Overview of Phage Defense Systems in Bacteria and Their Applications
Abstract
1. Introduction
2. Anti-Phage Mechanisms at the Single-Cell Level
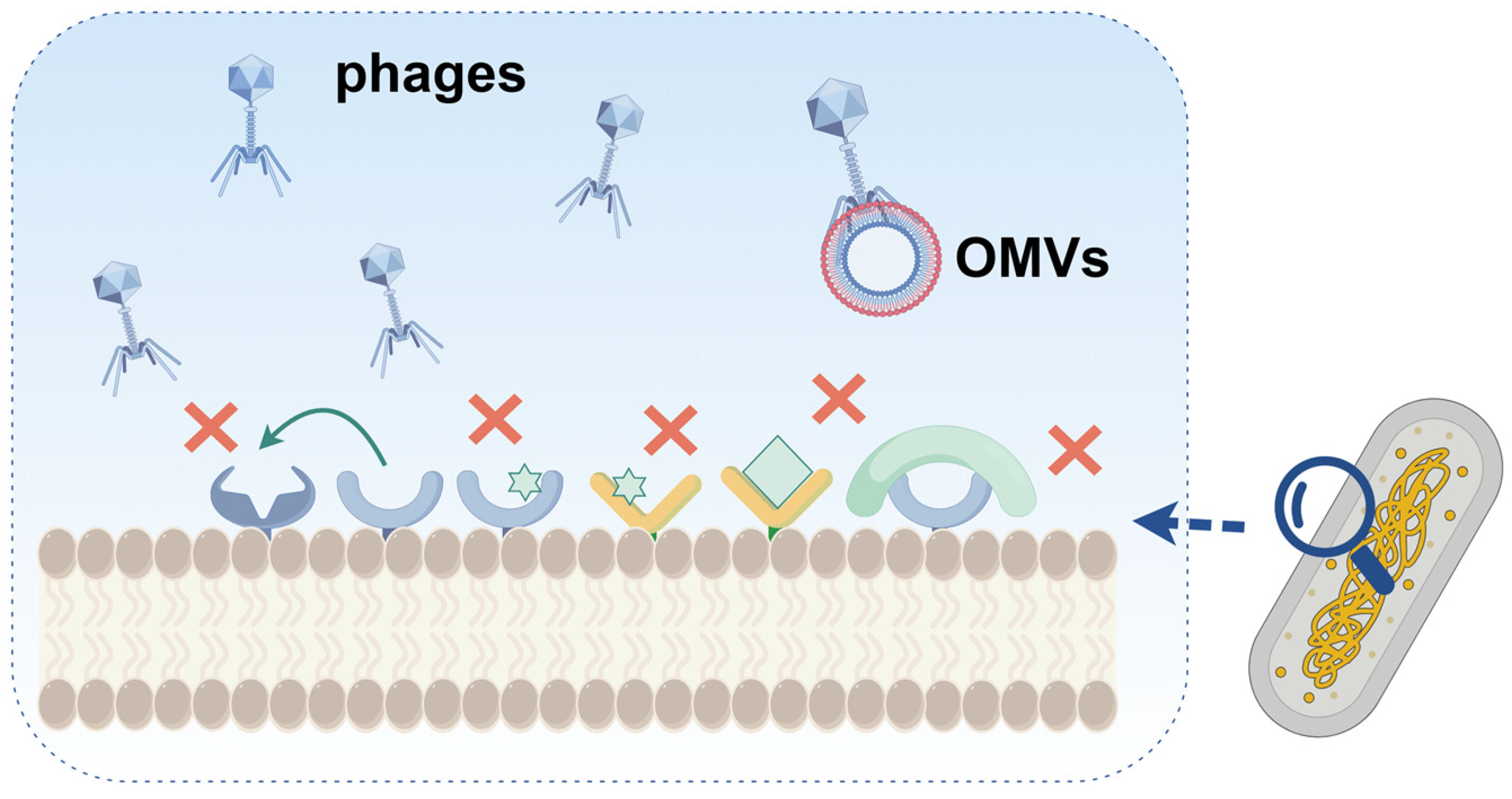
2.1. Adsorption Inhibition
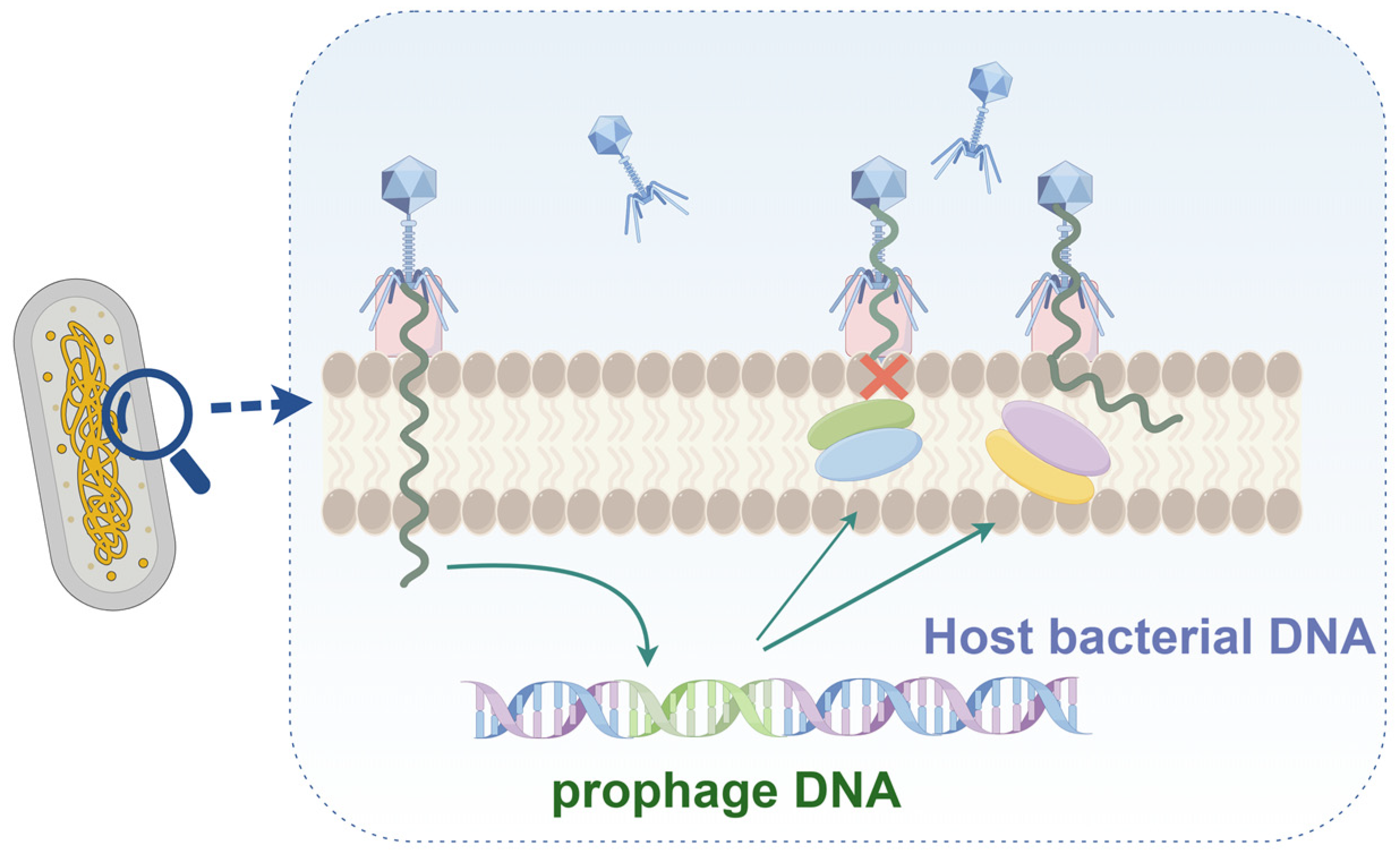
2.2. Blocking DNA Injection
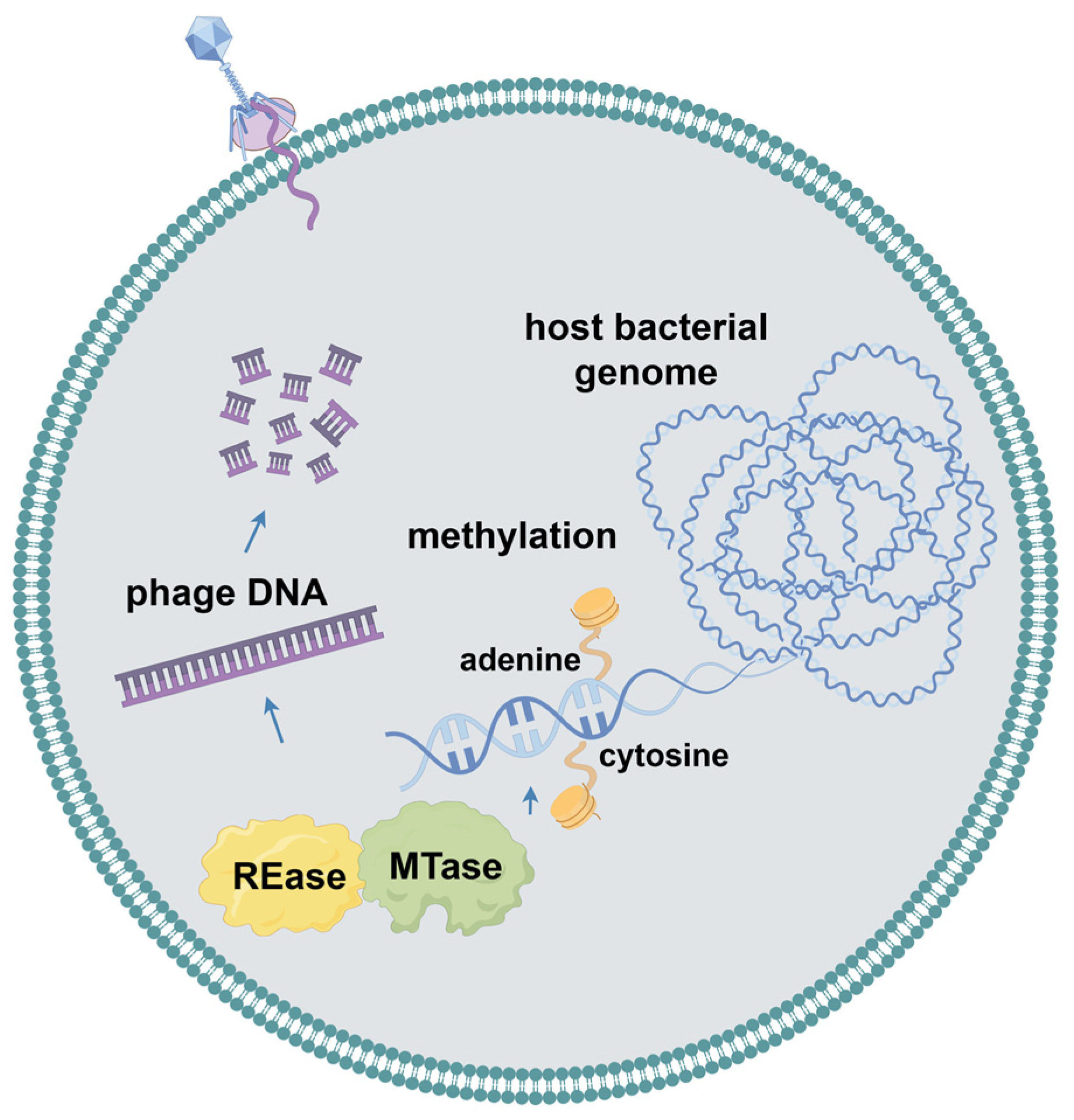
2.3. Restriction–Modification Systems
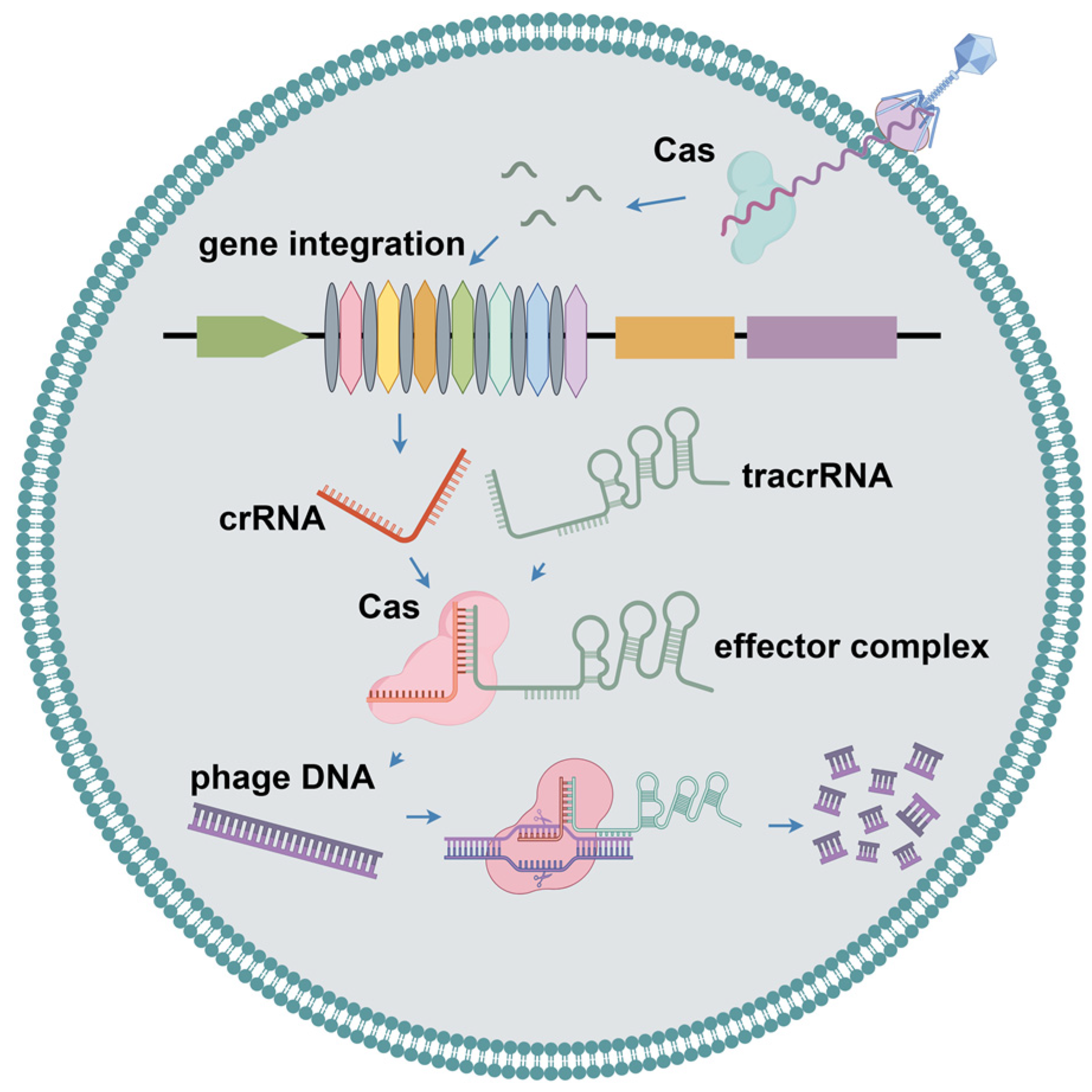
2.4. CRISPR-Cas Systems
2.5. Abortive Infection
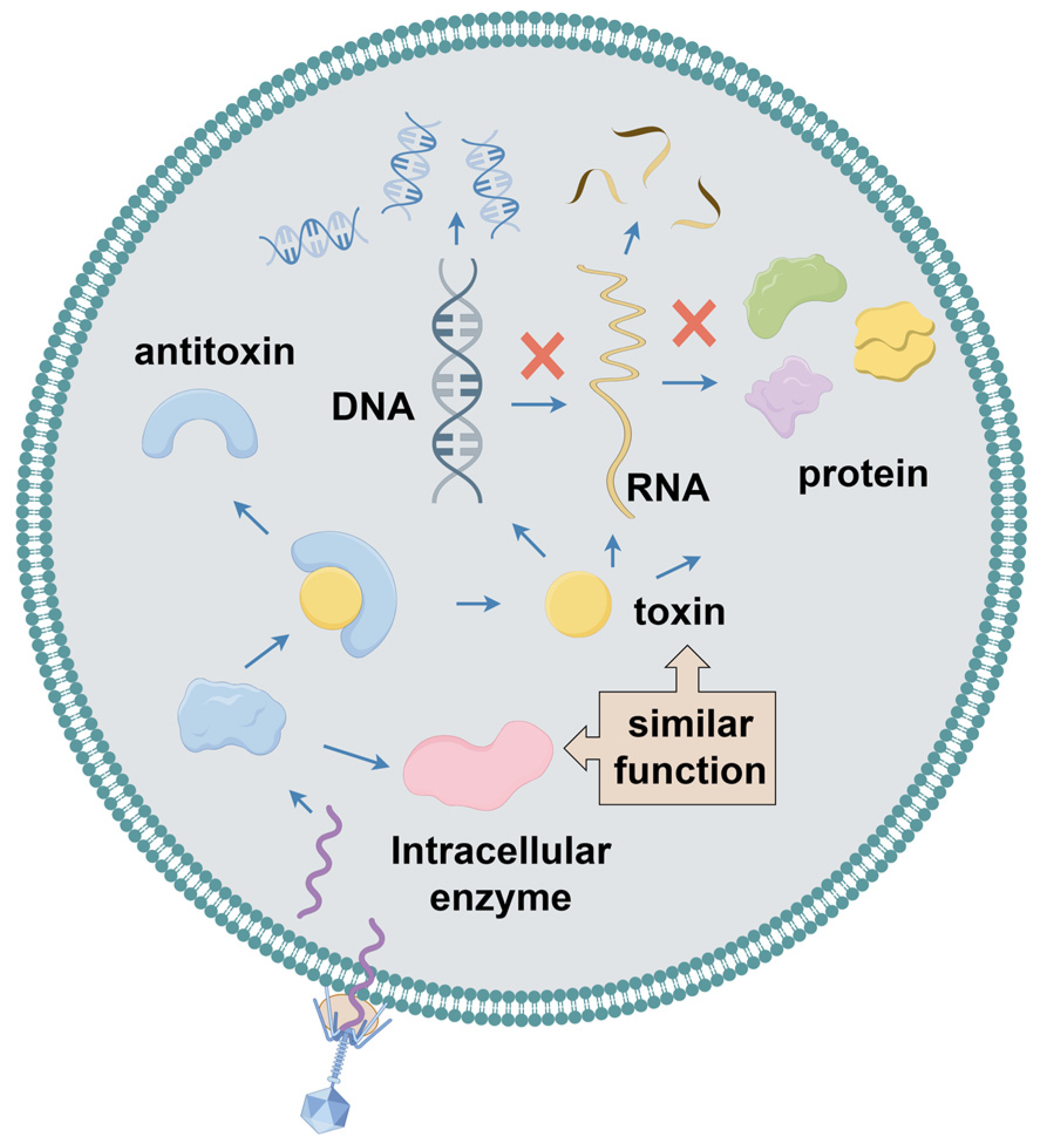
2.6. Toxin–Anti-Toxin System
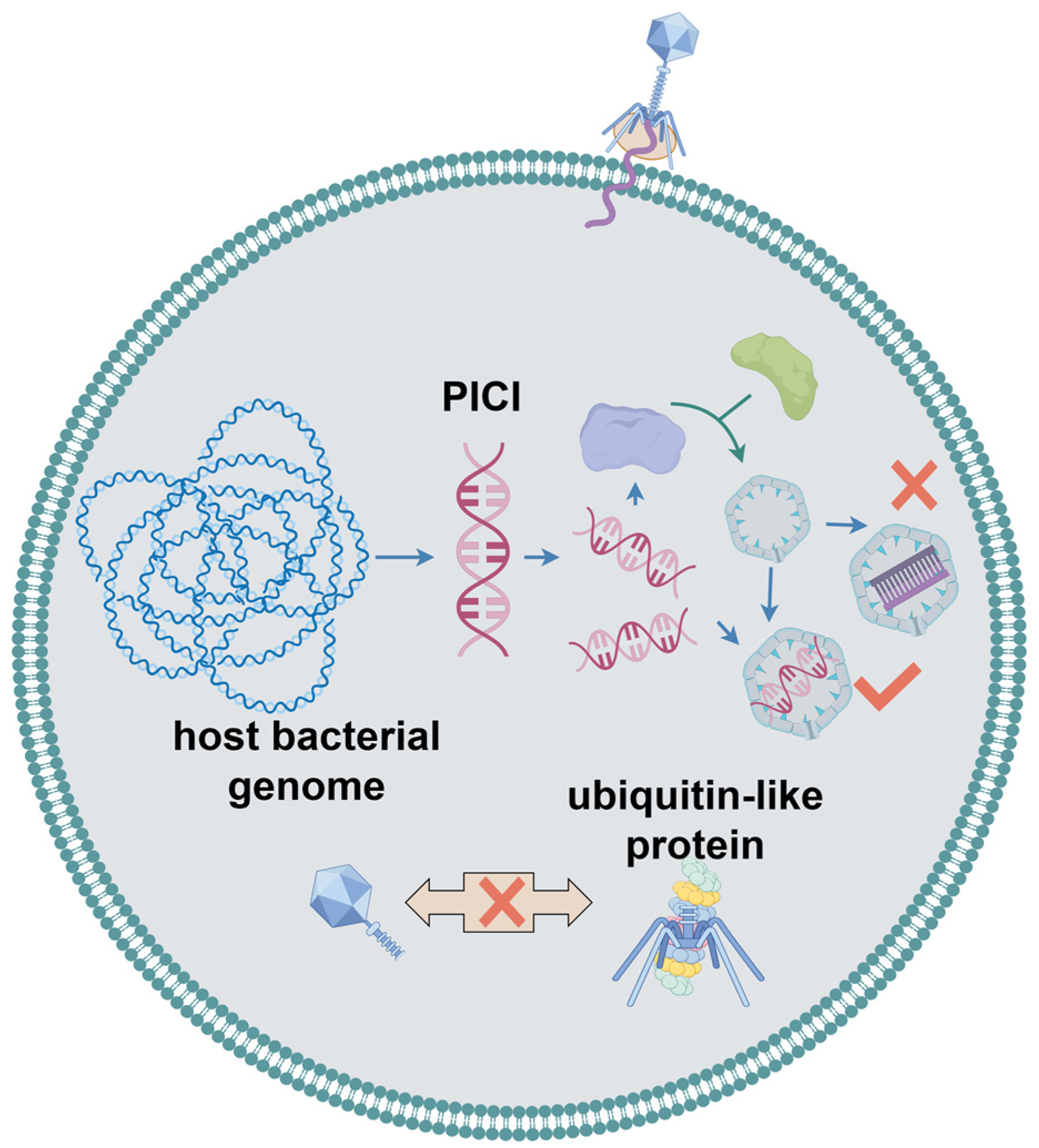
2.7. Bacteriophage Assembly Interference
2.8. DRT2 System
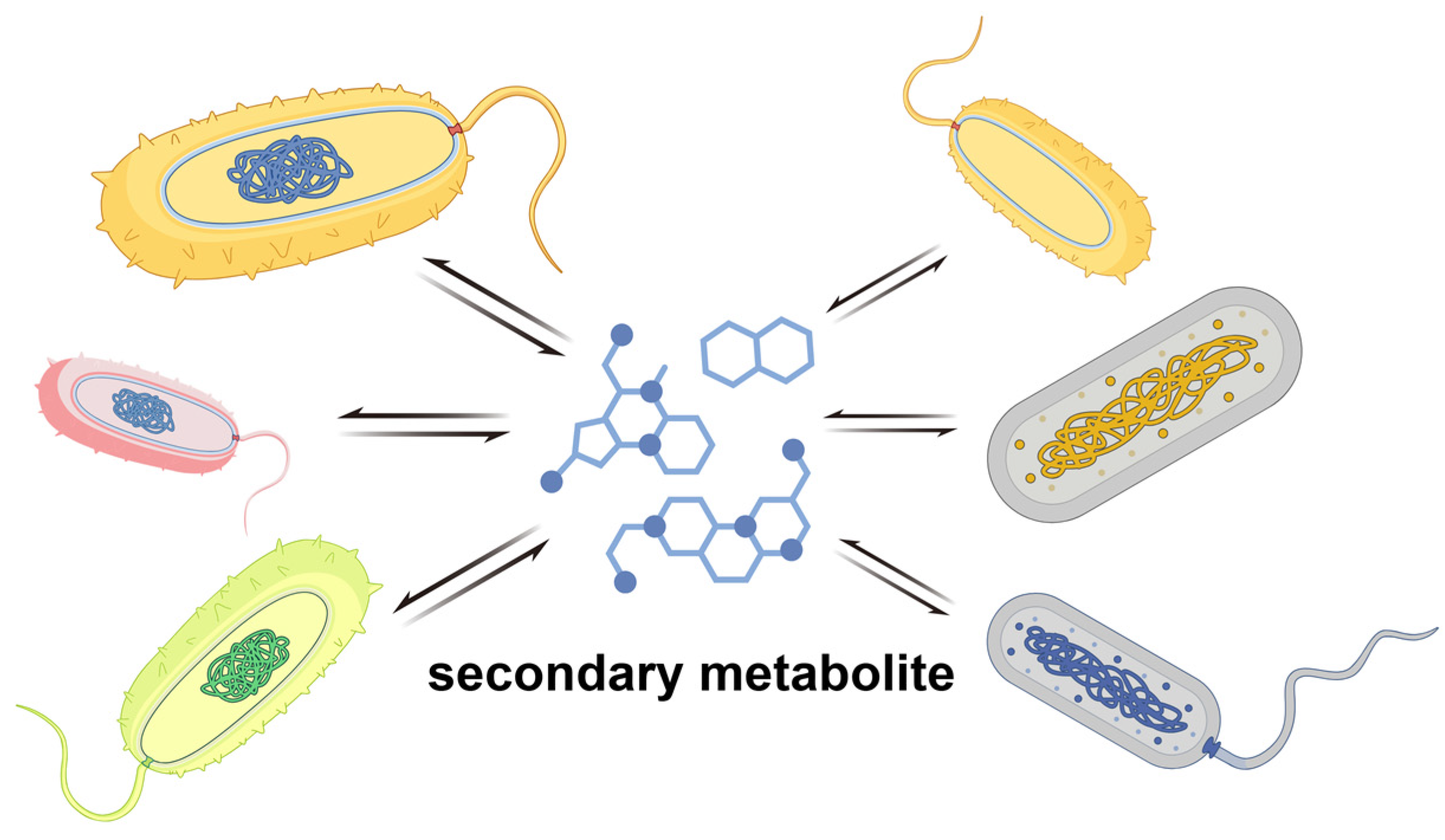
3. Anti-Phage Behavior at the Multicellular Level
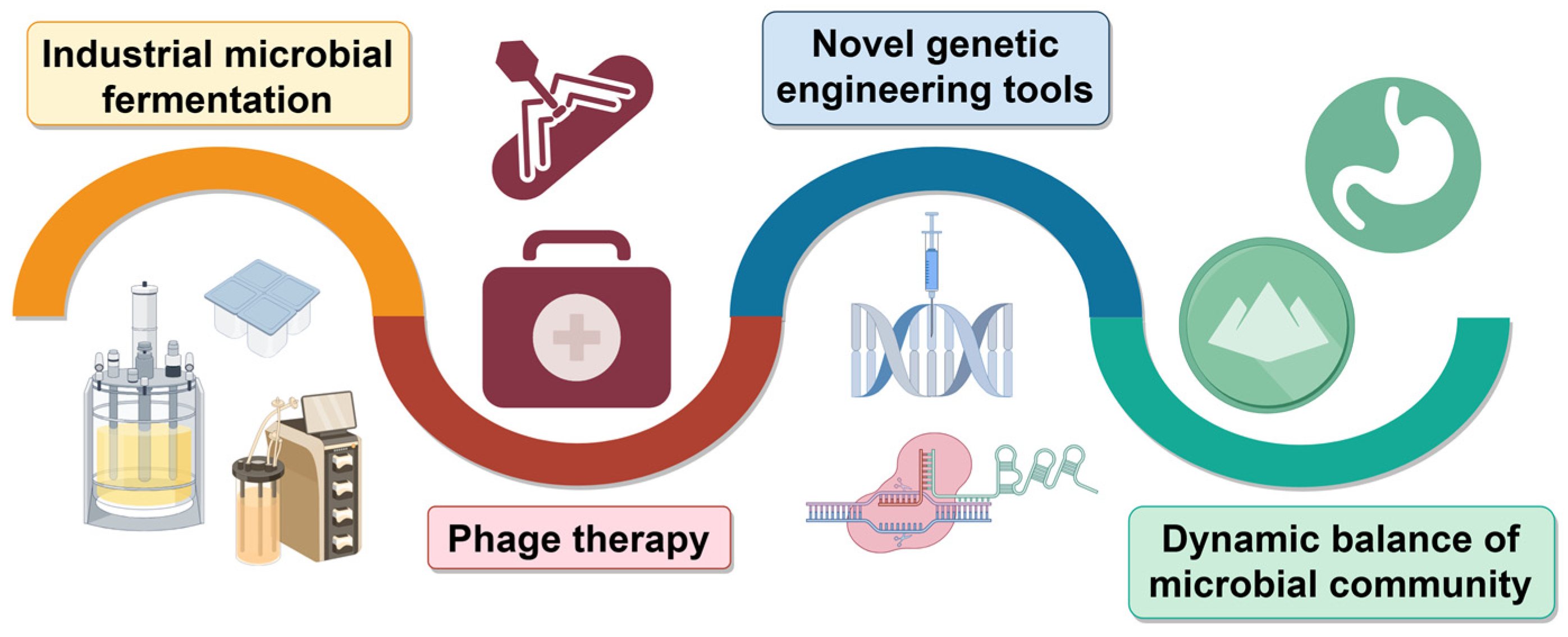
4. Application of Phage-Resistant Bacteria Strains
5. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Adriaenssens, E.M.; Zerbini, F.M.; Abrescia, N.G.A.; Aiewsakun, P.; Alfenas-Zerbini, P.; Bao, Y.; Barylski, J.; Drosten, C.; Duffy, S.; et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023, 21, e3001922. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Borra, J.; González, S.; López-Larrea, C. The origin of the bacterial immune response. Adv. Exp. Med. Biol. 2012, 738, 1–13. [Google Scholar] [PubMed]
- Vassallo, C.N.; Doering, C.R.; Littlehale, M.L.; Teodoro, G.I.C.; Laub, M.T. A functional selection reveals previously undetected anti-phage defence systems in the E. coli pangenome. Nat. Microbiol. 2022, 7, 1568–1579. [Google Scholar] [CrossRef]
- Luthe, T.; Kever, L.; Thormann, K.; Frunzke, J. Bacterial multicellular behavior in antiviral defense. Curr. Opin. Microbiol. 2023, 74, 102314. [Google Scholar] [CrossRef]
- Rostøl, J.T.; Marraffini, L. (Ph)ighting Phages: How Bacteria Resist Their Parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Arias, C.F.; Acosta, F.J.; Bertocchini, F.; Herrero, M.A.; Fernández-Arias, C. The coordination of anti-phage immunity mechanisms in bacterial cells. Nat. Commun. 2022, 13, 7412. [Google Scholar] [CrossRef]
- Zou, X.; Xiao, X.H.; Mo, Z.R.; Ge, Y.S.; Jiang, X.; Huang, R.L.; Li, M.X.; Deng, Z.X.; Chen, S.; Wang, L.R.; et al. Systematic strategies for developing phage resistant Escherichia coli strains. Nat. Commun. 2022, 13, 4491. [Google Scholar] [CrossRef]
- Karpov, D.S. CRISPR-Cas Systems and Genome Editing: Beginning the Era of CRISPR/Cas Therapies for Humans. Int. J. Mol. Sci. 2024, 25, 5292. [Google Scholar] [CrossRef]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front. Microbiol. 2019, 10, 954. [Google Scholar] [CrossRef]
- Domingo-Calap, P.; Mora-Quilis, L.; Sanjuán, R. Social Bacteriophages. Microorganisms 2020, 8, 533. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Robles, T.; Dillard, R.S.; Cairns, L.S.; Silva-Valenzuela, C.A.; Housman, M.; Ali, A.; Wright, E.R.; Camilli, A.; DiRita, V.J. Vibrio cholerae Outer Membrane Vesicles Inhibit Bacteriophage Infection. J. Bacteriol. 2018, 200, e00792-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, W.; Zhang, Q.; Wang, H.; Xu, X.; Diao, B.; Zhang, L.; Kan, B. The core oligosaccharide and thioredoxin of Vibrio cholerae are necessary for binding and propagation of its typing phage VP3. J. Bacteriol. 2009, 191, 2622–2629. [Google Scholar] [CrossRef]
- Pickard, D.; Toribio, A.L.; Petty, N.K.; van Tonder, A.; Yu, L.; Goulding, D.; Barrell, B.; Rance, R.; Harris, D.; Wetter, M.; et al. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica Serovar Typhi. J. Bacteriol. 2010, 192, 5746–5754. [Google Scholar] [CrossRef]
