The Identification and Characterization of WOX Family Genes in Coffea arabica Reveals Their Potential Roles in Somatic Embryogenesis and the Cold-Stress Response
Abstract
1. Introduction
2. Results
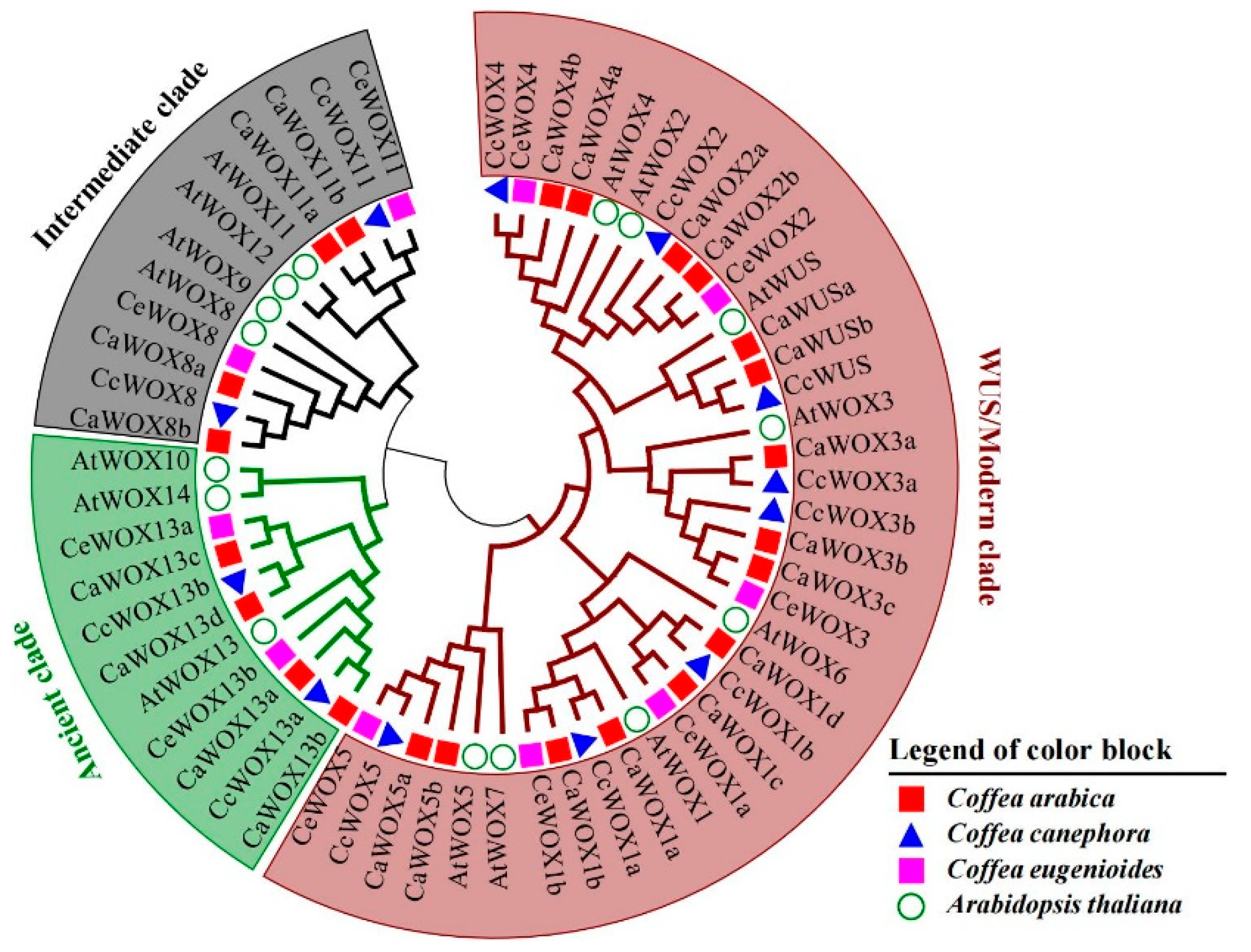
2.1. Identification and Phylogenetic Analysis of WOX Genes in Three Coffee Plants
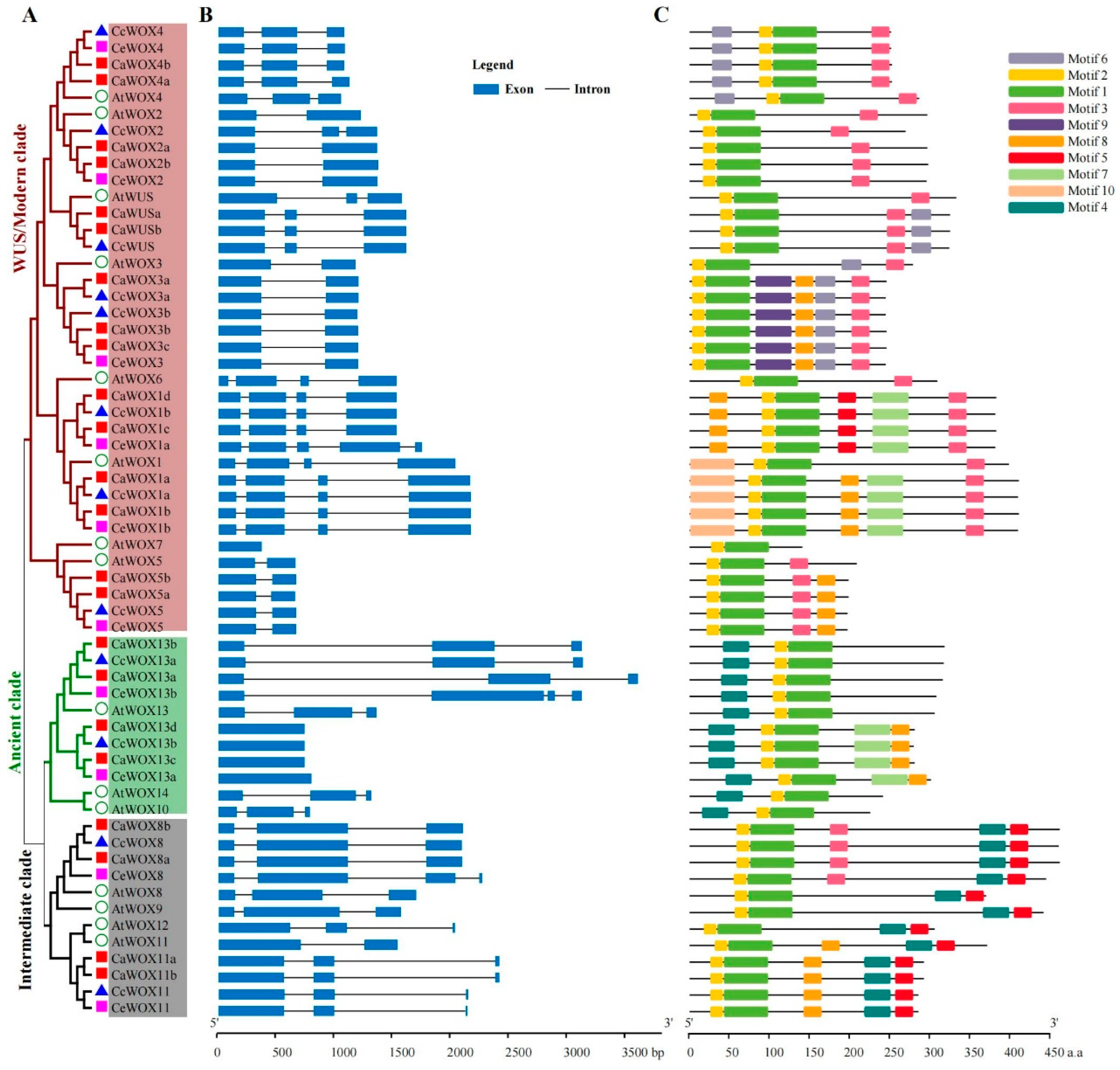
2.2. Gene Structure and Domain Analysis of CaWOXs
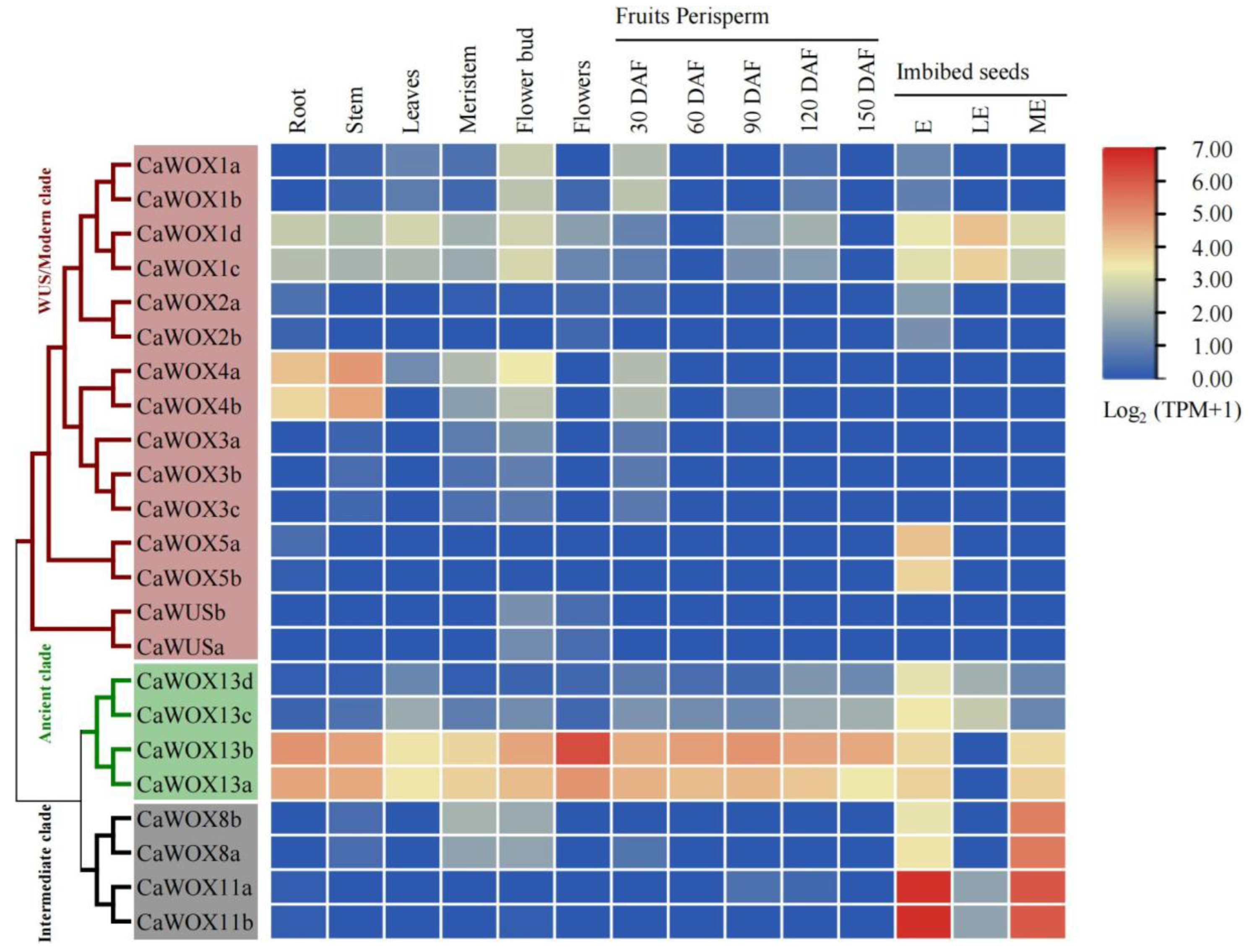
2.3. Expression Profiles of CaWOXs in Different Tissues
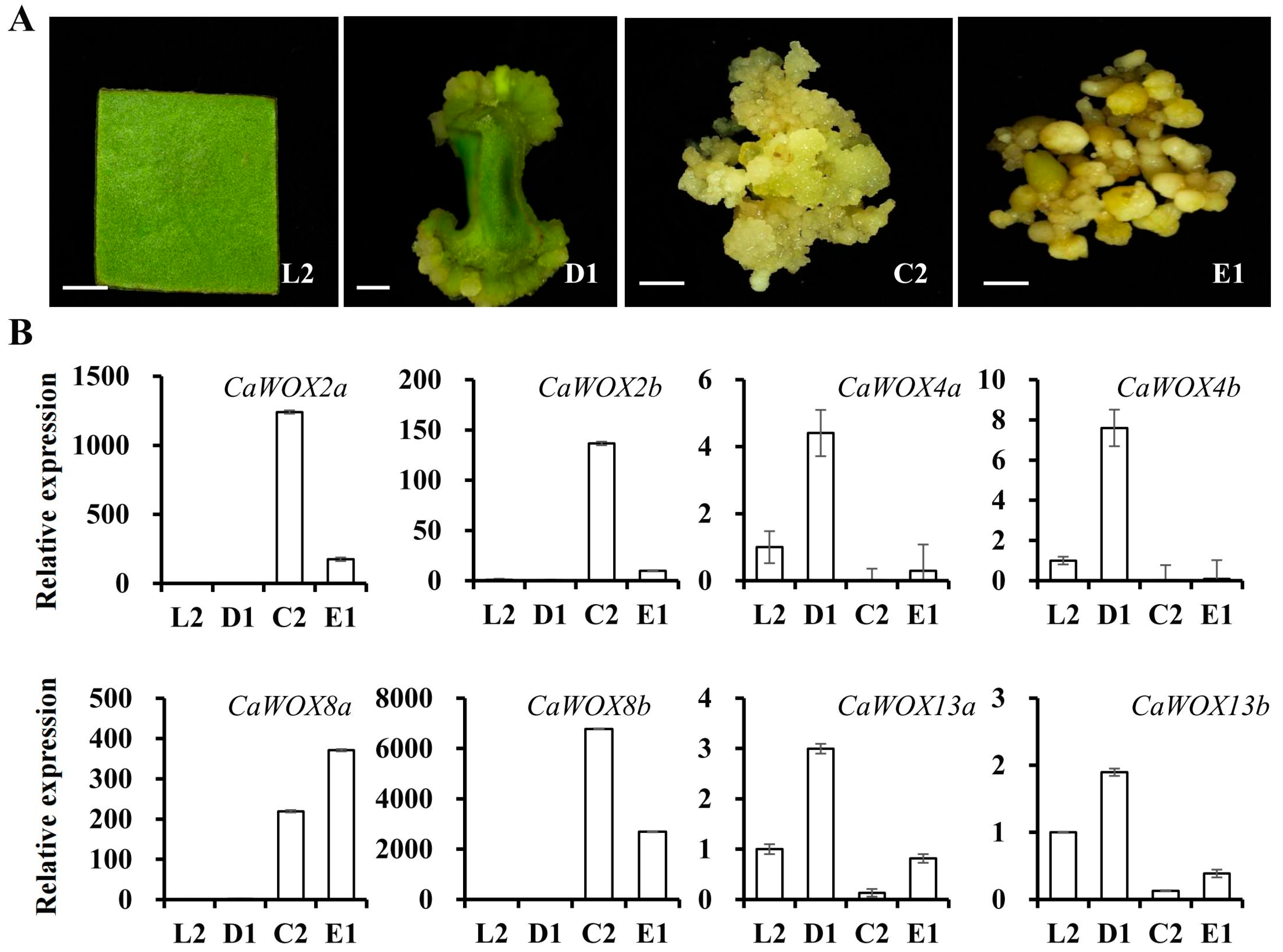
2.4. Expression Patterns of CaWOX Genes During Somatic Embryogenesis
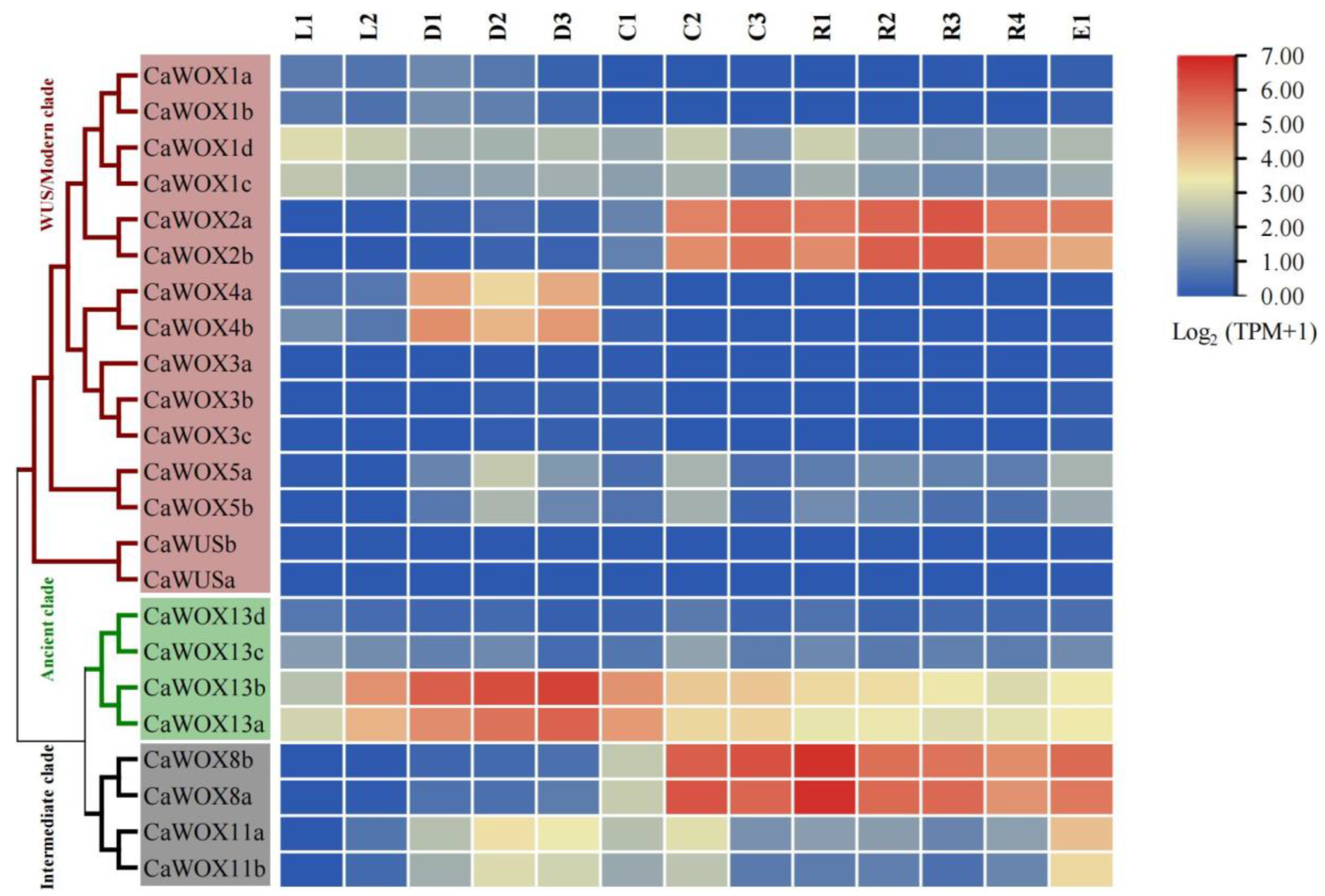
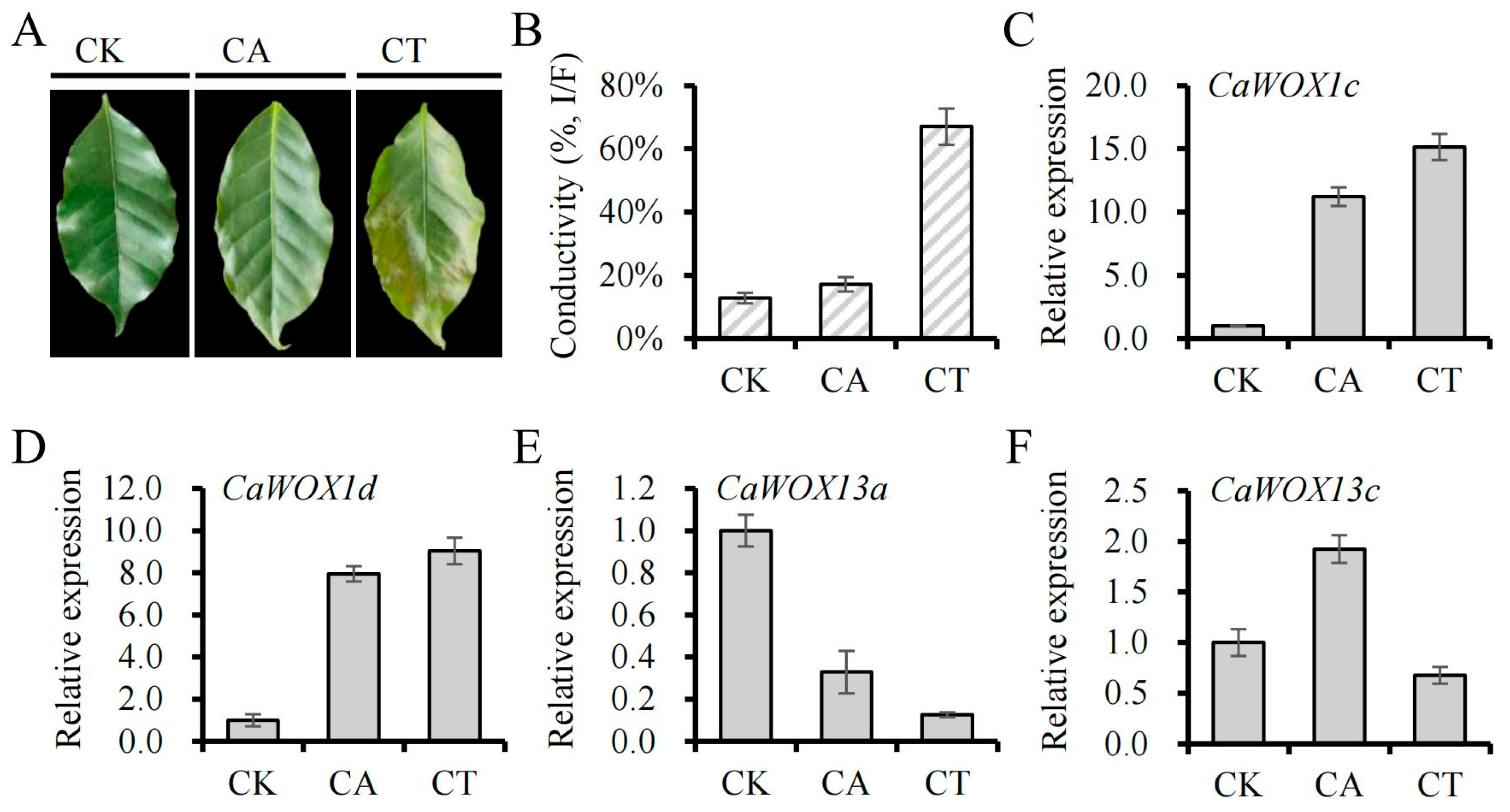
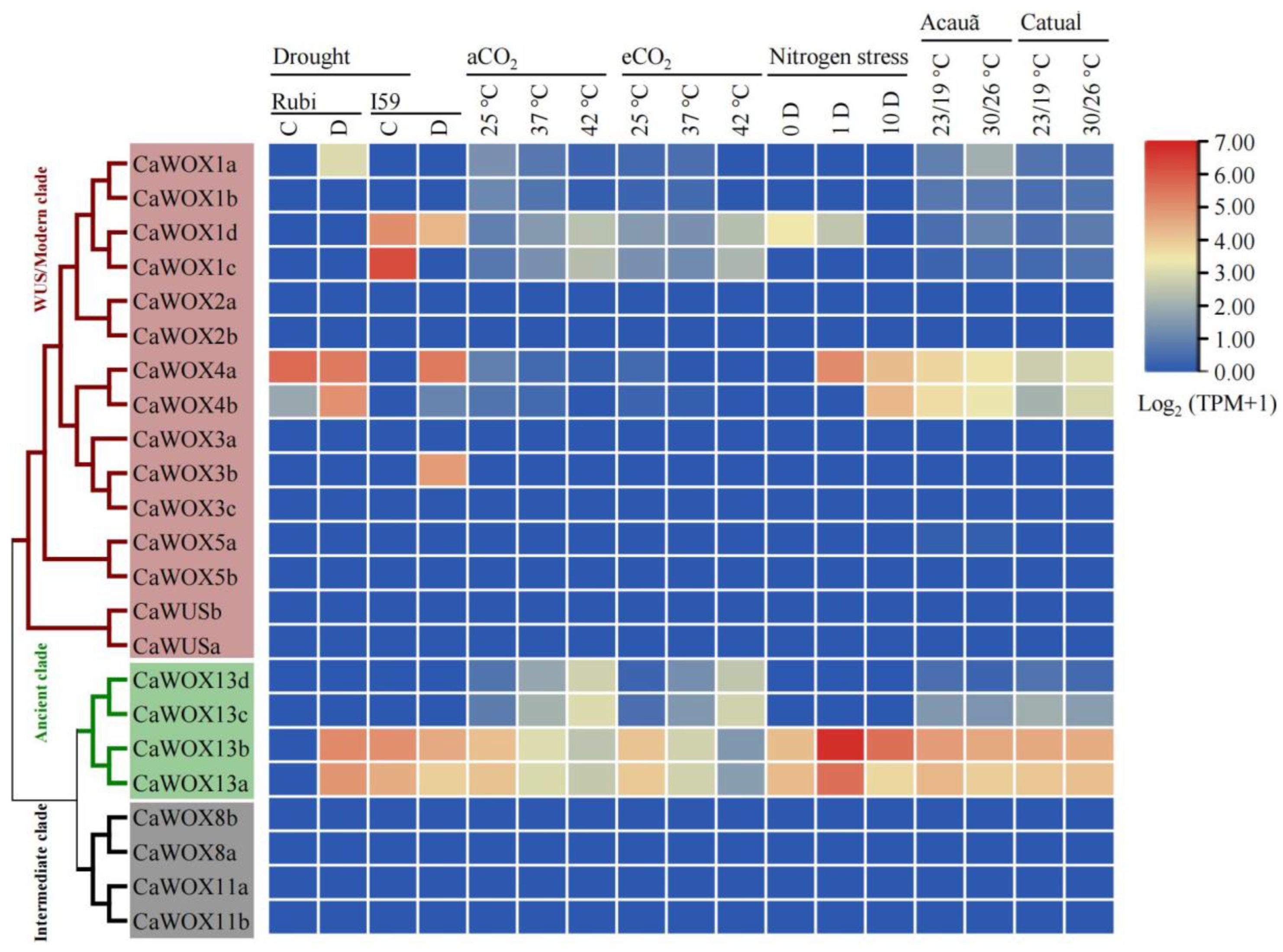
2.5. Expression Patterns of CaWOX Genes Under Cold Stress and Other Stress Treatments
3. Discussion
3.1. WOX Genes in C. arabica
3.2. CaWOX Functions
3.3. CaWOX and Somatic Embryogenesis
3.4. Potential Mechanisms of CaWOXs in Somatic Embryogenesis
3.5. CaWOX and Cold Stress
4. Materials and Methods
4.1. Identification of WOX Genes in Coffea Species
4.2. Phylogenetic and Bioinformatic Analysis of CaWOXs
4.3. Cold Treatment, Electrolyte Leakage Test, and Tissue Culture
4.4. Expression Analysis of CaWOX Genes Within RNA-Seq Data
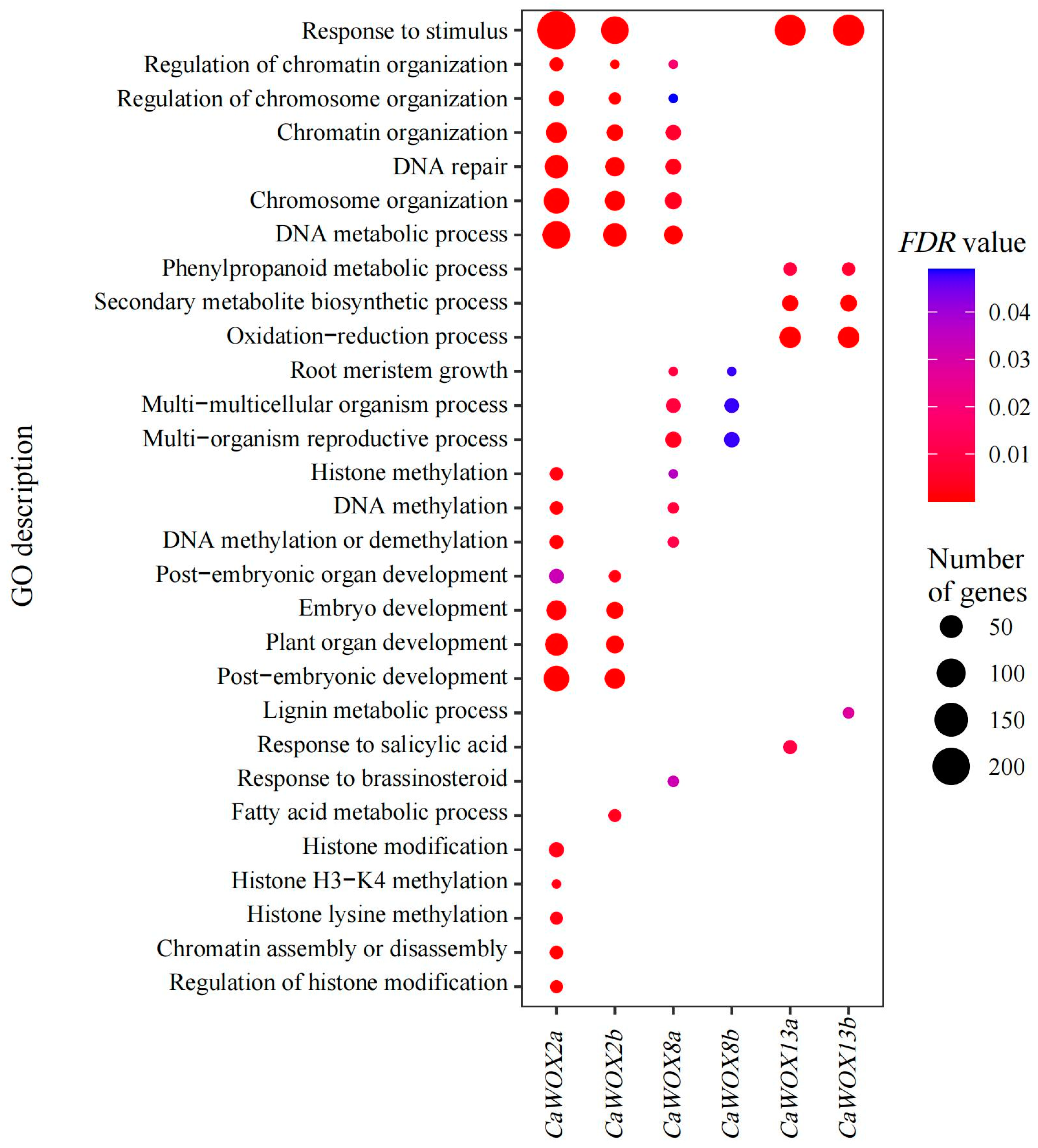
4.5. Co-Expression and GO Enrichment Analysis
4.6. RNA Isolation, cDNA Synthesis, and Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [PubMed]
- van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef] [PubMed]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.J.; Müller, M.; Affolter, M.; Percival-Smith, A.; Billeter, M.; Qian, Y.Q.; Otting, G.; Wüthrich, K. The structure of the homeodomain and its functional implications. Trends Genet. 1990, 6, 323–329. [Google Scholar] [CrossRef]
- Chen, W.; Yan, J.; Guan, Y.; Lou, H.; Wu, J. Genome-wide identification of WOX gene family and its expression pattern in rapid expansion of Torreya grandis ovulate and staminate strobili. Sci. Hortic. 2024, 330, 113050. [Google Scholar] [CrossRef]
- Yin, S.; Zhao, L.; Liu, J.; Sun, Y.; Li, B.; Wang, L.; Ren, Z.; Chen, C. Pan-genome Analysis of WOX Gene Family and Function Exploration of CsWOX9 in Cucumber. Int. J. Mol. Sci. 2023, 24, 17568. [Google Scholar] [CrossRef]
- Lian, G.; Ding, Z.; Wang, Q.; Zhang, D.; Xu, J. Origins and evolution of WUSCHEL-related homeobox protein family in plant kingdom. ScientificWorldJournal 2014, 2014, 534140. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Horstman, A.; Zethof, J.; Koes, R.; Rijpkema, A.S.; Gerats, T. Differential recruitment of WOX transcription factors for lateral development and organ fusion in Petunia and Arabidopsis. Plant Cell 2009, 21, 2269–2283. [Google Scholar] [CrossRef]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Zuo, J.; Niu, Q.-W.; Frugis, G.; Chua, N.-H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002, 30, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Herrera, A.; Ku Gonzalez, A.; Canche Moo, R.; Quiroz-Figueroa, F.R.; Loyola-Vargas, V.M.; Rodriguez-Zapata, L.C.; Burgeff D′Hondt, C.; Suárez-Solís, V.M.; Castaño, E. Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Cult. 2008, 94, 171–180. [Google Scholar] [CrossRef]
