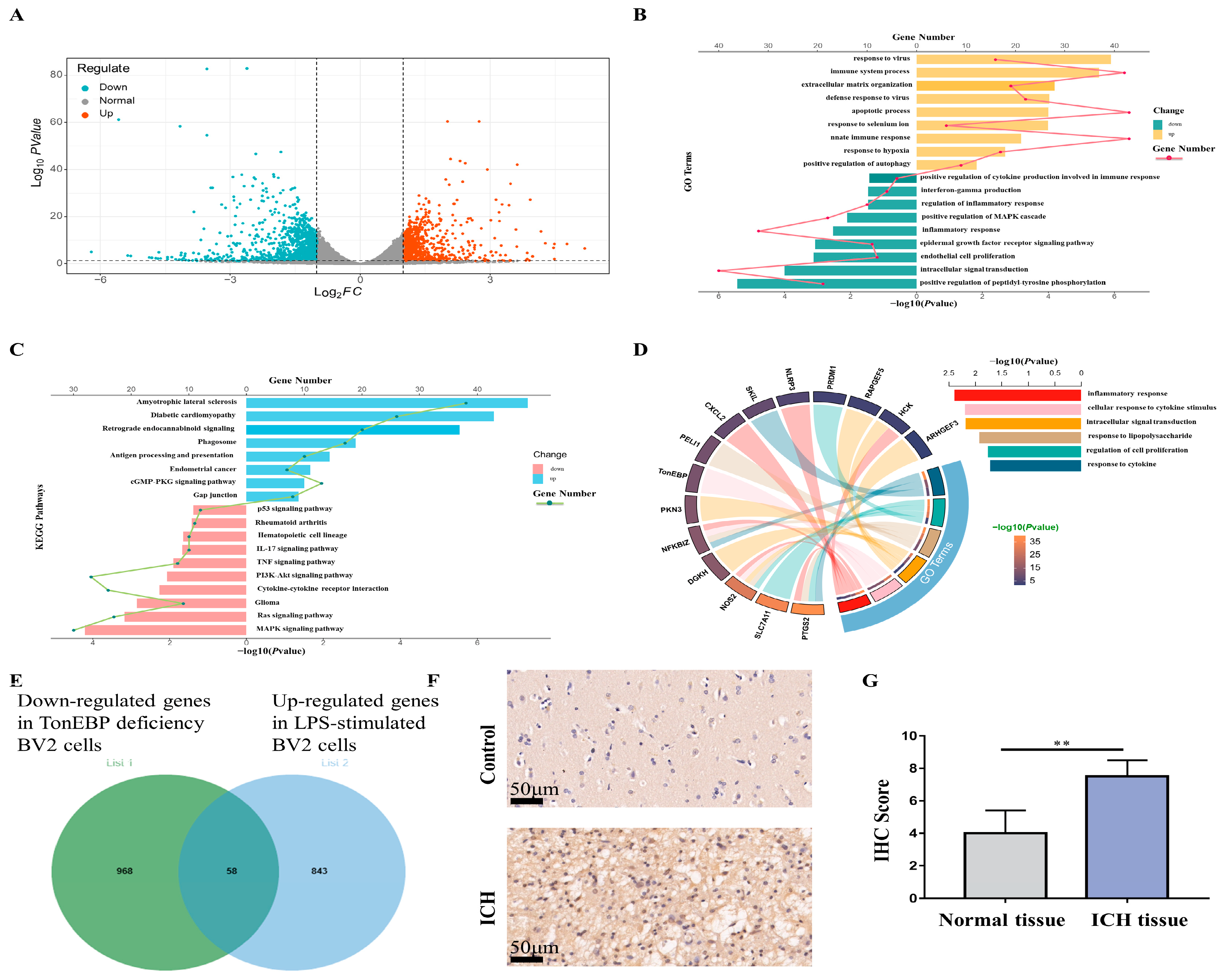
2.5. RNA-seq for Potential Targets of TonEBP
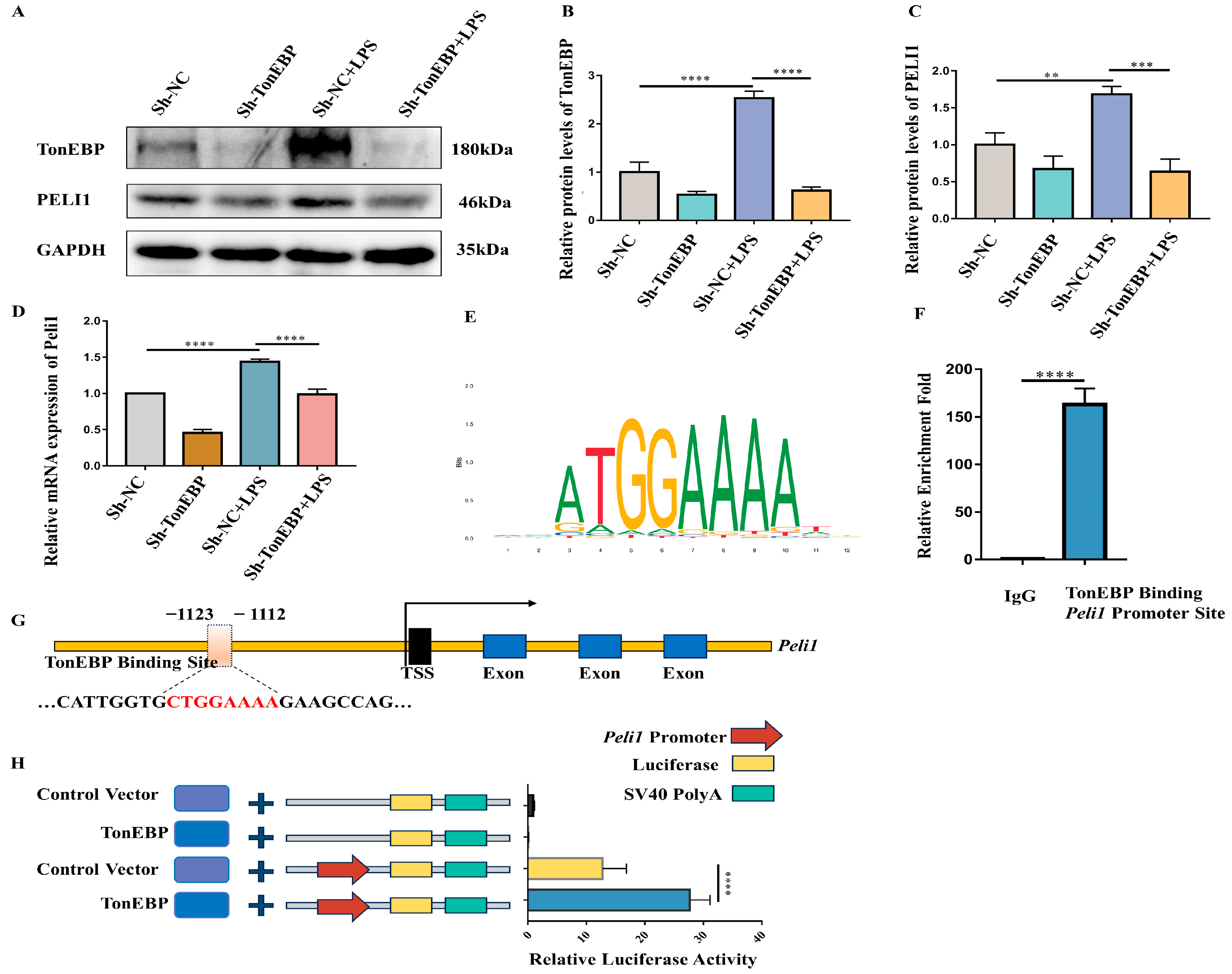
2.6. TonEBP Regulates PELI1 Expression at the Transcriptional Level
3. Discussion
4. Materials and Methods
4.1. Human Tissues
4.2. Animals
4.3. ICH Model
4.4. Cell Culture and LPS Treatment
4.5. RNA Extraction and Real-Time Quantitative PCR
4.6. ELISA Assay for Cytokines
4.7. Immunofluorescence
4.8. Immunohistochemical Staining
4.9. Construction and Transfection of Plasmids
4.10. Promoter Luciferase Reporter Assay
4.11. Cleavage under Targets and Release Using Nuclease (CUT&RUN) Assay
4.12. RNA Sequencing and Analysis
4.13. Lentivirus Transfection
4.14. Western Blot
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Puy, L.; Parry-Jones, A.R.; Sandset, E.C.; Dowlatshahi, D.; Ziai, W.; Cordonnier, C. Intracerebral haemorrhage. Nat. Rev. Dis. Primers 2023, 9, 14. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, J.C., III; Greenberg, S.M.; Anderson, C.S.; Becker, K.; Bendok, B.R.; Cushman, M.; Fung, G.L.; Goldstein, J.N.; Macdonald, R.L.; Mitchell, P.H.; et al. Guidelines for the Management of Spontaneous Intracerebral Hemorrhage: A Guideline for Healthcare Professionals From the American Heart Association/American Stroke Association. Stroke 2015, 46, 2032–2060. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, D.A.; Pandey, A.S.; Thompson, B.G.; Keep, R.F.; Hua, Y.; Xi, G. Injury mechanisms in acute intracerebral hemorrhage. Neuropharmacology 2018, 134 Pt B, 240–248. [Google Scholar] [CrossRef]
- Zheng, H.; Chen, C.; Zhang, J.; Hu, Z. Mechanism and Therapy of Brain Edema after Intracerebral Hemorrhage. Cerebrovasc. Dis. 2016, 42, 155–169. [Google Scholar] [CrossRef]
- Keep, R.F.; Hua, Y.; Xi, G. Intracerebral haemorrhage: Mechanisms of injury and therapeutic targets. Lancet. Neurol. 2012, 11, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Aronowski, J.; Zhao, X. Molecular pathophysiology of cerebral hemorrhage: Secondary brain injury. Stroke 2011, 42, 1781–1786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Khan, S.; Liu, Y.; Zhang, R.; Li, H.; Wu, G.; Tang, Z.; Xue, M.; Yong, V.W. Modes of Brain Cell Death Following Intracerebral Hemorrhage. Front. Cell. Neurosci. 2022, 16, 799753. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Z.; Lu, H.; Yang, Q.; Wu, H.; Wang, J. Microglial Polarization and Inflammatory Mediators after Intracerebral Hemorrhage. Mol. Neurobiol. 2017, 54, 1874–1886. [Google Scholar] [CrossRef]