- Harvey, H.; Bondy-Denomy, J.; Marquis, H.; Sztanko, K.M.; Davidson, A.R.; Burrows, L.L. Pseudomonas aeruginosa defends against phages through type IV pilus glycosylation. Nat. Microbiol. 2018, 3, 47–52. [Google Scholar] [CrossRef]
- Trudelle, D.M.; Bryan, D.W.; Hudson, L.K.; Denes, T.G. Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019, 84, 103239. [Google Scholar] [CrossRef] [PubMed]
- Scholl, D.; Adhya, S.; Merril, C. Escherichia coli K1’s capsule is a barrier to bacteriophage T7. Appl. Environ. Microbiol. 2005, 71, 4872–4874. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef] [PubMed]
- Xuan, G.; Lin, H.; Kong, J.; Wang, J. Phage Resistance Evolution Induces the Sensitivity of Specific Antibiotics in Pseudomonas aeruginosa PAO1. Microbiol. Spectr. 2022, 10, e0135622. [Google Scholar] [CrossRef]
- Endriss, F.; Braun, V. Loop deletions indicate regions important for FhuA transport and receptor functions in Escherichia coli. J. Bacteriol. 2004, 186, 4818–4823. [Google Scholar] [CrossRef]
- Riede, I.; Eschbach, M.L. Evidence that TraT interacts with OmpA of Escherichia coli. FEBS Lett. 1986, 205, 241–245. [Google Scholar] [CrossRef]
- Achtman, M.; Kennedy, N.; Skurray, R. Cell—Cell interactions in conjugating Escherichia coli: Role of traT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 1977, 74, 5104–5108. [Google Scholar] [CrossRef]
- Hor, J.; Wolf, S.G.; Sorek, R. Bacteria conjugate ubiquitin-like proteins to interfere with phage assembly. Nature 2024, 631, 850–856. [Google Scholar] [CrossRef] [PubMed]
- van Houte, S.; Buckling, A.; Westra, E.R. Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol. Mol. Biol. Rev. 2016, 80, 745–763. [Google Scholar] [CrossRef] [PubMed]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Wang, W.Q.; Li, Y.M.; Tang, K.H.; Lin, J.Z.; Gao, X.Y.; Guo, Y.X.; Wang, X.X. Filamentous prophage capsid proteins contribute to superinfection exclusion and phage defence in Pseudomonas aeruginosa. Environ. Microbiol. 2022, 24, 4285–4298. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Oakland, J.T.; Kurniawan, F.; Moeller, N.H.; Banerjee, S.; Aihara, H. Structural basis of superinfection exclusion by bacteriophage T4 Spackle. Commun. Biol. 2020, 3, 691. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Göhler, A.; Heller, K.J.; Neve, H. The ltp gene of temperate Streptococcus thermophilus phage TP-J34 confers superinfection exclusion to Streptococcus thermophilus and Lactococcus lactis. Virology 2006, 350, 146–157. [Google Scholar] [CrossRef]
- Illingworth, C.; Hunter, M.; Fusco, D. Superinfection exclusion: A viral strategy with short-term benefits and long-term drawbacks. PLoS Comput. Biol. 2022, 18, e1010125. [Google Scholar]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef] [PubMed]
- Tock, M.R.; Dryden, D.T.F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Teklemariam, A.D.; Al-Hindi, R.R.; Qadri, I.; Alharbi, M.G.; Ramadan, W.S.; Ayubu, J.; Al-Hejin, A.M.; Hakim, R.F.; Hakim, F.F.; Hakim, R.F.; et al. The Battle between Bacteria and Bacteriophages: A Conundrum to Their Immune System. Antibiotics 2023, 12, 381. [Google Scholar] [CrossRef] [PubMed]
- Dy, R.L.; Richter, C.; Salmond, G.P.C.; Fineran, P.C. Remarkable Mechanisms in Microbes to Resist Phage Infections. Annu. Rev. Virol. 2014, 1, 307–331. [Google Scholar] [CrossRef]