- Bouchabké-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Zhang, X.; Ma, D.; Liu, C. Identification and Evolutionary Analysis of Cotton (Gossypium hirsutum) WOX Family Genes and Their Potential Function in Somatic Embryogenesis. Int. J. Mol. Sci. 2023, 24, 11077. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, R.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.-J. Over-expression of WOX1 Leads to Defects in Meristem Development and Polyamine Homeostasis in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 493–506. [Google Scholar] [CrossRef]
- Breuninger, H.; Rikirsch, E.; Hermann, M.; Ueda, M.; Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 2008, 14, 867–876. [Google Scholar] [CrossRef]
- Shimizu, R.; Ji, J.; Kelsey, E.; Ohtsu, K.; Schnable, P.S.; Scanlon, M.J. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol. 2009, 149, 841–850. [Google Scholar] [CrossRef]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 Imparts Auxin Responsiveness to Cambium Cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 act downstream of the PXY receptor kinase to regulate plant vascular proliferation independently of any role in vascular organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde, E.S.N.; Kreis, M.; Deveaux, Y. WOX14 promotes bioactive gibberellin synthesis and vascular cell differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef]
- Kong, X.; Lu, S.; Tian, H.; Ding, Z. WOX5 is Shining in the Root Stem Cell Niche. Trends Plant Sci. 2015, 20, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Savina, M.S.; Pasternak, T.; Omelyanchuk, N.A.; Novikova, D.D.; Palme, K.; Mironova, V.V.; Lavrekha, V.V. Cell Dynamics in WOX5-Overexpressing Root Tips: The Impact of Local Auxin Biosynthesis. Front. Plant Sci. 2020, 11, 560169. [Google Scholar] [CrossRef] [PubMed]
- Park, S.O.; Zheng, Z.; Oppenheimer, D.G.; Hauser, B.A. The PRETTY FEW SEEDS2 gene encodes an Arabidopsishomeodomain protein that regulates ovule development. Development 2005, 132, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL Related Homeobox Protein WOX7 Regulates the Sugar Response of Lateral Root Development in Arabidopsis thaliana. Molocular Plant 2016, 9, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Blilou, I.; Grieneisen, V.A.; Sozzani, R.; Zamioudis, C.; Miskolczi, P.; Nieuwland, J.; Benjamins, R.; Dhonukshe, P.; et al. A Bistable Circuit Involving SCARECROW-RETINOBLASTOMA Integrates Cues to Inform Asymmetric Stem Cell Division. Cell 2012, 150, 1002–1015. [Google Scholar] [CrossRef]
- Wan, Q.; Zhai, N.; Xie, D.; Liu, W.; Xu, L. WOX11: The founder of plant organ regeneration. Cell Regen. 2023, 12, 1. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Ogura, N.; Sasagawa, Y.; Ito, T.; Tameshige, T.; Kawai, S.; Sano, M.; Doll, Y.; Iwase, A.; Kawamura, A.; Suzuki, T.; et al. WUSCHEL-RELATED HOMEOBOX 13 suppresses de novo shoot regeneration via cell fate control of pluripotent callus. Sci. Adv. 2023, 9, eadg6983. [Google Scholar] [CrossRef]