- Miyakawa, H.; Woo, S.K.; Dahl, S.C.; Handler, J.S.; Kwon, H.M. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc. Natl. Acad. Sci. USA 1999, 96, 2538–2542. [Google Scholar] [CrossRef]
- Yoo, E.J.; Oh, K.H.; Piao, H.; Kang, H.J.; Jeong, G.W.; Park, H.; Lee, C.J.; Ryu, H.; Yang, S.H.; Kim, M.G.; et al. Macrophage transcription factor TonEBP promotes systemic lupus erythematosus and kidney injury via damage-induced signaling pathways. Kidney Int. 2023, 104, 163–180. [Google Scholar] [CrossRef]
- Xu, J.; Gao, C.; He, Y.; Fang, X.; Sun, D.; Peng, Z.; Xiao, H.; Sun, M.; Zhang, P.; Zhou, T.; et al. NLRC3 expression in macrophage impairs glycolysis and host immune defense by modulating the NF-κB-NFAT5 complex during septic immunosuppression. Mol. Ther. J. Am. Soc. Gene Ther. 2023, 31, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeong, E.A.; Kim, K.E.; Yi, C.O.; Jin, Z.; Lee, J.E.; Lee, D.H.; Kim, H.J.; Kang, S.S.; Cho, G.J.; et al. TonEBP/NFAT5 haploinsufficiency attenuates hippocampal inflammation in high-fat diet/streptozotocin-induced diabetic mice. Sci. Rep. 2017, 7, 7837. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.R.; Im, S.K.; Bae, Y.H.; Park, E.S.; Jin, B.K.; Kwon, H.M.; Lee, B.J.; Bu, Y.; Hur, E.M.; Lee, B.D. Inflammatory signals induce the expression of tonicity-responsive enhancer binding protein (TonEBP) in microglia. J. Neuroimmunol. 2016, 295, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Tellechea, M.; Buxadé, M.; Tejedor, S.; Aramburu, J.; López-Rodríguez, C. NFAT5-Regulated Macrophage Polarization Supports the Proinflammatory Function of Macrophages and T Lymphocytes. J. Immunol. 2018, 200, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Liu, Y.G.; Huang, Q.B.; Wang, H.W.; Song, Y.; Xu, Z.K.; Li, F. Nuclear factor-κB activation in perihematomal brain tissue correlates with outcome in patients with intracerebral hemorrhage. J. Neuroinflamm. 2015, 12, 53. [Google Scholar] [CrossRef] [PubMed]