- Goldfarb, T.; Sberro, H.; Weinstock, E.; Cohen, O.; Doron, S.; Charpak-Amikam, Y.; Afik, S.; Ofir, G.; Sorek, R. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 2014, 34, 169–183. [Google Scholar] [CrossRef]
- Gordeeva, J.; Morozova, N.; Sierro, N.; Isaev, A.; Sinkunas, T.; Tsvetkova, K.; Matlashov, M.; Truncaitė, L.; Morgan, R.D.; Ivanov, N.V.; et al. BREX system of Escherichia coli distinguishes self from non-self by methylation of a specific DNA site. Nucleic Acids Res. 2019, 47, 253–265. [Google Scholar] [CrossRef]
- Ofir, G.; Melamed, S.; Sberro, H.; Mukamel, Z.; Silverman, S.; Yaakov, G.; Doron, S.; Sorek, R. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2017, 3, 90–98. [Google Scholar] [CrossRef]
- Weigele, P.; Raleigh, E.A. Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. Chem. Rev. 2016, 116, 12655–12687. [Google Scholar] [CrossRef]
- Sun, Y.; Kong, L.; Wu, G.; Cao, B.; You, D. DNA Phosphorothioate Modifications Are Widely Distributed in the Human Microbiome. Biomolecules 2020, 10, 1175. [Google Scholar] [CrossRef]
- Xiong, L.; Liu, S.; Chen, S.; Xiao, Y.; Zhu, B.; Gao, Y.; Zhang, Y.; Chen, B.; Luo, J.; Deng, Z.; et al. A new type of DNA phosphorothioation-based antiviral system in archaea. Nat. Commun. 2019, 10, 1688. [Google Scholar] [CrossRef]
- Xiong, X.; Wu, G.; Wei, Y.; Liu, L.; Zhang, Y.; Su, R.; Jiang, X.; Li, M.; Gao, H.; Tian, X.; et al. SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nat. Microbiol. 2020, 5, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wan, M.; Huang, R.; Zhang, Y.; Xie, Y.; Wei, Y.; Ahmad, M.; Wu, D.; Hong, Y.; Deng, Z.; et al. SspABCD-SspFGH Constitutes a New Type of DNA Phosphorothioate-Based Bacterial Defense System. mBio 2021, 12, e00613-21. [Google Scholar] [CrossRef] [PubMed]
- Tong, T.; Chen, S.; Wang, L.; Tang, Y.; Ryu, J.Y.; Jiang, S.; Wu, X.; Chen, C.; Luo, J.; Deng, Z. Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E2988–E2996. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Nami, Y.; Rostampour, M.; Panahi, B. CRISPR-Cas systems and diversity of targeting phages in Lactobacillus johnsonii strains; insights from genome mining approach. Infect. Genet. Evol. 2023, 114, 105500. [Google Scholar] [CrossRef]
- Deng, X.; Yuan, J.; Chen, L.; Chen, H.; Wei, C.; Nielsen, P.H.; Wuertz, S.; Qiu, G. CRISPR-Cas phage defense systems and prophages in Candidatus Accumulibacter. Water Res. 2023, 235, 11990. [Google Scholar] [CrossRef]
- Lin, P.; Pu, Q.; Shen, G.; Li, R.; Guo, K.; Zhou, C.; Liang, H.; Jiang, J.; Wu, M. CdpR Inhibits CRISPR-Cas Adaptive Immunity to Lower Anti-viral Defense while Avoiding Self-Reactivity. iScience 2019, 13, 55–68. [Google Scholar] [CrossRef]
- Cury, J.; Bernheim, A. CRISPR-Cas and restriction–modification team up to achieve long-term immunity. Trends Microbiol. 2022, 30, 513–514. [Google Scholar] [CrossRef]
- Lopatina, A.; Tal, N.; Sorek, R. Abortive Infection: Bacterial Suicide as an Antiviral Immune Strategy. Annu. Rev. Virol. 2020, 7, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Depardieu, F.; Didier, J.P.; Bernheim, A.; Sherlock, A.; Molina, H.; Duclos, B.; Bikard, D. A Eukaryotic-like Serine/Threonine Kinase Protects Staphylococci against Phages. Cell Host Microbe 2016, 20, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Garb, J.; Lopatina, A.; Bernheim, A.; Zaremba, M.; Siksnys, V.; Melamed, S.; Leavitt, A.; Millman, A.; Amitai, G.; Sorek, R. Multiple phage resistance systems inhibit infection via SIR2-dependent NAD(+) depletion. Nat. Microbiol. 2022, 7, 1849–1856. [Google Scholar] [CrossRef]