- Salaün, C.; Lepiniec, L.; Dubreucq, B. Genetic and Molecular Control of Somatic Embryogenesis. Plants 2021, 10, 1467. [Google Scholar] [CrossRef]
- Khan, F.S.; Goher, F.; Hu, C.G.; Zhang, J.Z. WUSCHEL-related homeobox (WOX) transcription factors: Key regulators in combating abiotic stresses in plants. Hortic. Adv. 2024, 2, 2. [Google Scholar] [CrossRef]
- Zhu, J.; Shi, H.; Lee, B.H.; Damsz, B.; Cheng, S.; Stirm, V.; Zhu, J.K.; Hasegawa, P.M.; Bressan, R.A. An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 9873–9878. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Huang, Y.; Zhu, N.; Zhao, Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene 2014, 549, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Rahman, Z.u.; Azam, S.M.; Liu, Y.; Yan, C.; Ali, H.; Zhao, L.; Chen, P.; Yi, L.; Priyadarshani, S.V.G.N.; Yuan, Q. Expression Profiles of Wuschel-Related Homeobox Gene Family in Pineapple (Ananas comosus L). Trop. Plant Biol. 2017, 10, 204–215. [Google Scholar] [CrossRef]
- Tang, F.; Chen, N.; Zhao, M.; Wang, Y.; He, R.; Peng, X.; Shen, S. Identification and Functional Divergence Analysis of WOX Gene Family in Paper Mulberry. Int. J. Mol. Sci. 2017, 18, 1782. [Google Scholar] [CrossRef]
- Hao, Q.; Zhang, L.; Yang, Y.; Shan, Z.; Zhou, X.-a. Genome-Wide Analysis of the WOX Gene Family and Function Exploration of GmWOX18 in Soybean. Plants 2019, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Guo, Y.; Chen, X.; Zheng, Y.; Sun, Y.; Yang, J.; Ye, N. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees 2019, 33, 1129–1142. [Google Scholar] [CrossRef]
- Chekol, H.; Warkineh, B.; Shimber, T.; Mierek-Adamska, A.; Dąbrowska, G.B.; Degu, A. Drought Stress Responses in Arabica Coffee Genotypes: Physiological and Metabolic Insights. Plants 2024, 13, 828. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Chen, Z.; Li, M.; Fan, L.; Zhang, M. Assessing the climate change impacts on Coffee arabica cultivation regions in China. Theor. Appl. Climatol. 2024, 155, 7773–7791. [Google Scholar] [CrossRef]
- Rigal, C.; Xu, J.; Hu, G.; Qiu, M.; Vaast, P. Coffee production during the transition period from monoculture to agroforestry systems in near optimal growing conditions, in Yunnan Province. Agric. Syst. 2020, 177, 102696. [Google Scholar] [CrossRef]
- Zaman, S.; Shan, Z. Literature Review of Proteomics Approach Associated with Coffee. Foods 2024, 13, 1670. [Google Scholar] [CrossRef]
- Salojärvi, J.; Rambani, A.; Yu, Z.; Guyot, R.; Strickler, S.; Lepelley, M.; Wang, C.; Rajaraman, S.; Rastas, P.; Zheng, C.; et al. The genome and population genomics of allopolyploid Coffea arabica reveal the diversification history of modern coffee cultivars. Nat. Genet. 2024, 56, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Denoeud, F.; Carretero-Paulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 2014, 345, 1181–1184. [Google Scholar] [CrossRef] [PubMed]
- Awada, R.; Lepelley, M.; Breton, D.; Charpagne, A.; Campa, C.; Berry, V.; Georget, F.; Breitler, J.-C.; Léran, S.; Djerrab, D.; et al. Global transcriptome profiling reveals differential regulatory, metabolic and hormonal networks during somatic embryogenesis in Coffea arabica. BMC Genom. 2023, 24, 41. [Google Scholar] [CrossRef] [PubMed]