- Kopitar-Jerala, N. Innate Immune Response in Brain, NF-Kappa B Signaling and Cystatins. Front. Mol. Neurosci. 2015, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Jin, W.; Chang, M.; Sun, S.C. Peli: A family of signal-responsive E3 ubiquitin ligases mediating TLR signaling and T-cell tolerance. Cell Mol. Immunol. 2012, 9, 113–122. [Google Scholar] [CrossRef]
- Mo, H.; Wang, Z.; He, Z.; Wan, J.; Lu, R.; Wang, C.; Chen, A.; Cheng, P. Decreased Peli1 expression attenuates osteoarthritis by protecting chondrocytes and inhibiting M1-polarization of macrophages. Bone Jt. Res. 2023, 12, 121–132. [Google Scholar] [CrossRef]
- Chen, H.; Hou, Y.; Zhai, Y.; Yang, J.; Que, L.; Liu, J.; Lu, L.; Ha, T.; Li, C.; Xu, Y.; et al. Peli1 deletion in macrophages attenuates myocardial ischemia/reperfusion injury by suppressing M1 polarization. J. Leukoc. Biol. 2023, 113, 95–108. [Google Scholar] [CrossRef]
- Xiao, Y.; Jin, J.; Chang, M.; Chang, J.H.; Hu, H.; Zhou, X.; Brittain, G.C.; Stansberg, C.; Torkildsen, Ø.; Wang, X.; et al. Peli1 promotes microglia-mediated CNS inflammation by regulating Traf3 degradation. Nat. Med. 2013, 19, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yin, C.; Liu, T.; Abdul, M.; Zhou, Y.; Cao, J.L.; Lu, C. Pellino1 regulates neuropathic pain as well as microglial activation through the regulation of MAPK/NF-κB signaling in the spinal cord. J. Neuroinflamm. 2020, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Jin, J.; Zou, Q.; Hu, H.; Cheng, X.; Sun, S.C. Peli1 negatively regulates type I interferon induction and antiviral immunity in the CNS. Cell Biosci. 2015, 5, 34. [Google Scholar] [CrossRef]
- Yeh, H.; Ikezu, T. Transcriptional and Epigenetic Regulation of Microglia in Health and Disease. Trends Mol. Med. 2019, 25, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Aronowski, J. Nrf2 to pre-condition the brain against injury caused by products of hemolysis after ICH. Transl. Stroke Res. 2013, 4, 71–75. [Google Scholar] [CrossRef]
- Karuppagounder, S.S.; Alim, I.; Khim, S.J.; Bourassa, M.W.; Sleiman, S.F.; John, R.; Thinnes, C.C.; Yeh, T.L.; Demetriades, M.; Neitemeier, S.; et al. Therapeutic targeting of oxygen-sensing prolyl hydroxylases abrogates ATF4-dependent neuronal death and improves outcomes after brain hemorrhage in several rodent models. Sci. Transl. Med. 2016, 8, 328ra29. [Google Scholar] [CrossRef]