- Zaremba, M.; Dakineviciene, D.; Golovinas, E.; Zagorskaitė, E.; Stankunas, E.; Lopatina, A.; Sorek, R.; Manakova, E.; Ruksenaite, A.; Silanskas, A.; et al. Short prokaryotic Argonautes provide defence against incoming mobile genetic elements through NAD(+) depletion. Nat. Microbiol. 2022, 7, 1857–1869. [Google Scholar] [CrossRef] [PubMed]
- Sather, L.M.; Zamani, M.; Muhammed, Z.; Kearsley, J.V.S.; Fisher, G.T.; Jones, K.M.; Finan, T.M. A broadly distributed predicted helicase/nuclease confers phage resistance via abortive infection. Cell Host Microbe 2023, 31, 343–355. [Google Scholar] [CrossRef]
- Mayo-Muñoz, D.; Smith, L.M.; Garcia-Doval, C.; Malone, L.M.; Harding, K.R.; Jackson, S.A.; Hampton, H.G.; Fagerlund, R.D.; Gumy, L.F.; Fineran, P.C. Type III CRISPR-Cas provides resistance against nucleus-forming jumbo phages via abortive infection. Mol. Cell 2022, 82, 4471–4486. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial Retrons Function In Anti-Phage Defense. Cell 2020, 183, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Bobonis, J.; Mitosch, K.; Mateus, A.; Karcher, N.; Kritikos, G.; Selkrig, J.; Zietek, M.; Monzon, V.; Pfalz, B.; Garcia-Santamarina, S.; et al. Bacterial retrons encode phage-defending tripartite toxin-antitoxin systems. Nature 2022, 609, 144–150. [Google Scholar] [CrossRef]
- Tal, N.; Millman, A.; Stokar-Avihail, A.; Fedorenko, T.; Leavitt, A.; Melamed, S.; Yirmiya, E.; Avraham, C.; Brandis, A.; Mehlman, T.; et al. Bacteria deplete deoxynucleotides to defend against bacteriophage infection. Nat. Microbiol. 2022, 7, 1200–1209. [Google Scholar] [CrossRef]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.W.; Bogard, R.W.; Young, T.S.; Mekalanos, J.J. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 2012, 149, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Lowey, B.; Whiteley, A.T.; Keszei, A.F.A.; Morehouse, B.R.; Mathews, I.T.; Antine, S.P.; Cabrera, V.J.; Kashin, D.; Niemann, P.; Jain, M.; et al. CBASS Immunity Uses CARF-Related Effectors to Sense 3′-5′- and 2′-5′-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection. Cell 2020, 182, 38–49. [Google Scholar] [CrossRef]
- Whiteley, A.T.; Eaglesham, J.B.; de Oliveira Mann, C.C.; Morehouse, B.R.; Lowey, B.; Nieminen, E.A.; Danilchanka, O.; King, D.S.; Lee, A.S.Y.; Mekalanos, J.J.; et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 2019, 567, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Bernheim, A.; Sorek, R. The pan-immune system of bacteria: Antiviral defence as a community resource. Nat. Rev. Microbiol. 2020, 18, 113–119. [Google Scholar] [CrossRef]
- Millman, A.; Melamed, S.; Amitai, G.; Sorek, R. Diversity and classification of cyclic-oligonucleotide-based anti-phage signalling systems. Nat. Microbiol. 2020, 5, 1608–1615. [Google Scholar] [CrossRef]
- Ye, Q.; Lau, R.K.; Mathews, I.T.; Birkholz, E.A.; Watrous, J.D.; Azimi, C.S.; Pogliano, J.; Jain, M.; Corbett, K.D. HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity. Mol. Cell 2020, 77, 709–722. [Google Scholar] [CrossRef]
- Duncan-Lowey, B.; McNamara-Bordewick, N.K.; Tal, N.; Sorek, R.; Kranzusch, P.J. Effector-mediated membrane disruption controls cell death in CBASS antiphage defense. Mol. Cell 2021, 81, 5039–5051. [Google Scholar] [CrossRef] [PubMed]