- Mofatto, L.S.; Carneiro, F.d.A.; Vieira, N.G.; Duarte, K.E.; Vidal, R.O.; Alekcevetch, J.C.; Cotta, M.G.; Verdeil, J.-L.; Lapeyre-Montes, F.; Lartaud, M.; et al. Identification of candidate genes for drought tolerance in coffee by high-throughput sequencing in the shoot apex of different Coffea arabica cultivars. BMC Plant Biol. 2016, 16, 94. [Google Scholar] [CrossRef]
- de Oliveira, R.R.; Ribeiro, T.H.C.; Cardon, C.H.; Fedenia, L.; Maia, V.A.; Barbosa, B.C.F.; Caldeira, C.F.; Klein, P.E.; Chalfun-Junior, A. Elevated Temperatures Impose Transcriptional Constraints and Elicit Intraspecific Differences Between Coffee Genotypes. Front. Plant Sci. 2020, 11, 1113. [Google Scholar] [CrossRef]
- Marques, I.; Fernandes, I.; Paulo, O.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. A Transcriptomic Approach to Understanding the Combined Impacts of Supra-Optimal Temperatures and CO(2) Revealed Different Responses in the Polyploid Coffea arabica and Its Diploid Progenitor C. canephora. Int. J. Mol. Sci. 2021, 22, 3125. [Google Scholar] [CrossRef]
- Xu, A.; Yang, J.; Wang, S.; Zheng, L.; Wang, J.; Zhang, Y.; Bi, X.; Wang, H. Characterization and expression profiles of WUSCHEL-related homeobox (WOX) gene family in cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2023, 23, 471. [Google Scholar] [CrossRef]
- Duan, L.; Hou, Z.; Zhang, W.; Liang, S.; Huangfu, M.; Zhang, J.; Yang, T.; Dong, J.; Che, D. Genome-wide analysis of the WOX gene family and function exploration of RhWOX331 in rose (R. ‘The Fairy’). Front. Plant Sci. 2024, 15, 1461322. [Google Scholar] [CrossRef]
- Li, J.J.; Qiu, X.Y.; Dai, Y.J.; Nyonga, T.M.; Li, C.C. Genome-Wide Identification and Co-Expression Networks of WOX Gene Family in Nelumbo nucifera. Plants 2024, 13, 720. [Google Scholar] [CrossRef]
- Daude, M.M.; Dos Santos Silva, T.W.; Freitas, N.C.; Ságio, S.A.; Paiva, L.V.; Barreto, H.G. Transcriptional analysis of WUSCHEL-related HOMEOBOX (WOX) genes in Coffea arabica L. Biologia 2020, 75, 1483–1495. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Liao, J.; Deng, B.; Cai, X.; Yang, Q.; Hu, B.; Cong, J.; Zhang, Y.; Wang, G.; Xin, G.; Li, Y.; et al. Time-course transcriptome analysis reveals regulation of Arabidopsis seed dormancy by the transcription factors WOX11/12. J. Exp. Bot. 2022, 74, 1090–1106. [Google Scholar] [CrossRef] [PubMed]
- Asghar, S.; Ghori, N.; Hyat, F.; Li, Y.; Chen, C. Use of auxin and cytokinin for somatic embryogenesis in plant: A story from competence towards completion. Plant Growth Regul. 2023, 99, 413–428. [Google Scholar] [CrossRef]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Hoerster, G.; Wang, N.; Ryan, L.; Wu, E.; Anand, A.; McBride, K.; Lowe, K.; Jones, T.; Gordon-Kamm, B. Use of non-integrating Zm-Wus2 vectors to enhance maize transformation. In Vitro Cell. Dev. Biol. –Plant 2020, 56, 265–279. [Google Scholar] [CrossRef]
- Wang, K.; Shi, L.; Liang, X.; Zhao, P.; Wang, W.; Liu, J.; Chang, Y.; Hiei, Y.; Yanagihara, C.; Du, L.; et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants 2022, 8, 110–117. [Google Scholar] [CrossRef]