- Gong, L.; Manaenko, A.; Fan, R.; Huang, L.; Enkhjargal, B.; McBride, D.; Ding, Y.; Tang, J.; Xiao, X.; Zhang, J.H. Osteopontin attenuates inflammation via JAK2/STAT1 pathway in hyperglycemic rats after intracerebral hemorrhage. Neuropharmacology 2018, 138, 160–169. [Google Scholar] [CrossRef]
- Laban, H.; Siegmund, S.; Zappe, M.; Trogisch, F.A.; Heineke, J.; Torre, C.; Fisslthaler, B.; Arnold, C.; Lauryn, J.; Büttner, M.; et al. NFAT5/TonEBP Limits Pulmonary Vascular Resistance in the Hypoxic Lung by Controlling Mitochondrial Reactive Oxygen Species Generation in Arterial Smooth Muscle Cells. Cells 2021, 10, 3293. [Google Scholar] [CrossRef]
- Shin, H.J.; Kim, H.; Heo, R.W.; Kim, H.J.; Choi, W.S.; Kwon, H.M.; Roh, G.S. Tonicity-responsive enhancer binding protein haplodeficiency attenuates seizure severity and NF-κB-mediated neuroinflammation in kainic acid-induced seizures. Cell Death Differ. 2014, 21, 1095–1106. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Sanada, S.; An, S.M.; Ye, B.J.; Lee, J.H.; Seo, Y.K.; Lee, C.; Lee-Kwon, W.; Küper, C.; Neuhofer, W.; et al. LPS-induced NFκB enhanceosome requires TonEBP/NFAT5 without DNA binding. Sci. Rep. 2016, 6, 24921. [Google Scholar] [CrossRef]
- Choi, S.Y.; Lim, S.W.; Salimi, S.; Yoo, E.J.; Lee-Kwon, W.; Lee, H.H.; Lee, J.H.; Mitchell, B.D.; Sanada, S.; Parsa, A.; et al. Tonicity-Responsive Enhancer-Binding Protein Mediates Hyperglycemia-Induced Inflammation and Vascular and Renal Injury. J. Am. Soc. Nephrol. JASN 2018, 29, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Taetzsch, T.; Levesque, S.; McGraw, C.; Brookins, S.; Luqa, R.; Bonini, M.G.; Mason, R.P.; Oh, U.; Block, M.L. Redox regulation of NF-κB p50 and M1 polarization in microglia. Glia 2015, 63, 423–440. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, M.; Bo, L.; Liu, W.; Liu, Q.; Chen, X.; Xu, D.; Li, Z.; Jin, F. NFAT5 participates in seawater inhalation-induced acute lung injury via modulation of NF-κB activity. Mol. Med. Rep. 2016, 14, 5033–5040. [Google Scholar] [CrossRef] [PubMed]