- Lau, R.K.; Ye, Q.; Birkholz, E.A.; Berg, K.R.; Patel, L.; Mathews, I.T.; Watrous, J.D.; Ego, K.; Whiteley, A.T.; Lowey, B.; et al. Structure and Mechanism of a Cyclic Trinucleotide-Activated Bacterial Endonuclease Mediating Bacteriophage Immunity. Mol. Cell 2020, 77, 723–733. [Google Scholar] [CrossRef]
- Gao, L.; Altae-Tran, H.; Böhning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Zhang, D.; Schäffer, D.E.; Iyer, L.M.; Aravind, L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015, 43, 10633–10654. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Gao, L.; Zhang, F.; Koonin, E.V. Unexpected connections between type VI-B CRISPR-Cas systems, bacterial natural competence, ubiquitin signaling network and DNA modification through a distinct family of membrane proteins. FEMS Microbiol. Lett. 2019, 366, fnz088. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Luo, X.; Li, P.; Li, Z.; Ye, F.; Liu, S.; Gao, P. Molecular basis of RADAR anti-phage supramolecular assemblies. Cell 2023, 186, 999–1012. [Google Scholar] [CrossRef]
- Duncan-Lowey, B.; Tal, N.; Johnson, A.G.; Rawson, S.; Mayer, M.L.; Doron, S.; Millman, A.; Melamed, S.; Fedorenko, T.; Kacen, A.; et al. Cryo-EM structure of the RADAR supramolecular anti-phage defense complex. Cell 2023, 186, 987–998. [Google Scholar] [CrossRef]
- Page, R.; Peti, W. Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 2016, 12, 208–214. [Google Scholar] [CrossRef]
- Lin, J.; Guo, Y.; Yao, J.; Tang, K.; Wang, X. Applications of toxin-antitoxin systems in synthetic biology. Eng. Microbiol. 2023, 3, 100069. [Google Scholar] [CrossRef]
- Song, S.; Wood, T.K. A Primary Physiological Role of Toxin/Antitoxin Systems Is Phage Inhibition. Front. Microbiol. 2020, 11, 1895. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhai, Y.; Wei, M.; Zheng, C.; Jiao, X. Toxin-antitoxin systems: Classification, biological roles, and applications. Microbiol. Res. 2022, 264, 127159. [Google Scholar] [CrossRef] [PubMed]
- Guegler, C.K.; Laub, M.T. Shutoff of host transcription triggers a toxin-antitoxin system to cleave phage RNA and abort infection. Mol. Cell 2021, 81, 2361–2373. [Google Scholar] [CrossRef]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.; Fineran, P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef]
- Cui, Y.; Su, X.; Wang, C.; Xu, H.; Hu, D.; Wang, J.; Pei, K.; Sun, M.; Zou, T. Bacterial MazF/MazE toxin-antitoxin suppresses lytic propagation of arbitrium-containing phages. Cell Rep. 2022, 41, 111752. [Google Scholar] [CrossRef]
- Songailiene, I.; Juozapaitis, J.; Tamulaitiene, G.; Ruksenaite, A.; Šulčius, S.; Sasnauskas, G.; Venclovas, Č.; Siksnys, V. HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation. Mol. Cell 2020, 80, 955–970. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; Sumby, P.; Smith, M.C.M. The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology 2015, 477, 100–109. [Google Scholar] [CrossRef]
- Czarnecki, J.; Dziewit, L.; Kowalski, L.; Ochnio, M.; Bartosik, D. Maintenance and genetic load of plasmid pKON1 of Paracoccus kondratievae, containing a highly efficient toxin-antitoxin module of the hipAB family. Plasmid 2015, 80, 45–53. [Google Scholar] [CrossRef]
- Chen, Z.; Yao, J.; Zhang, P.; Wang, P.; Ni, S.; Liu, T.; Zhao, Y.; Tang, K.; Sun, Y.; Qian, Q.; et al. Minimized antibiotic-free plasmid vector for gene therapy utilizing a new toxin-antitoxin system. Metab. Eng. 2023, 79, 86–96. [Google Scholar] [CrossRef]