- Jones, T.; Lowe, K.; Hoerster, G.; Anand, A.; Wu, E.; Wang, N.; Arling, M.; Lenderts, B.; Gordon-Kamm, W. Maize Transformation Using the Morphogenic Genes Baby Boom and Wuschel2. Methods Molocular Biol. 2019, 1864, 81–93. [Google Scholar]
- Hassani, S.B.; Trontin, J.F.; Raschke, J.; Zoglauer, K.; Rupps, A. Constitutive Overexpression of a Conifer WOX2 Homolog Affects Somatic Embryo Development in Pinus pinaster and Promotes Somatic Embryogenesis and Organogenesis in Arabidopsis Seedlings. Front Plant Sci. 2022, 13, 838421. [Google Scholar] [CrossRef]
- Ueda, M.; Zhang, Z.; Laux, T. Transcriptional Activation of Arabidopsis Axis Patterning Genes WOX8/9 Links Zygote Polarity to Embryo Development. Dev. Cell 2011, 20, 264–270. [Google Scholar] [CrossRef]
- Kyo, M.; Maida, K.; Nishioka, Y.; Matsui, K. Coexpression of WUSCHEL related homeobox (WOX) 2 with WOX8 or WOX9 promotes regeneration from leaf segments and free cells in Nicotiana tabacum L. Plant Biotechnol. 2018, 35, 23–30. [Google Scholar] [CrossRef]
- Zhou, X.; Han, H.; Chen, J.; Han, H. The emerging roles of WOX genes in development and stress responses in woody plants. Plant Sci. 2024, 349, 112259. [Google Scholar] [CrossRef] [PubMed]
- Magnani, E.; Jiménez-Gómez, J.M.; Soubigou-Taconnat, L.; Lepiniec, L.; Fiume, E. Profiling the onset of somatic embryogenesis in Arabidopsis. BMC Genom. 2017, 18, 998. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.K.; Mummadi, S.T.; Liu, S.; Wei, H. Regulation of regeneration in Arabidopsis thaliana. aBIOTECH 2023, 4, 332–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-X.; Shang, G.-D.; Wu, L.-Y.; Xu, Z.-G.; Zhao, X.-Y.; Wang, J.-W. Chromatin Accessibility Dynamics and a Hierarchical Transcriptional Regulatory Network Structure for Plant Somatic Embryogenesis. Dev. Cell 2020, 54, 742–757.e8. [Google Scholar] [CrossRef] [PubMed]
- Sivanesan, I.; Nayeem, S.; Venkidasamy, B.; Kuppuraj, S.P.; Rn, C.; Samynathan, R. Genetic and epigenetic modes of the regulation of somatic embryogenesis: A review. Biol. Futur. 2022, 73, 259–277. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Zhou, M.; Ceasar, S.A.; Ali, D.J.; Maharajan, T.; Vinod, K.K.; Sharma, A.; Ahmad, Z.; Wei, Q. Epigenetic modifications and miRNAs determine the transition of somatic cells into somatic embryos. Plant Cell Rep. 2023, 42, 1845–1873. [Google Scholar] [CrossRef]
- Fan, Y.; Tang, Z.; Wei, J.; Yu, X.; Guo, H.; Li, T.; Guo, H.; Zhang, L.; Fan, Y.; Zhang, C.; et al. Dynamic Transcriptome Analysis Reveals Complex Regulatory Pathway Underlying Induction and Dose Effect by Different Exogenous Auxin IAA and 2,4-D During in vitro Embryogenic Redifferentiation in Cotton. Front. Plant Sci. 2022, 13, 931105. [Google Scholar] [CrossRef]