- Roth, I.; Leroy, V.; Kwon, H.M.; Martin, P.Y.; Féraille, E.; Hasler, U. Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-kappaB activity. Mol. Biol. Cell 2010, 21, 3459–3474. [Google Scholar] [CrossRef]
- Xie, X.; Huang, C.; Xu, D.; Liu, Y.; Hu, M.; Long, J.; Fang, X. Elevation of hypertonicity-induced protein NFAT5 promotes apoptosis of human umbilical vein endothelial cells through the NF-κB pathway. Mol. Med. Rep. 2021, 23, 184. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.W.; Lee, H.H.; Lee-Kwon, W.; Kwon, H.M. Microglial TonEBP mediates LPS-induced inflammation and memory loss as transcriptional cofactor for NF-κB and AP-1. J. Neuroinflamm. 2020, 17, 372. [Google Scholar] [CrossRef] [PubMed]
- Buxadé, M.; Lunazzi, G.; Minguillón, J.; Iborra, S.; Berga-Bolaños, R.; Del Val, M.; Aramburu, J.; López-Rodríguez, C. Gene expression induced by Toll-like receptors in macrophages requires the transcription factor NFAT5. J. Exp. Med. 2012, 209, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Zha, S.; Shen, X.; Zhao, Y.; Li, L.; Yang, L.; Lei, M.; Liu, W. NFAT5 mediates hypertonic stress-induced atherosclerosis via activating NLRP3 inflammasome in endothelium. Cell Commun. Signal. 2019, 17, 102. [Google Scholar] [CrossRef] [PubMed]
- Johnson, Z.I.; Doolittle, A.C.; Snuggs, J.W.; Shapiro, I.M.; Le Maitre, C.L.; Risbud, M.V. TNF-α promotes nuclear enrichment of the transcription factor TonEBP/NFAT5 to selectively control inflammatory but not osmoregulatory responses in nucleus pulposus cells. J. Biol. Chem. 2017, 292, 17561–17575. [Google Scholar] [CrossRef]
- Humphries, F.; Moynagh, P.N. Molecular and physiological roles of Pellino E3 ubiquitin ligases in immunity. Immunol. Rev. 2015, 266, 93–108. [Google Scholar] [CrossRef]
- Moynagh, P.N. The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Rev. Immunol. 2014, 14, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Yuan, J.; Wang, Y.; Xu, J.; Mao, C.; Xiao, Y. Peli1 controls the survival of dopaminergic neurons through modulating microglia-mediated neuroinflammation. Sci. Rep. 2019, 9, 8034. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Jin, W.; Sun, S.C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 2009, 10, 1089–1095. [Google Scholar] [CrossRef]