- Bleriot, I.; Blasco, L.; Pacios, O.; Fernández-García, L.; Ambroa, A.; López, M.; Ortiz-Cartagena, C.; Cuenca, F.F.; Oteo-Iglesias, J.; Pascual, Á.; et al. The role of PemIK (PemK/PemI) type II TA system from Klebsiella pneumoniae clinical strains in lytic phage infection. Sci. Rep. 2022, 12, 4488. [Google Scholar] [CrossRef] [PubMed]
- LeRoux, M.; Srikant, S.; Teodoro, G.I.C.; Zhang, T.; Littlehale, M.L.; Doron, S.; Badiee, M.; Leung, A.K.L.; Sorek, R.; Laub, M.T. The DarTG toxin-antitoxin system provides phage defence by ADP-ribosylating viral DNA. Nat. Microbiol. 2022, 7, 1028–1040. [Google Scholar] [CrossRef]
- Hsueh, B.Y.; Severin, G.B.; Elg, C.A.; Waldron, E.J.; Kant, A.; Wessel, A.J.; Dover, J.A.; Rhoades, C.R.; Ridenhour, B.J.; Parent, K.N.; et al. Phage defence by deaminase-mediated depletion of deoxynucleotides in bacteria. Nat. Microbiol. 2022, 7, 1210–1220. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Tang, K.; Sit, B.; Gu, J.; Chen, R.; Lin, J.; Lin, S.; Liu, X.; Wang, W.; Gao, X.; et al. Dual control of lysogeny and phage defense by a phosphorylation-based toxin/antitoxin system. bioRxiv 2022, bioRxiv:2022.09.05.506569. [Google Scholar] [CrossRef]
- Mittal, P.; Sinha, A.K.; Pandiyan, A.; Kumari, L.; Ray, M.K.; Pavankumar, T.L. A type II toxin-antitoxin system is responsible for the cell death at low temperature in Pseudomonas syringae Lz4W lacking RNase R. J. Biol. Chem. 2024, 300, 107600. [Google Scholar] [CrossRef]
- Fillol-Salom, A.; Martínez-Rubio, R.; Abdulrahman, R.F.; Chen, J.; Davies, R.; Penadés, J.R. Phage-inducible chromosomal islands are ubiquitous within the bacterial universe. ISME J. 2018, 12, 2114–2128. [Google Scholar] [CrossRef] [PubMed]
- Penadés, J.R.; Christie, G.E. The Phage-Inducible Chromosomal Islands: A Family of Highly Evolved Molecular Parasites. Annu. Rev. Virol. 2015, 2, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Tormo-Más, M.A.; Mir, I.; Shrestha, A.; Tallent, S.M.; Campoy, S.; Lasa, I.; Barbé, J.; Novick, R.P.; Christie, G.E.; Penadés, J.R. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature 2010, 465, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Fillol-Salom, A.; Miguel-Romero, L.; Marina, A.; Chen, J.; Penadés, J.R. Beyond the CRISPR-Cas safeguard: PICI-encoded innate immune systems protect bacteria from bacteriophage predation. Curr. Opin. Microbiol. 2020, 56, 52–58. [Google Scholar] [CrossRef]
- Tang, S.; Conte, V.; Zhang, D.J.; Žedaveinytė, R.; Lampe, G.D.; Wiegand, T.; Tang, L.C.; Wang, M.; Walker, M.W.G.; George, J.T.; et al. De novo gene synthesis by an antiviral reverse transcriptase. Science 2024, 386, eadq0876. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Li, D.; Gao, A.; Macrae, R.K.; Zhang, F. Phage-triggered reverse transcription assembles a toxic repetitive gene from a noncoding RNA. Science 2024, 386, eadq3977. [Google Scholar] [CrossRef]
- Osterman, I.; Sorek, R. Tricking phages with a reverse move. Science 2024, 386, 25–26. [Google Scholar] [CrossRef]
- Høyland-Kroghsbo, N.M.; Paczkowski, J.; Mukherjee, S.; Broniewski, J.; Westra, E.; Bondy-Denomy, J.; Bassler, B.L. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc. Natl. Acad. Sci. USA 2017, 114, 131–135. [Google Scholar] [CrossRef]
- Kronheim, S.; Daniel-Ivad, M.; Duan, Z.; Hwang, S.; Wong, A.I.; Mantel, I.; Nodwell, J.R.; Maxwell, K.L. A chemical defence against phage infection. Nature 2018, 564, 283–286. [Google Scholar] [CrossRef]