- Olivares-García, C.A.; Mata-Rosas, M.; Peña-Montes, C.; Quiroz-Figueroa, F.; Segura-Cabrera, A.; Shannon, L.M.; Loyola-Vargas, V.M.; Monribot-Villanueva, J.L.; Elizalde-Contreras, J.M.; Ibarra-Laclette, E.; et al. Phenylpropanoids Are Connected to Cell Wall Fortification and Stress Tolerance in Avocado Somatic Embryogenesis. Int. J. Mol. Sci. 2020, 21, 5679. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, H.; Zhao, G.; Wang, N.; Guo, L.; Hou, X. Molecular mechanism of somatic embryogenesis in paeonia ostii ‘Fengdan’ based on transcriptome analysis combined histomorphological observation and metabolite determination. BMC Genom. 2023, 24, 665. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef]
- Liu, Y.; Dang, P.; Liu, L.; He, C. Cold acclimation by the CBF–COR pathway in a changing climate: Lessons from Arabidopsis thaliana. Plant Cell Rep. 2019, 38, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant. 2013, 147, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A Core Regulatory Pathway Controlling Rice Tiller Angle Mediated by the LAZY1-Dependent Asymmetric Distribution of Auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Wilkinson, E.G.; Sageman-Furnas, K.; Strader, L.C. Auxin and abiotic stress responses. J. Exp. Bot. 2023, 74, 7000–7014. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molocular Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Molocular Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Jiang, Y.; Yang, Y.; Xiao, Z.; Bai, X.; Gao, J.; Tan, S.; Hur, Y.; Hao, S.; He, F. Identification and Expression Analysis of the NAC Gene Family in Coffea canephora. Agronomy 2019, 9, 670. [Google Scholar] [CrossRef]
- Dong, X.; Yi, H.; Lee, J.; Nou, I.-S.; Han, C.-T.; Hur, Y. Global Gene-Expression Analysis to Identify Differentially Expressed Genes Critical for the Heat Stress Response in Brassica rapa. PLoS ONE 2015, 10, e0130451. [Google Scholar] [CrossRef]
- Ivamoto, S.T.; Reis, O.J.; Domingues, D.S.; dos Santos, T.B.; de Oliveira, F.F.; Pot, D.; Leroy, T.; Vieira, L.G.E.; Carazzolle, M.F.; Pereira, G.A.G.; et al. Transcriptome Analysis of Leaves, Flowers and Fruits Perisperm of Coffea arabica L. Reveals the Differential Expression of Genes Involved in Raffinose Biosynthesis. PLoS ONE 2017, 12, e0169595. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Awada, R.; Campa, C.; Gibault, E.; Déchamp, E.; Georget, F.; Lepelley, M.; Abdallah, C.; Erban, A.; Martinez-Seidel, F.; Kopka, J.; et al. Unravelling the Metabolic and Hormonal Machinery During Key Steps of Somatic Embryogenesis: A Case Study in Coffee. Int. J. Mol. Sci. 2019, 20, 4665. [Google Scholar] [CrossRef]
- dos Santos, T.B.; Soares, J.D.M.; Lima, J.E.; Silva, J.C.; Ivamoto, S.T.; Baba, V.Y.; Souza, S.G.H.; Lorenzetti, A.P.R.; Paschoal, A.R.; Meda, A.R.; et al. An integrated analysis of mRNA and sRNA transcriptional profiles in Coffea arabica L. roots: Insights on nitrogen starvation responses. Funct. Integr. Genom. 2019, 19, 151–169. [Google Scholar] [CrossRef]
- Marques, I.; Fernandes, I.; David, P.H.C.; Paulo, O.S.; Goulao, L.F.; Fortunato, A.S.; Lidon, F.C.; DaMatta, F.M.; Ramalho, J.C.; Ribeiro-Barros, A.I. Transcriptomic Leaf Profiling Reveals Differential Responses of the Two Most Traded Coffee Species to Elevated [CO2]. Int. J. Mol. Sci. 2020, 21, 9211. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
| Gene Name | Gene ID | Chromosome Location | gDNA Length (bp) | Protein Length (a.a.) | MW (KDa) | Isoelectric Point (pI) | Best BLAST Hit Within Arabidopsis | ||