- Xu, W.; Yang, T.; Lou, X.; Chen, J.; Wang, X.; Hu, M.; An, D.; Gao, R.; Wang, J.; Chen, X. Role of the Peli1-RIPK1 Signaling Axis in Methamphetamine-Induced Neuroinflammation. ACS Chem. Neurosci. 2023, 14, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Lu, J.; Mei, S.; Wu, H.; Sun, Z.; Fang, Y.; Xu, S.; Wang, X.; Shi, L.; Xu, W.; et al. Ceria nanoparticles ameliorate white matter injury after intracerebral hemorrhage: Microglia-astrocyte involvement in remyelination. J. Neuroinflamm. 2021, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Bao, K.; Zhou, X.; Deng, Y.; Li, X.; Zhang, J.; Lan, X.; Zhao, J.; Lu, D.; Xu, Y.; et al. PSMC5 regulates microglial polarization and activation in LPS-induced cognitive deficits and motor impairments by interacting with TLR4. J. Neuroinflamm. 2023, 20, 277. [Google Scholar] [CrossRef] [PubMed]
- Rauluseviciute, I.; Riudavets-Puig, R.; Blanc-Mathieu, R.; Castro-Mondragon, J.A.; Ferenc, K.; Kumar, V.; Lemma, R.B.; Lucas, J.; Chèneby, J.; Baranasic, D.; et al. JASPAR 2024: 20th anniversary of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2023, 52, D174–D182. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palahati, A.; Luo, Y.; Qin, L.; Duan, Y.; Zhang, M.; Gan, H.; Zhai, X. TonEBP: A Key Transcription Factor in Microglia Following Intracerebral Hemorrhage Induced-Neuroinflammation. Int. J. Mol. Sci. 2024, 25, 1438. https://doi.org/10.3390/ijms25031438
Palahati A, Luo Y, Qin L, Duan Y, Zhang M, Gan H, Zhai X. TonEBP: A Key Transcription Factor in Microglia Following Intracerebral Hemorrhage Induced-Neuroinflammation. International Journal of Molecular Sciences. 2024; 25(3):1438. https://doi.org/10.3390/ijms25031438
Chicago/Turabian StylePalahati, Ailiyaer, Yujia Luo, Le Qin, Yuhao Duan, Mi Zhang, Hui Gan, and Xuan Zhai. 2024. "TonEBP: A Key Transcription Factor in Microglia Following Intracerebral Hemorrhage Induced-Neuroinflammation" International Journal of Molecular Sciences 25, no. 3: 1438. https://doi.org/10.3390/ijms25031438
APA StylePalahati, A., Luo, Y., Qin, L., Duan, Y., Zhang, M., Gan, H., & Zhai, X. (2024). TonEBP: A Key Transcription Factor in Microglia Following Intracerebral Hemorrhage Induced-Neuroinflammation. International Journal of Molecular Sciences, 25(3), 1438. https://doi.org/10.3390/ijms25031438