- Hardy, A.; Kever, L.; Frunzke, J. Antiphage small molecules produced by bacteria-beyond protein-mediated defenses. Trends Microbiol. 2023, 31, 92–106. [Google Scholar] [CrossRef]
- Xuan, G.; Tan, L.; Yang, Y.; Kong, J.; Lin, H.; Wang, J. Quorum sensing autoinducers AHLs protect Shewanella baltica against phage infection. Int. J. Food Microbiol. 2023, 403, 110304. [Google Scholar] [CrossRef] [PubMed]
- Mienda, B.S.; Drager, A. Genome-Scale Metabolic Modeling of Escherichia coli and Its Chassis Design for Synthetic Biology Applications. Methods Mol. Biol. 2021, 2189, 217–229. [Google Scholar] [PubMed]
- Dong, X.R.; Liu, B.; Bao, Y.H.; Liu, W.F.; Tao, Y. Metabolic engineering of Escherichia coli for high-level production of violaxanthin. Microb. Cell Fact. 2023, 22, 115. [Google Scholar]
- Bao, Z.X.; Gao, Y.T.; Song, Y.T.; Ding, N.; Li, W.; Wu, Q.; Zhang, X.M.; Zheng, Y.; Li, J.M.; Hu, X.J. Construction of an Escherichia coli chassis for efficient biosynthesis of human-like N-linked glycoproteins. Front. Bioeng. Biotechnol. 2024, 12, 1370685. [Google Scholar] [CrossRef] [PubMed]
- Giesbers, C.A.P.; Fagan, J.; Parlindungan, E.; Palussiere, S.; Courtin, P.; Lugli, G.A.; Ventura, M.; Kulakauskas, S.; Chapot-Chartier, M.-P.; Mahony, J.; et al. Reduced synthesis of phospho-polysaccharide in Lactococcus as a strategy to evade phage infection. Int. J. Food Microbiol. 2023, 407, 110415. [Google Scholar] [CrossRef] [PubMed]
- Guérin, H.; Quénée, P.; Palussière, S.; Courtin, P.; André, G.; Péchoux, C.; Costache, V.; Mahony, J.; van Sinderen, D.; Kulakauskas, S.; et al. PBP2b Mutations Improve the Growth of Phage-Resistant Lactococcus cremoris Lacking Polysaccharide Pellicle. Appl. Environ. Microbiol. 2023, 89, e0210322. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Chen, X.; Xu, M.; Liu, R.; Lian, W.; Ma, Y.; Ibrahim, A.A. Selection and characterization of spontaneous phage-resistant mutant of Limosilactobacillus fermentum. Int. J. Food Microbiol. 2024, 423, 110833. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Lin, H.; Mi, Z.; Xing, S.; Tong, Y.; Wang, J. Screening of Polyvalent Phage-Resistant Escherichia coli Strains Based on Phage Receptor Analysis. Front. Microbiol. 2019, 10, 850. [Google Scholar] [CrossRef]
- Nagarajan, V.; Peng, M.; Tabashsum, Z.; Salaheen, S.; Padilla, J.; Biswas, D. Antimicrobial Effect and Probiotic Potential of Phage Resistant Lactobacillus plantarum and its Interactions with Zoonotic Bacterial Pathogens. Foods 2019, 8, 194. [Google Scholar] [CrossRef]
- Xu, Z.; Ding, Z.; Shi, L.; Xie, Y.; Zhang, Y.; Sao, S.; Wang, Q.; Liu, Q. Design combinations of evolved phage and antibiotic for antibacterial guided by analyzing the phage resistance of poorly antimicrobial phage. Microbiol. Spectr. 2023, 11, e0095823. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.N.; Zeng, Y.G.; Chen, L.G.; Li, L.L.; Yang, L.; Zhang, Y.Y.; Guo, M.Q.; Li, L.S.; Li, J.; et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.K.; Shen, M.M.; Liu, S.L.; Zhou, X. Characterization and resistance mechanism of phage-resistant strains of Salmonella enteritidis. Poult. Sci. 2024, 103, 103756. [Google Scholar] [CrossRef] [PubMed]
- Alseth, E.O.; Custodio, R.; Sundius, S.A.; Kuske, R.A.; Brown, S.P.; Westra, E.R. The impact of phage and phage resistance on microbial community dynamics. PLoS Biol. 2024, 22, e3002346. [Google Scholar] [CrossRef]