|---|---|---|---|---|---|---|---|---|---|
| At_ID | At_Name | E-Value | |||||||
| CaWOX3a | Cara001g022170 | Chr 01 | 1199 | 216 | 24.88 | 9.35 | AT2G28610.1 | AtWOX3 | 5.50 × 10−46 |
| CaWOX2a | Cara001g022960 | Chr 01 | 1360 | 261 | 29.45 | 8.79 | AT5G59340.1 | AtWOX2 | 9.46 × 10−52 |
| CaWOX3b | Cara002g013730 | Chr 02 | 1197 | 216 | 24.87 | 9.35 | AT2G28610.1 | AtWOX3 | 3.45 × 10−43 |
| CaWOX3c | Cara002g013760 | Chr 02 | 1197 | 216 | 24.87 | 9.35 | AT2G28610.1 | AtWOX3 | 3.45 × 10−43 |
| CaWOX2b | Cara002g014580 | Chr 02 | 1369 | 262 | 29.44 | 9.05 | AT5G59340.1 | AtWOX2 | 2.33 × 10−49 |
| CaWOX8b | Cara003g039800 | Chr 03 | 2097 | 407 | 45.00 | 8.75 | AT5G45980.1 | AtWOX8 | 1.39 × 10−78 |
| CaWOX5a | Cara003g048620 | Chr 03 | 656 | 174 | 19.70 | 8.76 | AT3G11260.1 | AtWOX5 | 2.89 × 10−60 |
| CaWOX5b | Cara004g007900 | Chr 04 | 666 | 174 | 19.70 | 8.76 | AT3G11260.1 | AtWOX5 | 2.89 × 10−60 |
| CaWOX8a | Cara004g016390 | Chr 04 | 2092 | 407 | 44.89 | 8.55 | AT5G45980.1 | AtWOX8 | 5.16 × 10−79 |
| CaWOX1a | Cara007g007070 | Chr 07 | 2160 | 362 | 40.90 | 6.18 | AT3G18010.1 | AtWOX1 | 5.26 × 10−60 |
| CaWOX13d | Cara007g012080 | Chr 07 | 741 | 247 | 28.26 | 5.43 | AT4G35550.1 | AtWOX13 | 2.86 × 10−49 |
| CaWOX13c | Cara008g014990 | Chr 08 | 741 | 247 | 28.26 | 5.54 | AT4G35550.1 | AtWOX13 | 2.31 × 10−49 |
| CaWOX1b | Cara008g019840 | Chr 08 | 2164 | 362 | 40.97 | 6.53 | AT3G18010.1 | AtWOX1 | 3.10 × 10−59 |
| CaWOX11a | Cara011g029950 | Chr 11 | 2412 | 257 | 27.90 | 5.61 | AT3G03660.1 | AtWOX11 | 2.40 × 10−63 |
| CaWOX11b | Cara012g008700 | Chr 12 | 2410 | 257 | 27.90 | 5.61 | AT3G03660.1 | AtWOX11 | 2.40 × 10−63 |
| CaWUSb | Cara013g011100 | Chr 13 | 1611 | 286 | 32.12 | 6.45 | AT2G17950.1 | AtWUS | 2.41 × 10−46 |
| CaWOX13b | Cara013g012460 | Chr 13 | 3119 | 280 | 31.00 | 5.56 | AT4G35550.1 | AtWOX13 | 9.66 × 10−87 |
| CaWOX13a | Cara014g017290 | Chr 14 | 3604 | 278 | 30.89 | 5.56 | AT4G35550.1 | AtWOX13 | 9.35 × 10−89 |
| CaWUSa | Cara014g018710 | Chr 14 | 1612 | 286 | 32.14 | 6.45 | AT2G17950.1 | AtWUS | 2.03 × 10−46 |
| CaWOX4a | Cara019g006470 | Chr 19 | 1122 | 222 | 25.44 | 9.51 | AT1G46480.1 | AtWOX4 | 1.00 × 10−76 |
| CaWOX4b | Cara020g021370 | Chr 20 | 1078 | 222 | 25.47 | 9.51 | AT1G46480.1 | AtWOX4 | 1.36 × 10−76 |
| CaWOX1d | Cara021g016240 | Chr 21 | 1526 | 337 | 38.23 | 7.15 | AT3G18010.1 | AtWOX1 | 1.03 × 10−31 |
| CaWOX1c | Cara022g010550 | Chr 22 | 1526 | 337 | 38.26 | 6.84 | AT3G18010.1 | AtWOX1 | 9.18 × 10−32 |
| CcWOX11 | Cc00g05100 | Chr un | 2503 | 251 | 27.45 | 5.84 | AT3G03660.1 | AtWOX11 | 9.05 × 10−65 |
| CcWOX3a | Cc00g26230 | Chr un | 1198 | 215 | 24.93 | 9.51 | AT2G28610.1 | AtWOX3 | 2.71 × 10−45 |
| CcWOX3b | Cc01g11960 | Chr 01 | 1191 | 215 | 24.87 | 9.57 | AT2G28610.1 | AtWOX3 | 8.83 × 10−42 |
| CcWOX2 | Cc01g12690 | Chr 01 | 1359 | 237 | 26.88 | 8.93 | AT5G59340.1 | AtWOX2 | 3.43 × 10−52 |
| CcWOX5 | Cc02g06840 | Chr 02 | 666 | 173 | 19.70 | 8.76 | AT3G11260.1 | AtWOX5 | 2.79 × 10−60 |
| CcWOX8 | Cc02g14220 | Chr 02 | 2087 | 406 | 45.00 | 8.75 | AT5G45980.1 | AtWOX8 | 1.35 × 10−78 |