- Zeng, X.; Liang, S.; Dong, J.; Gao, G.; Hu, Y.; Sun, Y. The trade-off of Vibrio parahaemolyticus between bacteriophage resistance and growth competitiveness. Front. Microbiol. 2024, 15, 1346251. [Google Scholar]
- Yuan, Y.; Peng, Q.; Zhang, S.; Liu, T.; Yang, S.; Yu, Q.; Wu, Y.; Gao, M. Phage Reduce Stability for Regaining Infectivity during Antagonistic Coevolution with Host Bacterium. Viruses 2019, 11, 118. [Google Scholar] [CrossRef]
- Su-Jin, P.; Han, J.-h.; Kim, Y.-K. Isolation of bacteriophage-resistant Pseudomonas tolaasii strains and their pathogenic characters. J. Appl. Biol. Chem. 2016, 59, 351–356. [Google Scholar]
- Sorensen, P.E.; Baig, S.; Stegger, M.; Ingmer, H.; Garmyn, A.; Butaye, P. Spontaneous Phage Resistance in Avian Pathogenic Escherichia coli. Front Microbiol. 2021, 12, 782757. [Google Scholar] [CrossRef]
- Thavalingam, A.; Cheng, Z.; Garcia, B.; Huang, X.; Shah, M.; Sung, W.; Wang, M.; Harrington, L.; Hwang, S.; Hidalgo-Reyes, Y.; et al. Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2. Nat. Commun. 2019, 10, 2806. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Yonesaki, T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 2012, 83, 669–681. [Google Scholar] [CrossRef]
- Costa, A.R.; van den Berg, D.F.; Esser, J.Q.; Muralidharan, A.; van den Bossche, H.; Bonilla, B.E.; van der Steen, B.A.; Haagsma, A.C.; Fluit, A.C.; Nobrega, F.L.; et al. Accumulation of defense systems in phage-resistant strains of Pseudomonas aeruginosa. Sci. Adv. 2024, 10, eadj0341. [Google Scholar] [CrossRef]
- Zhou, S.; Yuan, S.-F.; Nair, P.H.; Alper, H.S.; Deng, Y.; Zhou, J. Development of a growth coupled and multi-layered dynamic regulation network balancing malonyl-CoA node to enhance (2S)-naringenin biosynthesis in Escherichia coli. Metab. Eng. 2021, 67, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Schann, K.; Bakker, J.; Boinot, M.; Kuschel, P.; He, H.; Nattermann, M.; Paczia, N.; Erb, T.; Bar-Even, A.; Wenk, S. Design, construction and optimization of formaldehyde growth biosensors with broad application in biotechnology. Microb. Biotechnol. 2024, 17, e14527. [Google Scholar] [CrossRef] [PubMed]
- Diallo, K.; Dublanchet, A. Benefits of Combined Phage-Antibiotic Therapy for the Control of Antibiotic-Resistant Bacteria: A Literature Review. Antibiotics 2022, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Necel, A.; Bloch, S.; Topka-Bielecka, G.; Janiszewska, A.; Łukasiak, A.; Nejman-Faleńczyk, B.; Węgrzyn, G. Synergistic Effects of Bacteriophage vB_Eco4-M7 and Selected Antibiotics on the Biofilm Formed by Shiga Toxin-Producing Escherichia coli. Antibiotics 2022, 11, 712. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Gu, P. Overview of Phage Defense Systems in Bacteria and Their Applications. Int. J. Mol. Sci. 2024, 25, 13316. https://doi.org/10.3390/ijms252413316
Xu X, Gu P. Overview of Phage Defense Systems in Bacteria and Their Applications. International Journal of Molecular Sciences. 2024; 25(24):13316. https://doi.org/10.3390/ijms252413316
Chicago/Turabian StyleXu, Xiaomei, and Pengfei Gu. 2024. "Overview of Phage Defense Systems in Bacteria and Their Applications" International Journal of Molecular Sciences 25, no. 24: 13316. https://doi.org/10.3390/ijms252413316
APA StyleXu, X., & Gu, P. (2024). Overview of Phage Defense Systems in Bacteria and Their Applications. International Journal of Molecular Sciences, 25(24), 13316. https://doi.org/10.3390/ijms252413316