| CcWOX1a | Cc04g06330 | Chr 04 | 3252 | 361 | 40.97 | 6.05 | AT3G18010.1 | AtWOX1 | 3.52 × 10−60 |
| CcWOX13b | Cc04g10680 | Chr 04 | 740 | 246 | 28.26 | 5.43 | AT4G35550.1 | AtWOX13 | 2.77 × 10−49 |
| CcWUS | Cc07g10660 | Chr 07 | 1609 | 285 | 32.09 | 6.45 | AT2G17950.1 | AtWUS | 2.66 × 10−46 |
| CcWOX13a | Cc07g11890 | Chr 07 | 3819 | 279 | 31.00 | 5.56 | AT4G35550.1 | AtWOX13 | 9.32 × 10−87 |
| CcWOX4 | Cc10g04700 | Chr 10 | 2384 | 221 | 25.47 | 9.51 | AT1G46480.1 | AtWOX4 | 1.31 × 10−76 |
| CcWOX1b | Cc11g08460 | Chr 11 | 1525 | 336 | 38.23 | 7.15 | AT3G18010.1 | AtWOX1 | 1.01 × 10−31 |
| CeWOX2 | LOC113774651 | Chr 1 | 1429 | 260 | 29.35 | 9.05 | AT5G59340 | AtWOX2 | 1.22 × 10−50 |
| CeWOX3 | LOC113774781 | Chr 1 | 1196 | 215 | 24.87 | 9.35 | AT2G28610 | AtWOX3 | 3.34 × 10−43 |
| CeWOX4 | LOC113749539 | Chr 10 | 1482 | 221 | 25.47 | 9.51 | AT1G46480 | AtWOX4 | 1.31 × 10−76 |
| CeWOX1a | LOC113753065 | Chr 11 | 2362 | 336 | 38.27 | 7.6 | AT3G18010 | AtWOX1 | 9.75 × 10−32 |
| CeWOX5 | LOC113755443 | Chr 2 | 663 | 173 | 19.70 | 8.76 | AT3G11260 | AtWOX5 | 2.79 × 10−60 |
| CeWOX8 | LOC113764000 | Chr 2 | 2406 | 392 | 43.19 | 8.25 | AT5G45980 | AtWOX8 | 1.04 × 10−78 |
| CeWOX1b | LOC113768485 | Chr 4 | 3006 | 361 | 40.97 | 6.53 | AT3G18010 | AtWOX1 | 3.01 × 10−59 |
| CeWOX13a | LOC113769015 | Chr 4 | 797 | 265 | 30.71 | 5.66 | AT4G35550 | AtWOX13 | 4.76 × 10−49 |
| CeWOX13b | LOC113778647 | Chr 7 | 3557 | 271 | 30.26 | 5.56 | AT4G35550 | AtWOX13 | 2.39 × 10−86 |
| CeWOX11 | LOC113758086 | Chr un | 2155 | 251 | 27.45 | 5.84 | AT3G03660 | AtWOX11 | 9.05 × 10−65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.; Gao, J.; Jiang, M.; Tao, Y.; Chen, X.; Yang, X.; Wang, L.; Jiang, D.; Xiao, Z.; Bai, X.; et al. The Identification and Characterization of WOX Family Genes in Coffea arabica Reveals Their Potential Roles in Somatic Embryogenesis and the Cold-Stress Response. Int. J. Mol. Sci. 2024, 25, 13031. https://doi.org/10.3390/ijms252313031
Dong X, Gao J, Jiang M, Tao Y, Chen X, Yang X, Wang L, Jiang D, Xiao Z, Bai X, et al. The Identification and Characterization of WOX Family Genes in Coffea arabica Reveals Their Potential Roles in Somatic Embryogenesis and the Cold-Stress Response. International Journal of Molecular Sciences. 2024; 25(23):13031. https://doi.org/10.3390/ijms252313031
Chicago/Turabian StyleDong, Xiangshu, Jing Gao, Meng Jiang, Yuan Tao, Xingbo Chen, Xiaoshuang Yang, Linglin Wang, Dandan Jiang, Ziwei Xiao, Xuehui Bai, and et al. 2024. "The Identification and Characterization of WOX Family Genes in Coffea arabica Reveals Their Potential Roles in Somatic Embryogenesis and the Cold-Stress Response" International Journal of Molecular Sciences 25, no. 23: 13031. https://doi.org/10.3390/ijms252313031
APA StyleDong, X., Gao, J., Jiang, M., Tao, Y., Chen, X., Yang, X., Wang, L., Jiang, D., Xiao, Z., Bai, X., & He, F. (2024). The Identification and Characterization of WOX Family Genes in Coffea arabica Reveals Their Potential Roles in Somatic Embryogenesis and the Cold-Stress Response. International Journal of Molecular Sciences, 25(23), 13031. https://doi.org/10.3390/ijms252313031





